Abstract
We exploit the optical and spatial features of sub-wavelength nanostructures to examine individual receptors on the plasma membrane of living cells. Receptors were sequestered in portions of the membrane projected into zero-mode waveguides (ZMWs). Using single-step photobleaching of GFP incorporated into individual subunits, the resulting spatial isolation was used to measure subunit stoichiometry in α4β4 and α4β2 nicotinic acetylcholine and P2X2 ATP receptors. We also show that nicotine and cytisine have differential effects on α4β2 stoichiometry.
Keywords: membrane receptors, nanostructures, single-molecule imaging, nicotinic receptors, zero-mode waveguide
The advent of single-molecule imaging techniques has provided insight into the dynamics of many complex biological systems1-6, including membrane proteins. Advances in fluorescence-based imaging techniques have allowed for studying structural and dynamic characteristics of ion channels, receptors, and transporters7. However, single-molecule measurements of membrane receptors continue to be challenged by three major factors: diffusion of proteins at the surface of the cell (as revealed by single-particle tracking), natural accumulations of membrane receptors at local densities of 10 to 10,000 per μm2, and high levels of autofluorescence8,9. Studies utilizing existing imaging techniques such as total internal reflection fluorescence (TIRF) are best suited to densities on the order of one per 1-10 μm2, or two to five orders of magnitude less than typical physiological densities.
One path to mitigate the challenge of high receptor densities is to control the expression in the biological system of interest. Recent studies have utilized expression in Xenopus laevis oocytes, which produced the required low densities of membrane proteins that also exhibited limited diffusion. This provided the means to detect single-molecule bleaching steps of GFP labels associated with individual subunits10. Furthermore, fluorescent unnatural amino acid side chains can readily be incorporated into oocytes, expanding studies to include more favorable fluorophores11. These studies and subsequent examples of similar experiments in mammalian cells12 have demonstrated the power of single-molecule techniques to determine the stoichiometry of membrane receptors. However, applications for these techniques remain limited primarily due to receptor diffusion and because oocytes lack some cellular machinery present in mammalian cells. These methods often control protein densities but fail to limit receptor diffusion or aggregation at the plasma membrane. While TIRF microscopy limits the excitation volume in the axial direction (200 nm), intracellular organelles are still visible13 leading to high background fluorescence which further complicates single-molecule measurements.
Alternatively, the utilization of zero-mode waveguide (ZMW) nanostructures has shown promise for single-molecule applications near physiological concentrations14-17. ZMWs are sub-wavelength holes constructed in a ~100 nm layer of aluminum mounted on top of a glass substrate18 (Supplementary Figure 1). In addition to providing a rapidly attenuating evanescent field in the axial direction, the ZMW also restricts the lateral dimensions of the excitation volume to the size of the well. Single-molecule experiments of membrane receptors with ZMWs should offer two key advantages: ZMWs spatially isolate a small number (1 to a few) receptors allowing single-molecule measurement at or near physiological densities by limiting diffusion and aggregation, and reducing background fluorescence from nearby molecules. In fact previous studies have shown that cell membranes can extend into the ZMWs, allowing the detection of fluctuations associated with diffusion of fluorescent lipids19 and membrane resident proteins14, 20.
Nicotinic acetylcholine receptors (nAChRs) are an important class of cation-selective transmembrane receptor channels that express throughout the central and peripheral nervous systems and are assembled from various subunits (α1-α10 and β2-β4). 21. The correct assembly of alpha and beta subunits into pentameric structures is essential for proper subcellular localization, agonist sensitivity, and ion channel Ca2+ permeability22,23. The identity and stoichiometry of subunits in each nAChR are important factors in regulating intracellular receptor processing and trafficking. To date, TIRF-based experiments with single-receptor resolution in mammalian cells have been limited to α7 homomeric nAChRs, which are capable of binding fluorescently labeled α-bungarotoxin12. The ability to directly interrogate the subunit stoichiometry of heteromeric neuronal nAChRs in living mammalian cells has not been reported.
