Abstract
The human severe acute respiratory syndrome coronavirus (SARS-CoV) and the NL63 coronaviruses are human respiratory pathogens for which no effective antiviral treatment exists. The papain-like cysteine proteases encoded by the coronavirus (SARS-CoV: PLpro; NL63: PLP1 and PLP2) represent potential targets for antiviral drug development. Three recent inhibitor-bound PLpro structures highlight the role of an extremely flexible six-residue loop in inhibitor binding. The high binding site plasticity is a major challenge in computational drug discovery/design efforts. From conventional molecular dynamics and accelerated molecular dynamics (aMD) simulations, we find that with conventional molecular dynamics simulation, PLpro translationally samples the open and closed conformation of BL2 loop on a picosecond–nanosecond timescale but does not reproduce the peptide bond inversion between loop residues Tyr269 and Gln270 that is observed on inhibitor GRL0617 binding. Only aMD simulation, starting from the closed loop conformation, reproduced the 180° ϕ–ψ dihedral rotation back to the open loop state. The Tyr–Gln peptide bond inversion appears to involve a progressive conformational change of the full loop, starting at one side, and progressing to the other. We used the SARS-CoV apo X-ray structure to develop a model of the NL63-PLP2 catalytic site. Superimposition of the PLP2 model on the PLpro X-ray structure identifies binding site residues in PLP2 that contribute to the distinct substrate cleavage site specificities between the two proteases. The topological and electrostatic differences between the two protease binding sites also help explain the selectivity of non-covalent PLpro inhibitors.
Keywords: accelerated molecular dynamics, homology model, SARS-CoV PLpro, HCoV-NL63, loop dynamics
Abbreviations: SARS-CoV PLpro, severe acute respiratory syndrome coronavirus papain-like protease; nsp, non-structural protein; DUB, deubiquitinating; HCoV, human coronavirus; MSA, multiple sequence alignment; BtCoV, bat coronavirus; MD, molecular dynamics; cMD, conventional molecular dynamics; aMD, accelerated molecular dynamics; 3CLpro, chymotrypsin-like protease; 3D, three-dimensional; 2D, two-dimensional; PDB, Protein Data Bank
Graphical Abstract
Highlights
► aMD reproduces inhibitor induced-fit conformational change in PLpro. ► SARS-CoV PLpro and NL63-PLP2 differing substrate and inhibitor specificities are correlated with electrostatic and topological catalytic site differences. ► Broad-spectrum coronavirus papain-like protease inhibitor discovery must compensate for enzymatic structural and electrostatic variation.
Introduction
Coronaviruses are enveloped, single-stranded, positive-sense RNA viruses.1 The coronavirus responsible for severe acute respiratory syndrome (SARS-CoV) is probably the most studied human coronavirus (HCoV) and produces a unique pathogenesis because it causes both upper and lower respiratory tract infections and can also cause gastroenteritis.2 Although containment of the first SARS epidemic in 2003 succeeded through epidemiological and quarantine measures, there is still no definitive therapy for SARS or other coronaviral infections. Shortly after the SARS outbreak, in 2003, researchers also identified HCoV-NL632, 3 as another HCoV that causes respiratory infections and pneumonia. The virus is found worldwide and infects mainly young children, elderly, and immunodeficient patients. Both SARS-CoV and HCoV-NL63 use angiotensin-converting enzyme 2 as the cellular receptor to infect host cells.4, 5
Coronaviral genomic RNA is released in the cell cytoplasm after infection, which then translates into two long polyproteins pp1a and pp1ab.6 The replicase gene of coronaviruses often encodes two cysteine papain-like proteases, PLP1 and PLP2, and a cysteine chymotrypsin-like protease (3CLpro). SARS-CoV, avian infectious bronchitis virus, and some of the bat coronaviruses (BtCoVs) are distinct in that they encode only one papain-like protease domain.7, 8, 9 In the case of SARS-CoV, autocatalytic processing of the polyproteins by PLpro and 3CLpro generates up to 16 non-structural proteins (nsps). 3CLpro is the main protease that processes multiples sites in the replicase polyprotein and has been targeted for therapeutic development.10, 11 PLpro cleaves pp1a at three sites12 and has been shown to be essential for viral replication.13, 14, 15 The resulting nsps coalesce with the endoplasmic reticulum membrane to form the multifunctional replicase complex. This complex is instrumental in sub-genomic RNA synthesis and, thus, proliferation of infection.16, 17
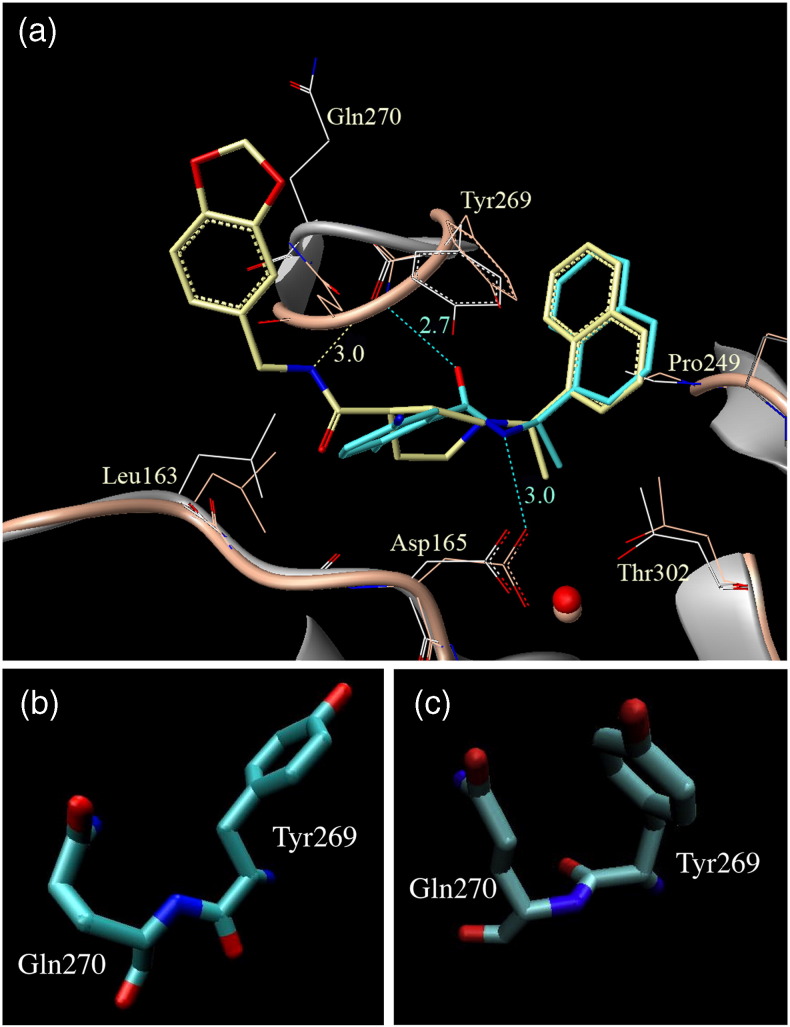
In recent work, we introduced two classes of SARS-CoV PLpro-specific non-covalent inhibitors that exhibit significant SARS antiviral efficacy.13, 14, 15 The crystal structure of inhibitor GRL0617 bound to the protein superimposed on the apo (open) X-ray structure (Fig. 1a ) indicates that group I inhibitors can induce major conformational changes in the binding site mostly dictated by the translation of the flexible BL2 loop (Gly267–Gly272) and the side chain of Leu163 in the BL1 loop.13, 15 In the ligand-bound form, the loop closes down on the ligand, and the peptide bond between loop residues Tyr269 and Gln270 rotates by 180°, enabling the backbone NH group of Tyr269 to make a favorable H-bonding interaction with the carbonyl oxygen in the carboxamide group of the inhibitor. The BL2 loop assumes a closed-inverted conformation compared to the open unbound X-ray structure. These compounds do not show any potency against either NL63-PLP2 or other human deubiquitinating (DUB) enzymes.15 Group II PLpro inhibitors, however, do not induce the peptide bond inversion of the loop residues upon binding.14 The BL2 loop still locks down over inhibitor 15g, which wraps around the loop similar to inhibitor GRL0617. Figure 1b and c shows the two distinct inhibitor-bound conformations of the protein backbone, with group I inhibitors inducing a peptide bond inversion in the loop, while group II inhibitors do not.13, 14, 15
Fig. 1.
Comparison of the BL2 loop from the three X-ray structures. (a) The purple ribbon diagram represents the 15g (shown in yellow stick figure)-bound crystal structure (PDB ID: 3mj5) conformation of SARS-CoV PLpro superimposed on the GRL0617S inhibitor (cyan stick figure) complex protein conformation (PDB ID: 3e9s) shown as gray ribbon. It also highlights the side-chain changes in some of the binding site residues and loop conformation differences between the two ligand-bound protein conformations. (b) A magnified image of the Tyr269–Gln270 peptide bond orientation (BL2 loop) in the apo (PDB ID: 2fe8) and 15g-bound protein X-ray crystal structure. (c) A magnified image of the same peptide bond between Tyr269 and Gln270 on the BL2 loop when bound to inhibitor GRL0617S. A 180° rotation of the peptide bond can be clearly observed between (b) and (c).
To establish a foundation for docking and other in silico screening approaches, we have used conventional molecular dynamics (cMD) followed by accelerated molecular dynamics (aMD) simulations18, 19, 20, 21, 22, 23 to reproduce the observed binding site conformational flexibility. We also examined the correlation between the movement in the inter-sheet loops of the finger domain and the motion of the BL2 loop. We find that peptide bond inversion between the open and closed states of the binding loop is not observed in the picosecond–nanosecond timescale using cMD. However, aMD allows the trajectory to find an appropriate way out of each state at an enhanced rate. This acceleration stems from the addition of a non-negative boost potential that raises the energy within the potential energy basins. This approach increases conformational sampling through modification of the energy landscape of the model system by lowering energy barriers through the application of a boost potential, ΔV(r), to the true potential surface, V(r). Hence, a trajectory propagated on this modified surface makes transitions from state to state with an accelerated rate.19 Starting from the GRL0617-bound BL2 loop conformation, we observe inversion of the peptide bond between residues Tyr269 and Gln270 to a conformation similar to that in the apo BL2 loop conformation in the last 5 ns of the aMD simulation, with the conformational transition propagating in this simulation from Gly267 at the beginning of the loop through the central Tyr269–Gln270 peptide bond to Gly272 at the end of the loop. The resulting detailed description of loop conformational flexibility will provide a solid foundation for future computational screening approaches to discover alternate molecular inhibitor scaffolds.
As noted above, the group I PLpro inhibitors show no activity against the very similar NL63-CoV PLP2 enzyme.15 To better understand molecular recognition in these viral papain-like proteases, and to provide guidance for development of broader-spectrum inhibitors, we have developed a homology model for the NL63-CoV PLP2 active-site structure using SARS-CoV PLpro structure as a template. Except for the anchoring glycines, the BL2 loop of NL63-PLP2 is composed of a completely different set of amino acids (G253–SFDN–G258) from that in SARS-CoV PLpro (G267–NYQC–G272). SARS-CoV PLpro recognizes the consensus sequence –LXGG–, which is similar to the sequence at the C-terminal tail of ubiquitin, thereby rendering it a viral DUB enzyme.12, 24 HCoV-NL63-PLP2 has also recently been identified as a DUB enzyme.25 However, in SARS-CoV, the cleavage of pp1a by PLpro occurs at three sites after Gly–Gly sequences, to release nsp1, nsp2, and nsp3.3 NL63-PLP2 recognizes similar substrate sequences, but the cleavage sites differ, with cleavage occurring between a Gly–Gly and Ala–Gly sequence instead of after Gly–Gly as in SARS-CoV PLpro.25
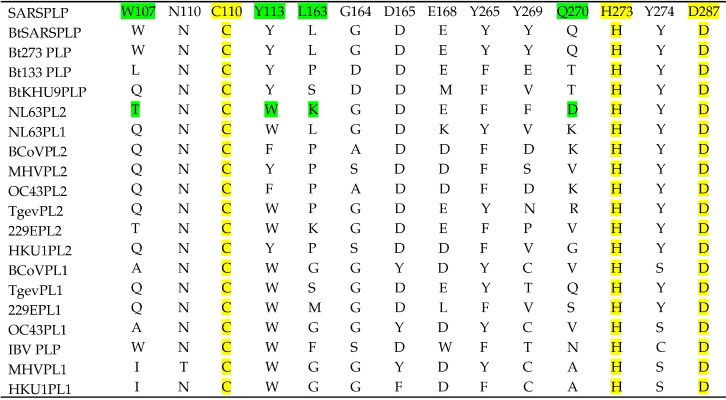
Following the approach of Sulea et al.,26 we constructed our own multiple sequence alignment (MSA) for 20 coronaviral PLP domains from both R and O groups of 13 coronaviruses including HCoV-SARS using a profile alignment strategy. The ubiquitin aldehyde-bound protein model proposed by Ratia et al. that proposed interactions between the substrate RLRGG in the active site of SARS-CoV PLpro provided information on the probable substrate binding pockets.12 We focused on 12 binding site and the two BL2 loop residues (Tyr269 and Gln270) that are instrumental in ligand binding from the SARS-CoV PLpro apo and two inhibitor-bound X-ray structures12, 14, 15 to identify the corresponding residues in the PLP domain of other organisms (Table 1 ). Based on the MSA, we built homology models for NL63-PLP1 and NL63-PLP2 using the experimental three-dimensional (3D) apo structure of SARS-CoV PLpro12 as a template. NL63-PLP1 was predicted to belong to the O group and lacks DUB activity and is hence less interesting for our comparative study here.26 In this study, we focus on the 3D structural superimposition of the model on the apo crystal structure to identify substrate cleavage site specificity determining residues between the binding pockets of SARS-CoV PLpro and NL63-PLP2 only. The electrostatic and topological differences between the binding sites of the two proteases defined by these residues, along with docking studies of our PLpro inhibitors, suggest mechanisms of inhibitor selectivity.
Table 1.
The 12 active-site residues and two BL2 loop (Y269 and Q270) residues of SARS-CoV PLpro and their corresponding aligned residues in the active sites of other coronaviral PLpros
The catalytic residues are highlighted in yellow. The residues highlighted in green are a few of the important residues that differ significantly between SARS-PLpro and NL63-PLP2.
Results and Discussion
Loop dynamics: Structural evolution along the course of the cMD trajectory
RMSD analysis from the 12-ns cMD trajectory was performed by fitting the backbone heavy atoms from the palm domain (Ser241–Lys307), excluding the two flexible loops (Gly267–Gly272 and Thr199–Glu204). The RMSD of the BL2 loop (Gly267–Gly272) was then calculated with respect to that of the crystal structures.
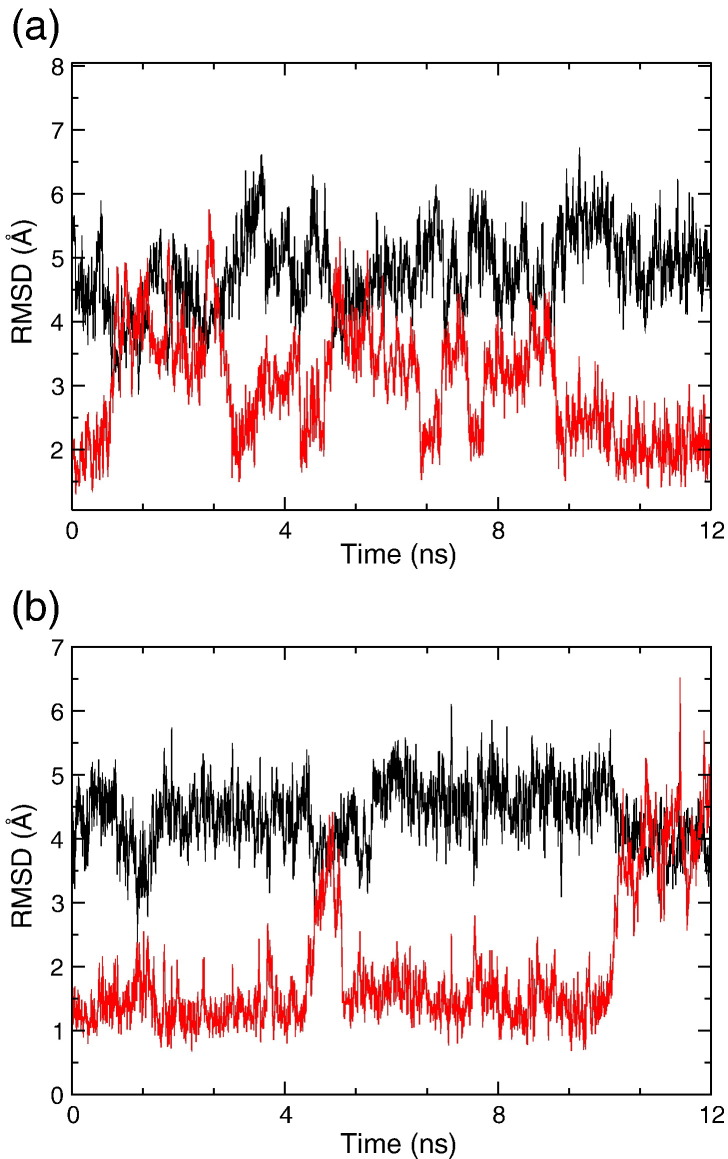
The RMSD profiles showed that the BL2 loop exhibits large picosecond–nanosecond flexibility. When started from the open form of the BL2 loop, the PLpro protein was able to translationally sample the closed form at a number of time points (∼ 4.5, 6, and 7.5 ns). Meanwhile, when the starting conformation of the BL2 loop was closed, the structure was able to translationally revert to the open form (∼ 5 and 10 ns). The analyses performed on both systems suggest that the BL2 loop preemptively samples both the open and the closed conformation in the absence of any ligand as seen in Fig. 2a and b.