We achieve spatial isolation of nanometer scale membrane areas within living cells by directly culturing Neuro-2a (N2a) neuroblastoma cells on ZMW arrays with defined diameters from 85 to 200 nm, corresponding to a 5.5 fold range in the membrane area that can potentially enter each ZMW (Figure 1). These cells grow neurite-like projections and generate large numbers of filopodia which may protrude into ZMWs making them ideally suited for the imaging of membrane resident proteins in ZMWs. Additionally, these cells do not contain endogenous nAChRs, which makes them suitable for the study of receptor assembly24. Imaging of cells transfected with plasma membrane localized monomeric cherry marker (PM-mcherry) showed that transfected cells rest directly above the ZMWs (Supplementary Figure 2). Importantly, we observed mcherry fluorescence in numerous adjacent ZMWs where the pattern created by the fluorescent wells resembled the footprint of cells cultured on a glass coverslip (Figure 2A and B). Clearly, the filopodia-like extensions can extend into ZMWs. This provides evidence that these nanostructures are capable of isolating small segments of the membrane which contain fluorescent protein markers.
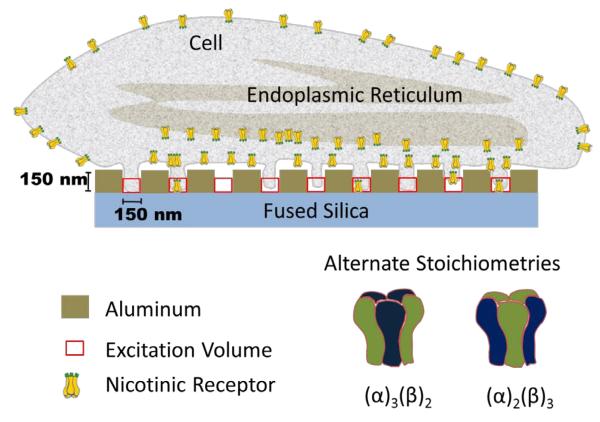
Figure 1. Membrane projection into ZMWs.
Schematic of a cell plated directly on the array with extensions of small portions of the membrane protruding into ZMWs. The schematic exaggerates the well size relative to spacing, in order to illustrate the entry of the cell membrane into the wells. An individual cell typically covers 75-100 wells. The excitation volume extends ~ 50 nm into the wells creating a ~ 250 zl excitation volume for a 120 nm ZMW, as depicted by the red squares (see supplemental material). Only a few nAChRs in the plasma membrane enter wells, probably in filopodia. Under many circumstances, the ER and other organelles contain a majority of the cell’s nAChR; but these do not enter wells. The diagram of the receptors illustrates that nicotinic receptors can potentially assemble into two stoichiometries: α3β2 or α2β3.
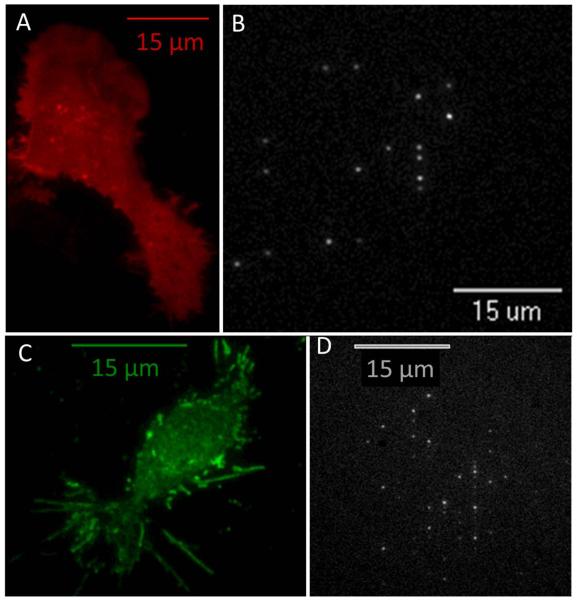
Figure 2. Isolation of membrane receptors in ZMWs.