Fig. 2.
RMSD showing the BL2 loop of PLpro can translationally sample both open and closed conformations during 12 ns of cMD. (a) RMSD profile of 12 ns simulation for system I (started from the apo form of PLpro). The BL2 loop can sample the translational closed conformation around 4.5, 6, and 7.5 ns. The RMSD of the apo form of the protein with respect to the apo X-ray structure is shown in black, whereas the RMSD with respect to the bound X-ray structure is shown in red. (b) RMSD profile of 12 ns simulation for system II (started from the bound form of PLpro). The closed form of the loop can sample the open state around 5 and 10 ns. The RMSD profile of the bound form of the protein over the simulation with respect to the apo X-ray structure is shown in black, whereas the RMSD with respect to the GRL0617 ligand-bound X-ray structure is shown in red.
RMSD-open versus RMSD-closed
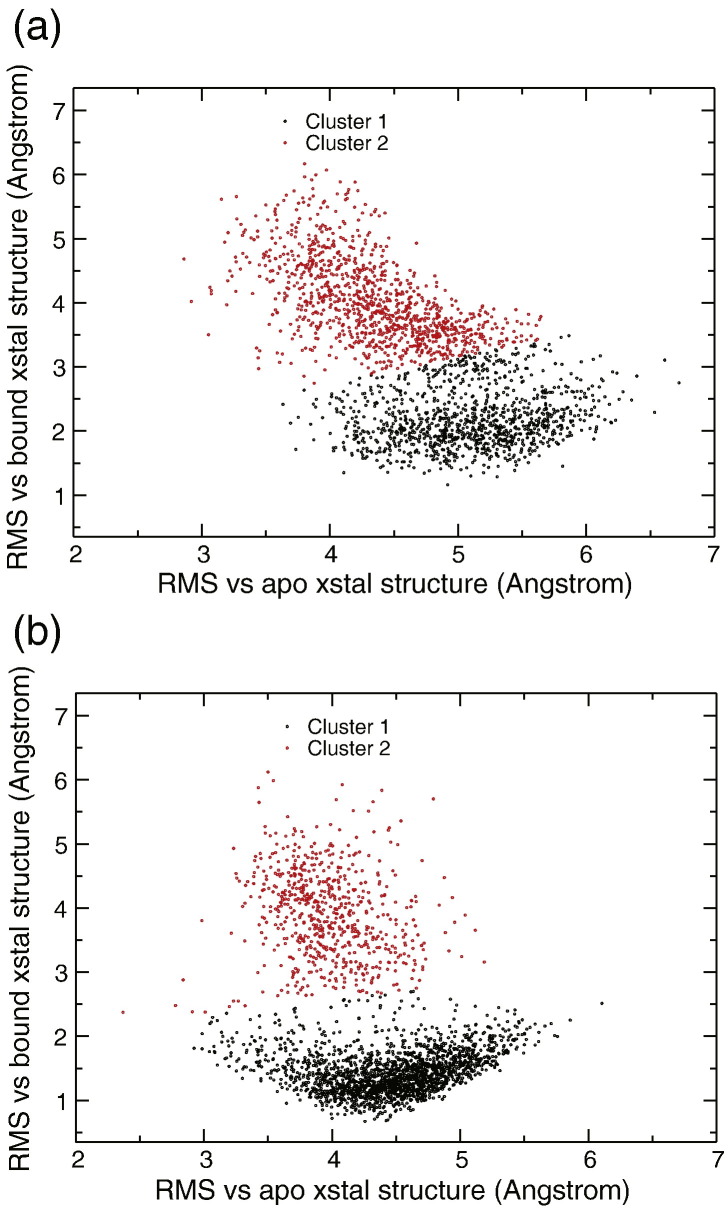
We performed k-means clustering,27 which shows that the RMS space can be categorized into two distinct clusters, as shown in Fig. 3a and b. For apo PLpro, the two clusters are centered at (5.09 Å, 2.14 Å) and (4.37 Å, 3.95 Å), respectively. For holo PLpro, the two clusters are centered at (4.48 Å, 1.39 Å) and (3.96 Å, 3.85 Å), respectively. One cluster shows low RMSD (∼ 1.2 Å) with respect to the closed loop form, whereas the other RMSD coordinate varies from 2.2 to 6.4 Å. The second cluster is more scattered but shows anti-linear behavior between the two RMSD coordinates. Regardless of whether starting from the apo or the holo structure, the shape and distribution of the data points are very similar, as noted in Fig. 3a and b.
Fig. 3.
(a) 2D RMS plot for the 12-ns-long MD trajectory starting from the apo structure. The black and red points represent the two clusters centered at (5.09 Å, 2.14 Å) and (4.37 Å, 3.95 Å), respectively. (b) 2D RMS plot for the 12-ns-long MD trajectory starting from the holo structure. The black and red points represent the two clusters centered at (4.48 Å, 1.39 Å) and (3.96 Å, 3.85 Å), respectively.
Loop dynamics: aMD to observe ϕ–ψ dihedral torsion inversion of the Tyr269–Gln270 peptide bond from bound to apo form
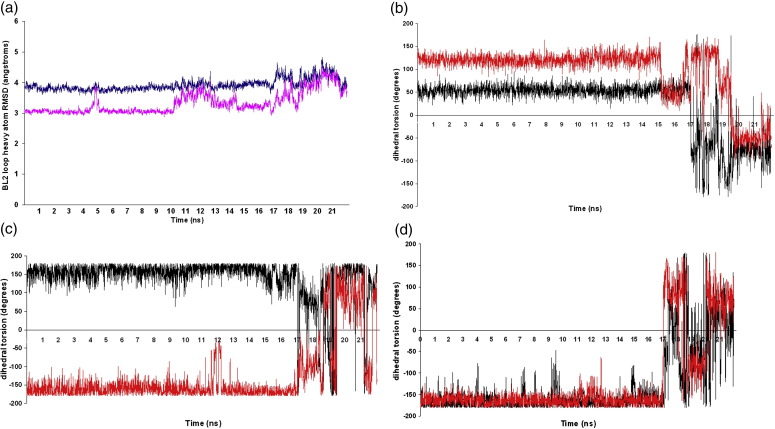
The 12-ns cMD translationally samples open and closed states of the BL2 loop reasonably well but does not replicate the experimentally observed dihedral torsion inversion between loop residues Tyr269 and Gln270 in the picosecond–nanosecond timescale, presumably due to the torsional energy barrier that must be overcome. Thus, the equilibrated system after 12 ns of cMD was subjected to 5 ns of whole-protein aMD, followed by 5 ns of aMD with a single boost to the potential on the 10 critical loop residues (Tyr265–Tyr274). After 19.5 ns of total simulation time, during the last 2.5 ns of aMD, we observed the peptide bond between Tyr269–Gln270 (the ψ of Tyr269 and ϕ of Gln270) invert from the GRL0617-bound protein conformation (X-ray dihedral angles of 124.9° and 72.2°) to the dihedral torsions of the apo open state (− 74.9 for ϕ of Gln270 and − 58.4 for ψ of Tyr269 for chain B, with the other two chains in the apo X-ray structure having comparable values). It is particularly interesting to note from Fig. 4a that starting from the closed form of the BL2 loop, although it translationally samples the open conformation around 5 ns, and again from ∼ 10 to 13 ns, it does not sample the dihedral rotation of the peptide bond during the same timescale without the additional boost applied through aMD just to the loop residues. In Fig. 4b, we show that not only does the BL2 loop sample both the open and the closed conformations, but the ψ–ϕ dihedral of the Tyr–Gln peptide bond also inverts from the starting closed state to the open form after 19.5 ns of total simulation time (12 ns cMD plus 7.5 ns aMD). During the last 2.5 ns, the closed BL2 loop continues to assume the open protein loop conformation (both translational and rotational) until the end of the simulation.
Fig. 4.
(a) The heavy-atom RMSD of the 10 amino acids from the BL2 loop (Gly262–Gly272) over 22 ns of total simulation time, starting from the bound closed form of the loop. The loop assumes the open state around 5, 10–13.5, 14.5, 18, and 19.5–22 ns. The RMSD profile of the bound form of the protein over the simulation with respect to the apo X-ray structure is shown in blue, whereas the RMSD with respect to the GRL0617 ligand-bound X-ray structure is shown in magenta. (b) The ϕ–ψ dihedral of the peptide bond between Tyr269–Gln270 inverts from the starting closed GRL0617-bound conformation (ϕ of Gln270, 72.2; ψ of Tyr269, 124.9) to the open unbound state (ϕ of Gln270, − 74.9; ψ of Tyr269, − 58.4) during the final 2.5 ns of simulation time. aMD on the 10 loop residues successfully sampled the ϕ–ψ dihedral rotation from one state to another. The ϕ angle of residue Gln270 is shown in black, and the ψ of Tyr269 is shown in red. (c and d) The dihedral torsion changes of two neighboring loop residues, Gly267 and Gly272, which eventually lead to the –Tyr–Gln– peptide bond inversion. The ψ angles (shown in red) of Gly267 as seen in (c) and of Gly272 as seen in (d) (shown in red) start increasing after 17 ns, which may allow the decrease of the ϕ of Gln270, and then around 19.5 ns, the ψ of Tyr269 starts decreasing until the Tyr–Gln peptide bond assumes the apo open loop conformation. The ϕ angles of the hinging glycine residues are shown in black and the ψ angles are shown in red.
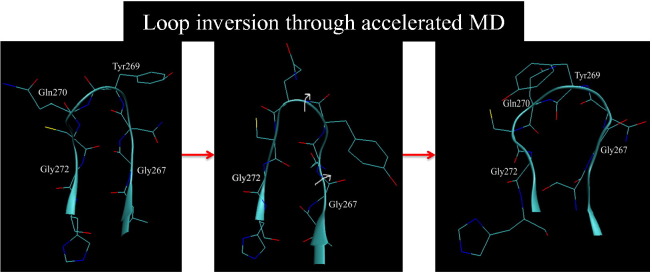
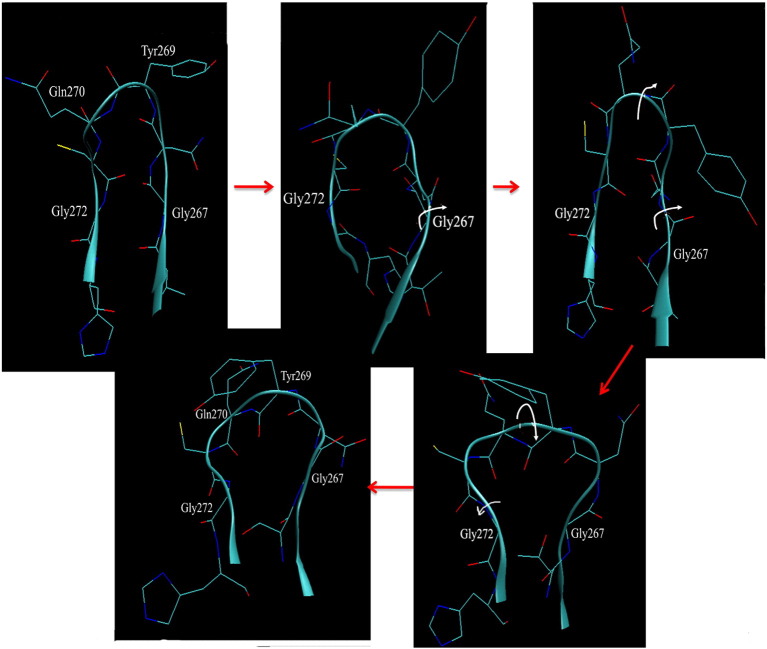
Some of the neighboring loop residues were studied to determine the order of torsion changes required to enable this peptide bond inversion. It is interesting to note that the BL2 loop is flanked by two Gly residues on each end that allow for considerable translational and rotational degrees of freedom for the loop by acting as hinges for the extensive loop motion. The ψ of Asn268 partially rotates first and stabilizes during 10–17 ns, then after 17 ns, it goes back to its original bound conformational torsion. The ϕ–ψ dihedral plots of Gly267 and Gly272 and Asn268 in Fig. 4c and d show that the ψ angles of both Gly267 and Gly272 start increasing and eventually do a complete 180° inversion after 17 ns. This inversion of the Gly residues is probably correlated with the Gln270 ϕ inversion to its apo conformation at nearly the same time. These inverted dihedrals remain stable for another 2 ns, after which the ψ of Tyr269 also rotates by 180° around 19.5 ns. At this point, both the Gln270 ϕ and the Tyr269 ψ sample the apoprotein dihedrals for the next 2 ns and the BL2 loop assumes the open state. In the last 2 ns of the simulation, the Gly267 ψ fluctuates; however, the Gly272 ψ stabilizes in the inverted state. This transition is summarized in more detail in Fig. 5 , where it can be seen that the conformational transition starts on the Gly267 side of the loop, moves across, and ends on the Gly272 side. The apo crystal structure demonstrates that the Gly272 ψ indeed inverts from − 144.8 in the GRL0617-bound form to + 134 (average of three chains 135, 154, and 113), whereas the ϕ remains unchanged. Similarly, the ϕ–ψ of Gly267, which can readily invert back to its starting dihedral torsions after the peptide bond flip, agrees well with the Gly267 ϕ–ψ dihedral values from the apo X-ray structure. Hence, we can conclude that the inversion of at least the ψ angle of one Gly residue is required for the peptide bond inversion between Tyr269 and Gln270. However, it should be noted that this transitional order is observed from a single simulation and that more extensive averaging over trajectories would be required to confirm the preferred inter-conversion pathway.
Fig. 5.
aMD simulation snapshots that capture the transition of the peptide bond inversion between Tyr269 and Gln270, starting from the bound (GRL0617S-bound) to the apo form from left to right, top to bottom. The white arrows point to the dihedral rotation of the neighboring glycine residues; first, Gly267, which partially enables the loop to rotate around Tyr269 and Gln270, turns and, finally, the ψ of Gly272 inverts completely, allowing the complete inversion of the peptide bond. The ψ angles observed from the crystal structures of the apo (PDB ID: 2fe8) and the 15g-bound loop conformations (PDB ID: 3mj5) further confirm this ψ inversion of Gly272 when compared to the ψ angle of Gly272 in the starting 617-bound conformation denoting a 180° rotation.