A. N2a cells plated on a glass coverslip transfected with a plasma membrane localized monomeric cherry marker (PM-mcherry) imaged with TIRF microscopy illustrating the extent of membrane expression in both the soma and the processes. B. N2a cells cultured directly on the ZMW array and transfected with the same membrane marker as in (a), and imaged from the glass side of the array. Fluorescent spots indicate the portions of the membrane containing the PM-mcherry marker that have entered the ZMW and lie within the excitation volume. C. N2a cells plated on a glass coverslip transfected with α4-GFP β4-wt and imaged with TIRF microscopy. The image shows that α4β4 nAChRs are primarily localized on the plasma membrane13 and also shows the presence of many filopodia (5 – 15 μm in length) containing fluorescent nAChRs. D. N2a cells cultured directly on the array and transfected in the same manner as in (c). Fluorescent spots indicate that portions of the membrane, probably filopodia, containing fluorescently labeled α4β4 nAChRs extend into the ZMWs. The size of the pattern made by the fluorescent wells is consistent with the size of a typical cell.
We utilized ZMW-mediated optical confinement to determine the stoichiometry of ligand-gated receptor channels. Integrating nanostructure based imaging with the conventional tactic of observing single-molecule bleaching steps10 provided the capability to examine individual fluorescent protein-tagged subunits in heteromeric nAChRs. We previously achieved physiologically relevant plasma membrane densities (50-100/μm2 based on TIRF measurements) of fluorescent α4β4 nAChRs by transfection in N2a cells13. We therefore considered this system well-suited for initial analyses on subunit fluorescence of assembled receptors in ZMWs. The relatively high concentration of nAChRs at the cell membrane increases the probability that GFP-labeled nAChRs will extend into the sub-wavelength nanostructures. Because the fluorescent protein is incorporated in the M3-M4 loop of all nAChRs and the evanescent excitation field only extends 50 nm into the wells, all visualized individual receptors are completely within the bottom half of the well. Additionally, only assembled pentameric receptors are trafficked from the ER to the plasma membrane13, 25. Thus, measurements of stoichiometry in these experiments are restricted to assembled receptors at the plasma membrane. The footprint of nAChR fluorescence in transfected N2a cells plated on standard glass coverslips resembles the shape and pattern of fluorescent wells for cells transfected on ZMW arrays (Figure 2C and D). Even at the same density that yields a clear ensemble fluorescence signal (Figure 2C) with no discernible single molecule features in TIRF, expression on ZMWs of >144 nm diameter leads to the observation of individual receptors exhibiting GFP fluorescence. Roughly half the number of wells that exhibit PM-mcherry signals, which are associated with the penetration of the cell membrane into ZMWs, show GFP fluorescence.
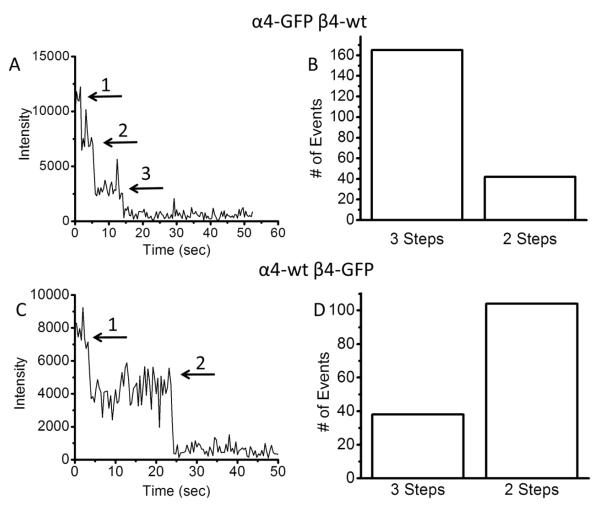