Overall electrostatic potential maps for SARS-CoV PLpro, NL63-PLP2, and NL63-PLP1 active sites
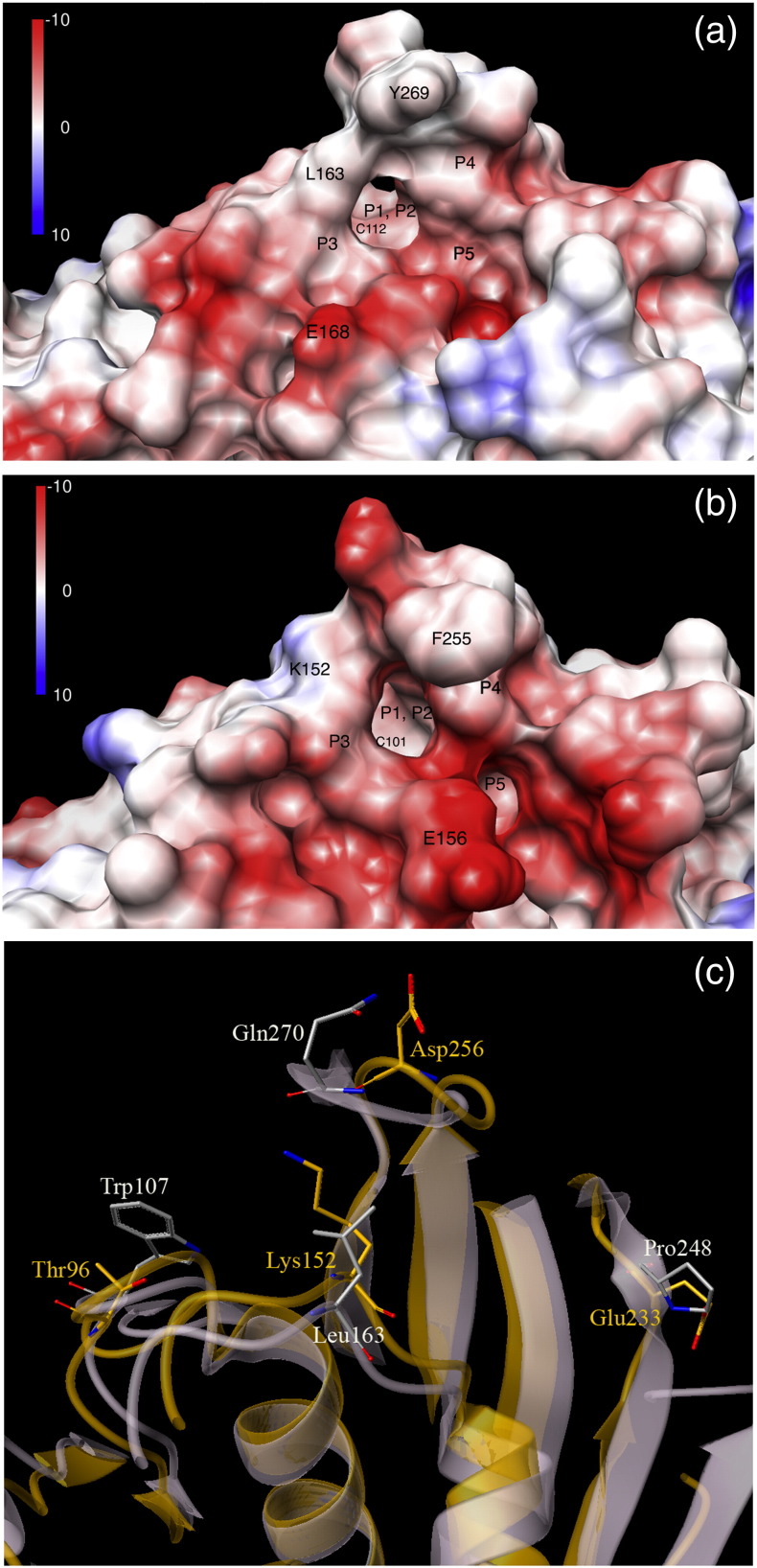
Figure 6b depicts the solvent-exposed electrostatic contour map of the active site of the HCoV-NL63-PLP2 model, which is compared to that of the X-ray structure of SARS-CoV PLpro in Fig. 6a. The active site in SARS-CoV PLpro is broadly electronegative, the P1 and P2 sites are mainly neutral if not slightly electropositive, the P3 pocket is fairly electronegative, while the P4 and P5 pockets are quite basic.
Fig. 6.
(a) Surface electrostatic maps of the active sites of SARS-CoV PLpro. (b) Surface electrostatic maps of the active sites of the NL63-PLP2 model. The substrate binding pockets and binding site residues analogous to Glu168 and Leu163 in SARS-CoV PLpro are labeled as Glu156 and Lys152. (c) Ribbon diagram of superimposed structures in (a) and (b). The superimposition highlights the differing electrostatic and lipophilic nature dictated by residues between the NL63-PLP2 model (in orange) and SARS-CoV PLpro apo crystal structure conformations (in white). The color gradation from red to white to blue in (a) and (b) shows the changing electrostatic potential from negative to neutral to positive (scale from − 10 to 0 to + 10 kcal/mol⁎e) at 298 K.
In our PLP2 model, the P1 and P2 sites are neutral, with dense electropositive patches in the neighboring sites. The residues around the P3 site of the enzyme HCoV-NL63-PLP2 follow a KGD (K152–D154) and F251 pattern instead of an LGD (L163–D165) and Y265 as in SARS-CoV PLpro; this generates a slightly more electropositive environment in HCoV-NL63-PLP2 than in SARS-CoV PLpro. The P4 and P5 pockets are fairly electronegative, as in SARS-CoV PLpro. In the P4 pocket of SARS-CoV PLpro, Pro248, Pro249, Tyr265, and Thr302 create a small hydrophobic crevice where the naphthalene rings of inhibitors fit in favorably. In the PLP2 model, the nonpolar hydrophobic Pro248 analogous residue is an acidic glutamic acid (Glu233) that mildly changes the nature of the pocket. The P4 and P5 pockets in both proteases also look topologically different. Figure 6c shows the superimposition of SARS-CoV PLpro apo crystal structure with the PLP2 model highlighting the differing binding site residues.
Intriguingly, the P5 pocket of HCoV-NL63-PLP1 is likely to be electropositive, in contrast to other enzymes, due to the presence of K163 at a position analogous to that of Glu168 (Glu156) in SARS-CoV PLpro (NL63-PLP2), and NL63-PLP1 is not reported to have DUB activity.25 Glu168 in SARS-CoV PLpro is in position to form a salt bridge with the S5 arginine side chain in the –LRGG– natural substrate of SARS-CoV PLpro. From a structural perspective, this appears important in determining DUB activity for the enzymes, since a change in surface electrostatic potential around the mouth of the active site would likely disrupt the binding of the substrate (C-terminal tail of ubiquitin, RLRGG) for cleavage and therefore produce loss of the enzyme's DUB functionality. Consequently, we predict that mutating residue Glu156 to Lys in NL63-PLP2 should lead to loss of DUB activity. The electrostatic characteristics of the binding site do not appear to be significantly influenced by dynamics. Four representative snapshots generated by the ptraj clustering utility from the most diverse clusters based on RMSD diversity showed no significant pocket reorganization during the cMD simulation.
Differences in substrate specificity between SARS-CoV PLpro and HCoV-NL63-PLP2 due to P3 pocket electrostatic potential
To compare sites, we first examined and analyzed the hydrogen-bonding interactions between GRL0617 and P1–P5 sites over the cMD simulation with a distance cutoff for hydrogen bonds set at 3.7 Å, and an angle cutoff set at 120°, with the results shown in Table 2a, from which it can be seen that the hydrogen-bonding interaction occupancies between PLpro and GRL0617 vary from ∼ 7% to 16%. Similarly, the hydrophobic interactions were qualitatively measured as the distance between the centroid of interacting aromatic pairs (and the five-membered rings of proline, in the case of hydrophobic interactions other than π–π stacking). The π–π stacking cutoff was set at 7 Å, whereas the cutoff for hydrophobic interaction was set at 5 Å. These interactions are described in Table 2b, from which it can be seen that only the hydrophobic interaction between the inhibitor naphthyl ring and Pro249 exhibits significant occupancy. No salt bridge interactions were measured, as there are no highly polar/charged groups in GRL0617. Table 2c then lists the number of hydrogen-bond donors and acceptors in the P1–P5 sub-pockets of each of these enzymes. The numbers of hydrogen-bond donors and acceptors can only be an estimate, as we have limited knowledge of how many of the H-bonds are made or broken within the protein due to changes in the side-chain rotamers of the residues upon substrate binding. Nonetheless, clear differences can be observed among HCoV-NL63-PLP1, HCoV-NL63-PLP2, and SARS-CoV PLpro.
Table 2.
(a) PLpro inhibitor hydrogen-bonding interactions, (b) PLpro inhibitor hydrophobic/π–π interactions, and (c) characterization of overall electrostatic potential (EP) in the substrate binding pockets of three enzymes
| (a) | ||||
|---|---|---|---|---|
| Donor | Acceptor | Distance (Å) | Occupancy (%) | Site |
| GRL0617 N2 | TYR 265 OH | 3.64 | 7.12 | BL2 loop |
| GRL0617 O1 | TYR 274 OH | 3.31 | 5.12 | Near BL2 loop |
| GRL0617 N2-H18 | ASP 165 OH1 | 3.23 | 17.25 | P3 |
| GRL0617 N2-H19 | ASP 165 OH2 | 3.22 | 15.92 | P3 |
| (b) | ||||
|---|---|---|---|---|
| Donor | Acceptor | Distance (Å) | Occupancy (%) | Site |
| GRL0617 (benzene ring) | TYR 269 (side chain) | 6.13 | 8.04 | BL2 loop |
| GRL0617 (naphthalene ring) | PRO248 (proline ring) | 4.74 | 7.88 | P4 |
| GRL0617 (naphthalene ring) | PRO249 (proline ring) | 4.82 | 35.8 | P4 |
| (c) | ||||
|---|---|---|---|---|
| Enzyme | P1 and P2 |
P3 |
P4 |
P5 |
| D/Aa | D/Aa | D/Aa | D/Aa | |
| SARS-CoV PLpro overall EP | 3/1 weakly (+) | 1/1 weakly (−)/neutral | 1/2 (−) | 1/3 (−) |
| NL63-PLP2 model overall EP | 2/1 weakly (+)/neutral | 3/2 weakly (+)/mix of neutral patches | 1/3 (−) | 1/3 (−) |
| NL63-PLP1 model overall EP | 3/1 weakly (+) | 2/1 weakly (+) | 1/2 (−) | 4/3 (+) |
Number of hydrogen-bond donors (D) and acceptors (A) in the binding pockets of SARS-PLpro crystal structure and the homology-based models of NL63Plp1 and NL63Plp2. The (+) and (−) signs indicate positive and negative potentials, respectively.
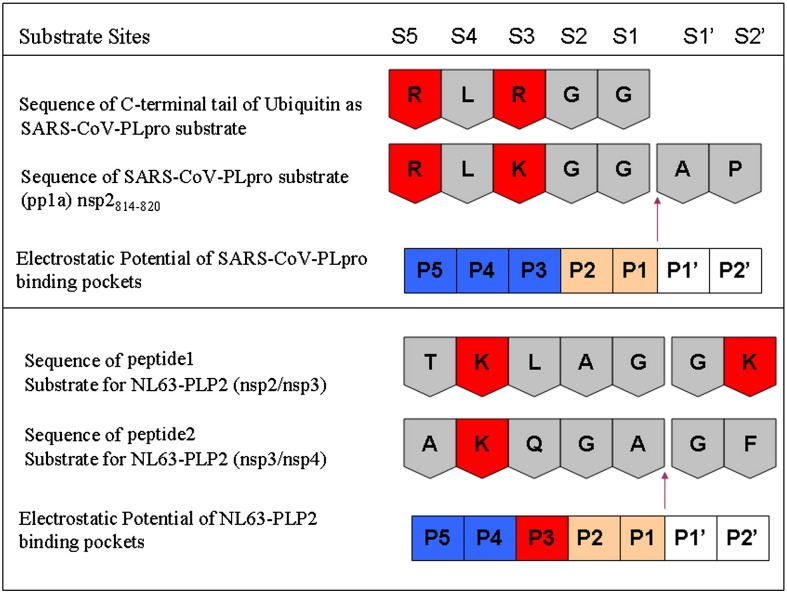
Chen et al. have recently shown that the substrate specificities differ between SARS-CoV PLpro and NL63-PLP2.25 NL63-PLP2 has DUB activity; both SARS-CoV PLpro and NL63-PLP2 cleave the LRGG tail of ubiquitin. Both proteins cleave similar substrates, but at different sites. NL63-PLP2 cleaves between nsp2 and nsp3 (FTKLAG↓GKISFS) and between nsp3 and nsp4 (VAKQGA↓GFKRTY),25 whereas SARS-CoV PLpro cleaves at LNGG↓AVT, LKGG↓API, and LKGG↓KV to release nsp1, nsp2, and nsp3.12 Figure 7 summarizes the difference in the substrate cleavage sites (between S1 and S1′) between the two enzymes. Considering the consensus sequence of LXGG, SARS-CoV PLpro cleaves after the Gly–Gly sequence whereas NL63-PLP2 cleaves between the two glycines, a one-position shift to the left in the cleavage site.
Fig. 7.
Substrate specificity and substrate cleavage sites for the enzymes SARS-CoV PLpro and NL63-PLP2 are summarized here. The electrostatic potential in the five pockets (P1–P5) of the enzymes, SARS-CoV PLpro, and NL63-PLP2 are color coded: red for positive charge, blue for negative charge, and gray for neutral. Two substrates are shown for each enzyme, also color coded in a similar fashion. The arrow denotes the cleavage site in the substrates by their respective proteases. In NL63-PLP2, P3 is positively charged (red) as compared to P3 in SARS-CoV PLpro; this forces the substrate in NL63-PLP2 to shift one residue and accommodate the cationic residue at S3 position in the P4 pocket.
We suggest that the difference in substrate specificity may result in part from the difference in the environment of the catalytic pocket in the P3 region. From Table 2, we observe that Leu163 in SARS-CoV PLpro is substituted by Lys152 in NL63-PLP2. Clearly, the presence of a cationic residue (Lys or Arg) at the substrate S3 position will not be accommodated well by the Lys (K152) in the active site of NL63-PLP2, which partly determines the P3 pocket specificity. Instead, the substrate Lys is more likely accommodated in the P4 pocket of NL63-PLP2, which is broadly electronegative. This results in cleavage of the substrate of NL63-PLP2, not after the Gly–Gly sequence in FTKLAGG↓K unlike that of SARS-CoV PLpro, but shifting the cleavage site by one residue to the left as in FTKLAG↓GK. The same applies for the substrate VAKQGA↓GF, as the cleavage occurs in a way that still allows only small residues to fit into the narrow tunnel around the catalytic triad but allows accommodation of the lysine in the P4 position instead of P3, unlike the suggested substrate orientation12, 26 in the active site of SARS-CoV PLpro.
Validating inhibitor specificity from docking studies
Structural analysis of the PLP2 model leads us to believe that some of the SARS-CoV PLpro inhibitors, especially the ones that bind through strong H-bonding interactions, are unlikely to inhibit NL63-PLP2 due to the differing electrostatic and stereochemical properties of their active sites. Structural models presented here correlate well with current experimental data, with recent work showing that the SARS-CoV PLpro inhibitor GRL0617 is inactive against NL63-PLP2.15 The compound docks in the P3–P4 sub-sites of the SARS-CoV PLpro enzyme as shown in Fig. 1a, with the naphthalene rings favorably sandwiched between Tyr269 and Pro249 along with the Tyr265 in the bottom of the highly hydrophobic P4 pocket (occupied by Leu in the –LRGG– substrate sequence).
From Table 1, Table 2, we note that a basic Lys residue in PLP2 substitutes the nonpolar hydrophobic Leu in the P3 pocket of SARS-CoV PLpro. The polar uncharged loop residue Gln270 in SARS-CoV PLpro is replaced by an acidic Asp residue in PLP2, which partly changes the electrostatic characteristics of the loop. The loop in PLP2 that corresponds to the BL2 loop in SARS-CoV PLpro is composed of a completely different set of amino acids (G267–NYQC–G272 in SARS-CoV PLpro versus G253–SFDN–G258 in PLP2). Additionally, the nonpolar hydrophobic Pro248 residue in the P4/P5 pocket is substituted by an acidic Glu in PLP2.
These residue substitutions not only change the electrostatic nature of the binding pockets but also change the topology between SARS-CoV PLpro and NL63-PLP2. GRL0617S is not very polar, and only the amide nitrogen stabilizes binding via two hydrogen bonds with residues Asp165 and Gln270.15 Thus, it is likely that the efficacy of GRL0617 against NL63-PLP2 is substantially limited due to both topological and electrostatic inconsistencies between the active sites of the targets. We attempted to dock GRL0617 in the active site of NL63-PLP2 using the commercial docking software GOLD v4.0 (all settings were set to default values) and compared it to the GRL0617-bound SARS-CoV PLpro structure. The resulting docked model in Fig. 8a shows that the H-bonding interaction otherwise made by the amide group is lost here (unless we assume the loop similarly inverts as in PLpro to enable an H-bonding interaction); the core of the molecule is not stabilized by any strong interaction. In the SARS-CoV PLpro-bound GRL0617S structure, the P4 pocket hydrophobic interactions are much stronger, which probably dictates the alignment of the molecule to form H-bonding with the loop residues, while for PLP2, the limited hydrophobicity and difference in topology in the P3 and P4 pockets prevent GRL0617 from binding optimally in the active site of the PLP2 model. This is further confirmed by poor docking scores for the 10 retained ligand conformations when compared to docking scores of GRL0617 in the active site of SARS-CoV PLpro.
Fig. 8.