The time course of fluorescence intensity from an individual ZMW can be analyzed to measure the number of GFP molecules resident in that ZMW. While α4 and β4 subunits are known to coexpress in the medial habenula and may participate in some behavioral responses to nicotine, little is known about the stoichiometry of their assembly. We conducted experiments with either α4-GFP + unlabeled (wt) β4, or with unlabeled α4 + β4-GFP. Because a single GFP molecule is genetically encoded into each labeled subunit, individual bleaching steps in the fluorescence intensity level indicate one subunit. On each ZMW array, 10-20% of the fluorescent wells showed bleaching events consistent with single receptors; a similar percentage of informative puncta is generally observed in TIRF-based studies of ion channels26. The remaining 80% of the wells exhibit fluorescence decays not consistent with individual molecules (see supplemental material). The accumulated data for α4-GFP + β4 revealed that ~80% individually isolated receptors exhibited three bleaching steps while ~20% exhibited two bleaching steps (Figure 3A and B). Three bleaching steps correspond to (α4)3(β4)2 subunit stoichiometry, while two bleaching steps correspond to the complementary (α4)2β4)3 stoichiometry. When we studied nAChRs formed from α4 + β4-GFP subunits, we found that <30% of the wells had three bleaching steps; 70% had two bleaching steps (Figure 3C and D). These two independent data sets indicate that the majority of α4β4 receptors assemble in a stoichiometry of (α4)3(β4)2. The complementary stoichiometry of (α4)2(β4)3 occurs at a lower frequency (20-30%). The self-consistent nature of these results, using either labeled α4 or β4, rules out many possible artifacts such as undetectable bleaching of the initial fluorophore or disassembly of the heteropentamer. The majority of the noninformative fluorescent ZMWs exhibited an exponential decay with no discernible steps (70% of wells), indicating either the presence of multiple receptors or transitions obscured by fluorophore blinking (Supplementary figure 3). A small fraction (10%) of the wells that exhibited step-like photobleaching was best fit to a single step. This is most likely due to simultaneous bleaching of two fluorophores (see supplementary information).
Figure 3. Single step bleaching of labeled subunits in N2a cells cultured on ZMW arrays.
A. Time course of fluorescence intensity from a ZMW in a dish containing N2a cells transfected with α4-GFP + β4-wt. The trace shows three bleaching steps as indicated by the arrows. The different fluorescence levels are separated by clear steps until all the GFP molecules bleach to the background level. B. Number of three-step and two-step bleaching events observed for α4-GFP + β4-wt showing predominantly three bleaching steps. This indicates primarily (α4)3(β4)2 stoichiometry. C. Time course of fluorescence intensity from a well in a dish transfected with α4-wt + β4-GFP. The time trace shows two bleaching steps as indicated by the arrows. D. Number of three-step and two-step bleaching events observed for α4-wt + β4-GFP showing predominantly two bleaching steps. This also indicates primarily (α4)3(β4)2 stoichiometry.
We also validated our stoichiometry results by showing consistency with an ion channel of known subunit number. P2X2 receptors are known to assemble as trimers27. In studies on P2X2-GFP receptors, we observed 3 bleaching steps in 85% of measurable wells (supplemental figure 4).