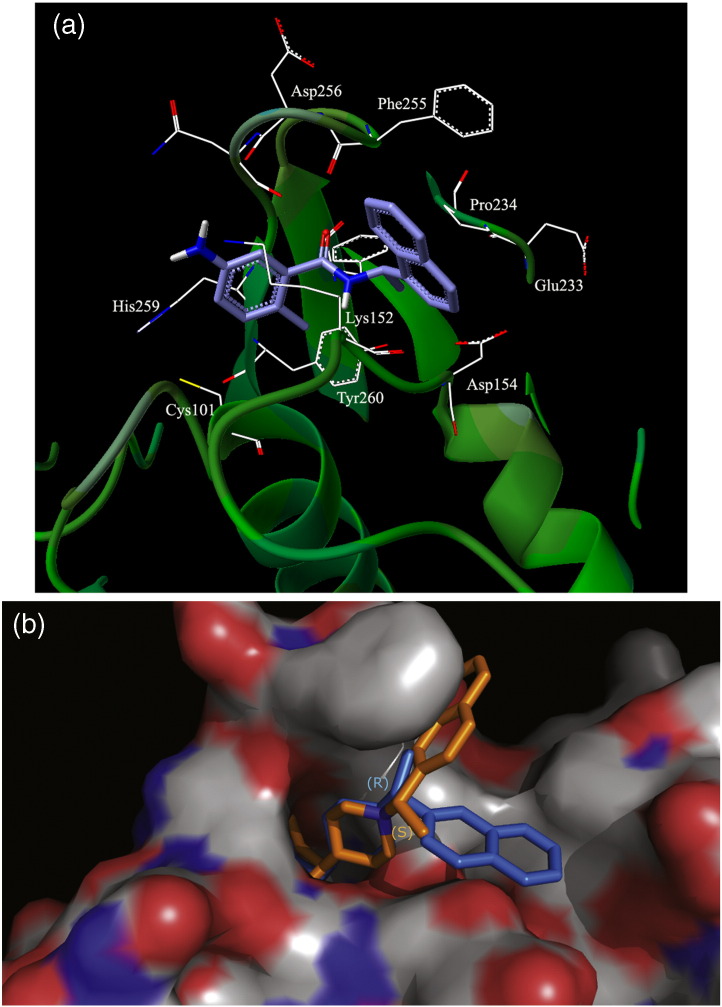
(a) The docked model of the best scoring GRL0617 conformation in the active site of the PLP2 model. The PLP2 model is represented by a green ribbon diagram, and the ligand is shown in blue. The ligand does not make any H-bonding interactions in the binding site. (b) The PLP2 model is represented as a van der Waals surface with hydrophobic regions in gray and polar residues in red (acceptor) and blue (donor) regions. The docked conformations of the previously published group II compounds 15g and 15h are shown in orange and blue, respectively. The (S)-Me stereoisomer, 15h, allows a π–π stacking with residue Phe255 on the loop that is lost in the case of the (R)-Me conformation in 15g.
However, it is important to note that, although this class of non-covalent inhibitors is not active against NL63-PLP2, other compounds that do not rely on the P3 and P4 pocket interactions but instead take advantage of the similarities between the two active sites (P1, P2, and P5 pockets) could become common inhibitors. Larger compounds that can be anchored in the binding site by the two ends (e.g., with biphenyl rings on one end) could be stable in the P5 pocket making aromatic–hydrophobic interactions, whereas long meta- or para-substituents to a phenyl ring around P2 could possibly make favorable H-bonding/hydrophobic interactions on the other end. In Fig. 8b, we show the docked conformation of 15g14 compared to the docked orientation of 15h,14 the (S)-Me stereoisomer, in the active site of NL63-PLP2. The naphthyl rings in 15h [in the (S)-Me conformation] can form a favorable π–π stacking interaction with the Phe255 in the protein that is lost for 15g in the (R)-Me conformation. Hence, in the absence of crystal structures, our NL63 models can provide valuable insight for further broad-spectrum drug development and design studies.
Conclusions
The recently solved inhibitor-bound crystal structure in complex with SARS-CoV PLpro13, 15 shows major conformational changes in comparison to the apo PLpro structure,12 with the binding site BL2 loop closing down on the inhibitor and undergoing a peptide bond inversion upon inhibitor binding. We studied the dynamics of the highly flexible BL2 loop via an initial 12 ns of cMD simulations of both the apo and the bound forms of the protein. PLpro translationally samples the open and closed conformation of the BL2 loop on the nanosecond–picosecond timescale. Two-dimensional (2D) RMSD plots also showed two-state distributions of the loop structures. The BL2 loop is most correlated with the inter-β-sheet loop in the finger domain, despite a separation of 40 Å. However, surmounting the energy barrier to reproduce the peptide bond inversion from the inhibitor GRL0617-bound protein conformation to the apo form could only be achieved through aMD where we supply a constant non-negative boost to the system to enhance its escape rate from one state to another. The resulting transition is not a simple inversion but begins with a torsional transition in the anchoring Gly at one end of the loop, proceeds across the loop, and ends with a torsional transition in the anchoring Gly at the other end of the loop. The characterization of PLpro active-site dynamics resulting from this work will provide a solid foundation for the development of future docking approaches to be used for computational screening to identify alternate scaffolds for future inhibitor development.
To better understand the potential for development of broad-spectrum PLpro/PLP inhibitors, we built a model for HCoV-NL63-PLP2, using the crystal structure of SARS-CoV PLpro as the template for threading. Structural superimposition of the two proteases suggested residues that may influence molecular recognition and differentiate one protease from another. We showed that electrostatic forces within the substrate binding pockets play an important role in differentiating the substrate specificities of SARS-CoV PLpro from those of NL3-PLP2. Single-residue substitutions in the active sites among the coronaviral proteases can be readily identified from Table 1. However, the impact of these substitutions is not clear from the 2D sequence alignment alone. Structural models provide additional insights into the stereochemical characteristics and topology of the active site that help determine specific substrate–protein interactions as seen through Fig. 6a–c.
We suggest that the shift in substrate (LRLRGG) specificity between NL63-PLP2 and SARS-CoV PLpro is mainly due to the replacement of the residue analogous to Leu163 in SARS-CoV PLpro by a cationic lysine in the P3 pocket region of NL63-PLP2. Clearly, the presence of a cationic residue (Lys or Arg) at the substrate S3 position will not be accommodated well in the P3 pocket of NL63-PLP2 but will be better accommodated in the broadly electronegative P4 pocket. This results in a one-residue shift in the alignment of the substrate in the active site of NL63-PLP2, as shown in Fig. 7. Thus, NL63-PLP2 cleavage occurs between the Gly–Gly sequences in LXGG instead of after the Gly–Gly sequence, as in SARS-CoV PLpro.
Our work thus indicates that the active sites of SARS-CoV PLpro, NL63-PLP2 and NL63-PLP1 are distinct. Our models suggest that the electrostatics contribute significantly to the difference in substrate specificity between the two enzymes, which can be probably addressed through further mutation studies. Identification of residues that cause the distinction will aid in future drug development efforts. Our current SARS-CoV PLpro inhibitors do not inhibit NL63-PLP2, and it appears likely that development of broad-spectrum papain-like protease inhibitors will require structural changes to accommodate the differing topologies and binding site residues highlighted in the binding site of HCoV-NL63-PLP2.
Methods
cMD simulation
MD simulations were carried out with the AMBER10 suite of programs28 for 12 ns using the ff99 force field.29 During the density equilibration, the pressure relaxation time was 0.2 ps and the temperature coupling parameter was 2.0. SHAKE constraints were applied so that bonds involving hydrogen were constrained.30, 31 During constant energy simulation, the pressure relaxation time was 2.0 ps and the temperature coupling parameter was 5.0. Throughout all simulations, the time step was set to 2 fs, and the particle mesh Ewald method was employed32, 33, 34, 35 accordingly with a direct sum tolerance of 0.000001. For minimization, the solvent was first minimized with the solute fixed with a 10-kcal/mol·Å constraint, where 1010 steps of steepest descent minimization were performed followed by at most 50,000 steps of conjugate gradient or until the RMS gradient was less than 10− 4 kcal/mol·Å. Then, the constraints on the solute were released, and 100 steps of steepest descent minimization were performed followed by at most 50,000 steps of conjugate gradient or until the RMS gradient was less than 10− 4 kcal/mol·Å. Two model systems were studied, the first of which was directly adapted from the apo (open) crystal structure [Protein Data Bank (PDB) ID: 2fe8],12 with the second system extracted from the closed complex structure of GRL0617 bound to PLPro (PDB ID: 3e9s)15 with the GRL0617 inhibitor removed. These structures show that the thumb, finger, and palm sub-domains of PLpro are in close contact with each other. In contrast, the ubiquitin-like domain is loosely connected to the rest of the catalytic domain and is separated from the active site by 30–50 Å. Hence, we considered the ubiquitin-like domain to be less relevant to the BL2 (Gly267–Gly272) loop movement and excluded it from our study. These modified structures, with open and closed BL2 loops, were used as starting points of our MD simulations. Each of the model systems was solvated in a 10-Å octahedral waterbox of TIP3P waters36 (total, 43,410 atoms). Adding one sodium counterion neutralized the systems. Minimization was carried out in two steps, with water molecules further than 10 Å from the protein minimized first for 50,000 steps followed by minimization of the entire system for another 5000 steps. Periodic boundary conditions were applied with a 10-Å non-bonded cutoff. The systems were first heated from 50 K to 300 K using the NVT ensemble in 50 ps. Then, the systems were density equilibrated for 2.95 ns using the NPT ensemble. The production run was performed in a microcanonical ensemble for 12 ns (NVE). NVE was used to permit obtaining information about the time dependence and equilibrium behavior of the system.