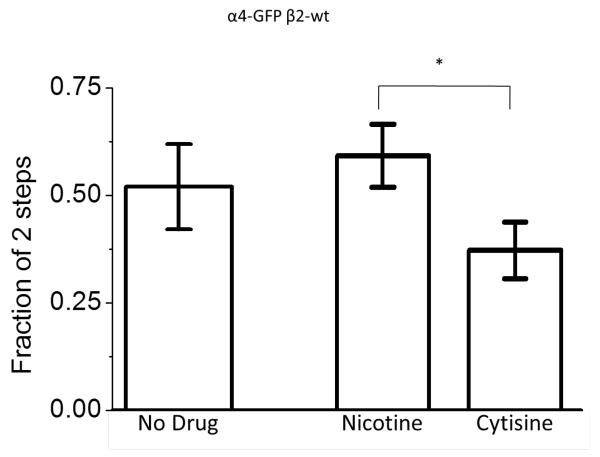
While studies of α4β4 illustrate the suitability of this ZMW based method to determine the previously unknown stoichiometry of a subtype of heteromeric receptor, we also examined the more widely expressed α4β2 nAChRs to demonstrate that the ZMW based method can detect pharmacologically induced changes in stoichiometry. Chronic exposure to nicotine and other nAChR ligands upregulates α4β2 nAChR density on the plasma membrane of neurons and cultured cell lines28-30; upregulation may underlie both nicotine dependence and the apparent neuroprotective effect of nicotine in Parkinson’s disease23. Upregulation is suggested to proceed via pharmacological chaperoning13, 31-33, and indirect evidence suggests that upregulation also involves changes in the stoichiometry of plasma membrane α4β2 nAChR stoichiometry30, 34. Most α4β2 receptors are retained intracellularly, which complicates biochemical attempts to demonstrate altered plasma membrane stoichiometry. Additionally, the large population of intracellular receptors produces intolerably high background in TIRF-based measurements of plasma membrane resident receptors. We applied the ZMW measurements to α4-GFP β2-wt nAChRs. In the absence of applied drug, we rarely detected α4β2 nAChR in ≤ 144 nm diameter ZMWs. However, the increased area provided by the larger ZMWs (~200 nm diameter) allowed us to gather single-nAChR photobleaching measurements without interference from intracellular fluorescence. Thus, an additional advantage of the ZMW arrays used in these measurements is the ability to choose a ZMW diameter appropriate to the receptor density. Comparing bleaching steps of α4-GFP yielded 2-step bleaching in 52% of the ZMWs (figure 4). This confirms previous suggestions, based on indirect measurements, that plasma membrane (α4)3(β2)2 and (α4)2(β2)3 nAChRs exist in roughly similar numbers13, 35—quite different from the measurements with α4β4 nAChRs which heavily favored the stoichiometry with three α4 subunits.
Figure 4. Stoichiometry of α4β2 receptors.
Fraction of the informative wells exhibiting 2 bleaching steps for α4-GFP + β2-wt exposed to no drug (n= 81), nicotine (n=179), and cytisine (n=117). The error bars show the relative standard error of the mean of the wells exhibiting 2-step bleaching. *, significant at P < 0.05.
After upregulation of plasma membrane α4β2 nAChRs by 24 hours of exposure to nicotine (500 nM) or cytisine (500 nM), we found, as expected, more frequent single-molecule events (~10% of PM-mCherry ZMWs), even in ~ 144 nm diameter ZMWs. Interestingly, these two drugs produced significantly different effects on nAChR stoichiometry: 60% (α4)2(β2)3 for nicotine, but only 38% for cytisine (figure 4). This confirmed that nicotine induced the preferential transport of the (α4)2(β2)3 stoichiometry to the plasma membrane30, 33, 34. In contrast, cytisine induced a plasma membrane population favoring the (α4)3(β2)2 stoichiometry. This effect of cytisine is consistent with measurements that cytisine activates nAChRs formed by microinjecting Xenopus oocytes with more α4 than β2 cRNA34, as well as with very recent macroscopic FRET-based measurements of intracellular α4β2 stoichiometry33. Thus, ZMW-based measurements of (α4)2(β2)3 stoichiometry confirm several existing ideas but more importantly also extend our knowledge of pharmacological chaperoning to the single-molecule level. Additionally, the shift in stoichiometry of α4β2 under different conditions indicates that there is no bias toward the trafficking of a particular stoichiometry to the terminal parts of filopodia.
The isolation of receptors in filopodia that have extended into ZMWs appears to limit the diffusion to such an extent that receptors can be imaged for 10s of seconds. Similar experiments in HEK cells did not yield a similar confinement but instead resulted in only transient fluorescence (<400 ms) indicating the diffusion of receptors in and out of the well. This suggests that the advantage of using cells that generate neurite like projections, such as N2a cells, is that they can be optimized for interactions with nanostructures for the isolation of receptors.
Single-molecule experiments on membrane proteins have previously required very low densities of fluorophores. This can be accomplished with low, non-physiological protein densities. Experiments with higher surface densities commonly use low-efficiency labeling of target proteins, but such schemes label only a small percentage of subunits preventing counting or full analysis within individual receptors. Due to concerns that artificially low concentrations of receptors, such as those in most single-molecule methods, can alter the dynamics of assembly and trafficking of receptors, we sought to develop the present method, allowing the isolation of proteins at physiological densities. We now show that the integration of ZMW nanostructures with N2a cells provides the capability to isolate individual receptors even at physiological membrane densities. We believe that ZMWs would be appropriate even at densities 10 fold greater than studied here.
At the opposite end of the density range, even rather sparse α4β2 receptors are experimentally accessible with larger-diameter ZMWs. This technique provides the tools to study the properties of individual membrane receptors in living cells, as controlled by pharmacological, pathological, and developmental processes. This technique could also provide the means to extend simultaneous single channel electrophysiology and fluorescence studies36. We believe that this integration of nanostructured devices with live cells can be widely applied to study a variety of membrane-resident proteins, including ion channels, transporters, and other receptors.
Supplementary Material
Acknowledgement
We thank A. Fernandez and M. Boitano for assistance in preparing ZMW arrays. Supported by a Beckman Fellowship and an NIH NRSA to C. I. R., by a California Tobacco-Related Disease Research Fellowship to R. S., and by the US National Institutes of Health (NS34407).
Footnotes
Supporting Information. Detailed methods and supplemental figures. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1).Sako Y, Minoghchi S, Yanagida T. Nat. Cell Bio. 2000;2(3):168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 2).Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313(5793):1642–5. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 3).Dani A, Huang B, Bergan J, Dulac C, Zhuang XW. Neuron. 2010;68(5):843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Kim SY, Miller EJ, Frydman J, Moerner WE. J. Mol. Biol. 2010;401(4):553–563. doi: 10.1016/j.jmb.2010.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Kelly Christopher V., Baird Barbara A., Craighead Harold G. Biophys. J. 2011;100(7):L34–L36. doi: 10.1016/j.bpj.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Iyer G, Michalet X, Chang Y-P, Pinaud FF, Matyas SE, Payne G, Weiss S. Nano Letters. 2008;8(12):4618–4623. doi: 10.1021/nl8032284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Taraska JW, Zagotta WN. Neuron. 2010;66(2):170–89. doi: 10.1016/j.neuron.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Fernandes CC, Berg DK, Gomez-Varela D. J. Neurosci. 2010;30(26):8841–8851. doi: 10.1523/JNEUROSCI.6236-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Hille B. Ionic Channels of Excitable Membranes. 3rd ed Sinauer; Sunderland, MA: 2001. [Google Scholar]
- 10).Ulbrich MH, Isacoff EY. Nat Methods. 2007;4(4):319–21. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Pantoja R, Rodriguez EA, Dibas MI, Dougherty DA, Lester HA. Biophys J. 2009;96(1):226–37. doi: 10.1016/j.bpj.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Simonson PD, DeBerg HA, Ge PH, Alexander JK, Jeyifous O, Green WN, Selvin PR. Biophys. J. 2010;99(10):L81–L83. doi: 10.1016/j.bpj.2010.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Srinivasan R, Pantoja R, Moss FJ, Mackey EDW, Son C, Miwa J, Lester HA. J. Gen .Physiol. 2011;137:59–79. doi: 10.1085/jgp.201010532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Moran-Mirabal JM, Torres AJ, Samiee KT, Baird BA, Craighead HG. Nanotechnology. 2007;18(19) [Google Scholar]
- 15).Moran-Mirabal JM, Craighead HG. Methods. 2008;46(1):11–17. doi: 10.1016/j.ymeth.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 16).Aouani H, Mahboub O, Bonod N, Devaux E, Popov E, Rigneault H, Ebbesen TW, Wenger J. Nano Lett. 2011;11(2):637–644. doi: 10.1021/nl103738d. [DOI] [PubMed] [Google Scholar]
- 17).Aouani H, Mahboub O, Devaux E, Rigneault H, Ebbesen TW, Wenger J. Nano Lett. 2011;11(6):2400–2406. doi: 10.1021/nl200772d. [DOI] [PubMed] [Google Scholar]
- 18).Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Science. 2003;299(5607):682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 19).Edel JB, Wu M, Baird B, Craighead HG. Biophys J. 2005;88(6):L43–5. doi: 10.1529/biophysj.105.061937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Wenger J, Conchonaud F, Dintinger J, Wawrezinieck L, Ebbesen TW, Rigneault H, Marguet D, Lenne PF. Biophys J. 2007;92(3):913–9. doi: 10.1529/biophysj.106.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Graham AJ, Ray MA, Perry EK, Jaros E, Perry RH, Volsen SG, Bose S, Evans N, Lindstrom J, Court JA. J. Chem. Neuroanat. 2003;25(2):97–113. doi: 10.1016/s0891-0618(02)00100-x. [DOI] [PubMed] [Google Scholar]
- 22).Tapia L, Kuryatov A, Lindstrom J. Mol Pharmacol. 2007;71(3):769–76. doi: 10.1124/mol.106.030445. [DOI] [PubMed] [Google Scholar]
- 23).Miwa JM, Freedman R, Lester HA. Neuron. 2011;70(1):20–33. doi: 10.1016/j.neuron.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Xiao C, Srinivasan R, Drenan RM, Mackey EDW, McIntosh JM, Lester HA. Biochem. Pharmacol. 2011;82(8):852–861. doi: 10.1016/j.bcp.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Richards CI, Srinivasan R, Xiao C, Mackey EDW, Miwa JM, Lester HA. J. Biol. Chem. 2011;286(36):31241–31249. doi: 10.1074/jbc.M111.256024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Ji W, Xu P, Li Z, Lu J, Liu L, Zhan Y, Chen Y, Hille B, Xu T, Chen L. Proc Natl Acad Sci U S A. 2008;105(36):13668–73. doi: 10.1073/pnas.0806499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Kawate T, Michel J, Birdsong W, Gouaux E. Nature. 2009;460(7255):592–598. doi: 10.1038/nature08198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Marks MJ, Burch JB, Collins AC. J Pharmacol Exp Ther. 1983;226(3):817–25. [PubMed] [Google Scholar]
- 29).Schwartz RD, Kellar KJ. J Neurochem. 1985;45(2):427–33. doi: 10.1111/j.1471-4159.1985.tb04005.x. [DOI] [PubMed] [Google Scholar]
- 30).Buisson B, Bertrand D. J Neurosci. 2001;21(6):1819–29. doi: 10.1523/JNEUROSCI.21-06-01819.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Kuryatov A, Luo J, Cooper J, Lindstrom J. Mol Pharmacol. 2005;68(6):1839–51. doi: 10.1124/mol.105.012419. [DOI] [PubMed] [Google Scholar]
- 32).Sallette J, Pons S, Devillers-Thiery A, Soudant M, Prado de Carvalho L, Changeux JP, Corringer PJ. Neuron. 2005;46(4):595–607. doi: 10.1016/j.neuron.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 33).Srinivasan R, Richards CI, Xiao C, Rhee D, Pantoja R, Dougherty DA, Miwa JM, Lester HA. Mol. Pharmacol. 2012 doi: 10.1124/mol.112.077792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Moroni M, Zwart R, Sher E, Cassels BK, Bermudez I. Mol Pharmacol. 2006;70(2):755–68. doi: 10.1124/mol.106.023044. [DOI] [PubMed] [Google Scholar]
- 35).Nelson ME, Kuryatov A, Choi CH, Zhou Y, Lindstrom J. Mol Pharmacol. 2003;63(2):332–41. doi: 10.1124/mol.63.2.332. [DOI] [PubMed] [Google Scholar]
- 36).Schmauder R, Kosanic D, Hovius R, Vogel H. ChemBioChem. 2011;12(16):2431–2434. doi: 10.1002/cbic.201100302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.