Accelerated molecular dynamics
aMD simulations were carried out using a modified version of the sander module in AMBER9. The aMD method has been previously described.18, 20, 21, 22, 23, 37, 38, 39 This method promotes sampling of infrequent events without prior knowledge of the locations of wells and barriers on the potential energy surface. Our implementation used the aMD version developed by Hamelberg and McCammon.38 In this implementation, when the true potential V(r) of the model system falls below a chosen threshold boost energy E, the simulation is performed on the modified potential V∗(r) = V(r) + ΔV(r), where ΔV(r) is a non-negative boost potential function and is given by ΔV(r) = (E − V(r))2/(α + (E − V(r)). This leads to an enhanced escape rate for V∗(r). Since the barriers in the dihedral potential energy term predominantly govern the sampling of torsional or rotameric states in proteins, the boost potential was applied only to the total dihedral energy term of the potential energy function. After 12 ns of conventional NVE production run, we performed 10 ns of aMD in two steps. The MD runs were performed in series. When no notable structural loop reorganizations were noted from the cMD run of 12 ns, we applied aMD in progressive phases. The 10 ns of aMD was split into two halves, with each 5-ns simulation run using a slightly different procedure. The experimental nature of this simulation exercise resulted in the aMD run being only 2 ns less than the cMD, which we recognize is somewhat unconventional. During each of the 5-ns aMD runs, only a single boost was used, with the difference being in the protein target area.
Specifically, the first 5 ns of aMD was conducted on the whole protein to see if we could reproduce the ϕ–ψ dihedral rotation by increasing the torsional energy basin of the entire system. The structures with both open and closed BL2 loops were treated equally for the cMD and 5 ns of whole-protein aMD simulations. The average dihedral energy V(r) of the open and closed systems after 12 ns of NVE was 2240 kcal/mol and 2268 kcal/mol, respectively. For the apo simulation, the threshold E apo value was set to 2300 [60 kcal/mol above V(r)apo], and the bound simulation E bound value was set to 2400 [132 kcal/mol above V(r)bound], with the α for both simulations set to 20% of their respective E values. After 5 ns of simulation, the average dihedral energy of the apo and bound proteins were 2285 and 2357 kcal/mol, respectively, but no ϕ–ψ dihedral inversion was observed. It should be noted that the inversion of the peptide bond in the loop from the bound form to the apo form would be expected to be much easier to capture than vice versa in the absence of a ligand that actually induces the peptide bond inversion. Hence, we continued the next 5 ns of aMD on a subset of loop residues for only the bound structure of the protein with ligand removed. aMD was employed on the 10 loop residues (Tyr265 to Tyr274, 165 atoms) with an α of 50% of E bound-post-5 ns applied to the average dihedral energy of these amino acids. Alternate α values of 25% and 35% were tested prior to the high boost of 50%. However, since α values of 25% or 35% were not able to overcome the energy barrier, we only show the results from an α value of 50%. The rest of the protein was simulated without any restraints. Within the next 1.5 ns (after 19.5 ns of total simulation time), we readily observed peptide dihedral torsion inversions from the bound closed form to the open apoprotein loop conformation. There was a 10-kcal/mol gain in the average dihedral energy term for the 10 amino acids over the 5 ns of aMD on the loop. The simulations were run in blocks of 2 ns, carrying out 11 MD runs in explicit water. These were run in series and the velocities were propagated with each run.
Homology modeling
The SARS-CoV PLpro (1540–1854) amino acid sequence was compared with the papain-like protease domain of 12 other coronaviruses (Table S1). Seven of these nine coronaviruses, namely, MHV strain JHM,40, 41 bovine CoV,42 HCoV-HKU1,43, 44 TGEV,45 HCoV-NL63,2, 3 HCoV-229E,46, 47 and HCoV-OC43,48 encode two papain-like protease domains, termed PLP1 and PLP2. SARS-CoV,17, 49 avian infectious bronchitis virus,50 and the four BtCoVs [i.e., Bat-SARS-CoV (National Center for Biotechnology Information accession: AAZ67050),8, 51 BtCoV/273/2005 (ABG47059),52 BtCoV/133/2005 (Q0Q4F3),52 and BtCoV-HKU9 (A3EXG5)53] encode only one papain-like protease domain termed PLpro.
Selected binding site residues of SARS-CoV PLpro for this study
Based on the SARS-CoV PLpro apo X-ray structure, 11 substrate binding site amino acids and the catalytic triad residues were chosen to represent the entire selected set of residues. All sequences are numbered according to the 315-amino-acid catalytic core domain of PLpro (actual amino acid sequence numbers). A brief description of the 14 selected residues follows. The protease activity is mediated by the catalytic triad of Cys112–His273–Asp287,12, 53, 54 assisted by the indole ring or amino group of usually a few consistent amino acids, W107 in SARS-CoV PLpro (Gln or Asn among the other PLP2s), to support the oxyanion hole.12 Coronavirus PLpro domains also have a signature substrate binding site that supports DUB activity.12, 24, 26, 54, 55 The residues instrumental in maintaining the structural signature for the strict specificity of the substrate –LXGG– in SARS-CoV PLpro include Tyr113, Tyr274, Asn110, Leu163, Gly164, Asp165, Glu168, Tyr265, and the blocking loop residues at the mouth of the active site beginning at Gly267 through Gly272 with Tyr269 and Gln270 being instrumental in ligand binding.12, 15, 26 Details of the roles of these 14 residues were previously described.12, 15 Table 1 highlights these key residues aligned with the corresponding binding site amino acids of 19 other coronaviral papain-like protease domains.
Based on the comprehensive MSA (Supplemental Material, Table S2), we built and minimized a homology model for NL63-PLP2 using the SARS-CoV PLpro 1.85-Å crystal structure12 as the template with modeling software DS-modeller.56, 57 3D structure superposition of the PLP2 core domain with the crystal structure of SARS-CoV PLpro helped determine the corresponding binding site residues in PLP2. The PyMOL program (DeLano Scientific)58 was used for structural superposition. All hydrogens were added to the PLP2 model using Sybyl 8.1 (Tripos, Inc.). The model thus generated was analyzed to characterize the physicochemical properties (hydrophobic and electrostatic patterns) of the substrate binding site using the Chimera software.59 The electrostatic surface potential was calculated using Coulomb's law as implemented in Chimera. The surface maps were generated using the default color scale ranging from red (− 10 kcal/mol∗e) to white (0) to blue (+ 10 kcal/mol∗e) at a temperature of 298 K. Tripos 3D Benchware60 was used to view hydrogen donors and acceptors in the active site of SARS-CoV PLpro and the PLP2 model. The donor and acceptor atoms for each pocket in each structure were counted by visual inspection.
Docking
Docking calculations used the genetic algorithm in GOLD v4.0;61 inhibitor GRL0617 was docked into the active site of the PLP2 model using the GoldScore scoring function with all default parameters and settings. Ten conformations were retained, and the best ranking conformation is shown in Fig. 8.
Acknowledgements
We thank Professor Donald Hamelberg of Georgia State University for providing the aMD routines. This work was supported in part by National Institutes of Health grants P01 AI060915, R56 AI089535 and R01 GM57513 and by the National Science Foundation through TeraGrid resources provided by the Abe/Queen Bee and Steele resources under grant numbers TG- MCB090040 and TG-MCB090168 (M.E.J.).
Edited by M. Levitt
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2011.09.030
Appendix A.
Supplementary Material
References
- 1.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 2.van der Hoek L., Pyrc K., Jebbink M., Vermeulen-Oost W., Berkhout R., Wolthers K. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fouchier R., Hartwig N., Bestebroer T. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl Acad. Sci. USA. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl Acad. Sci. USA. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofmann H., Simmons G., Rennekamp A., Chaipan C., Gramberg T., Heck E. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 2006;80:8639–8652. doi: 10.1128/JVI.00560-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziebuhr J. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 2004;7:412–419. doi: 10.1016/j.mib.2004.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau S., Woo P., Li K., Huang Y., Tsoi H., Wong B. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein H. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 9.Thiel V., Ivanov K., Putics A., Hertzig T., Schelle B., Bayer S. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 10.Bacha U., Barrila J., Velazquez-Campoy A., Leavitt S., Freire E. Identification of novel inhibitors of the SARS coronavirus main protease 3CLpro. Biochemistry. 2004;43:4906–4912. doi: 10.1021/bi0361766. [DOI] [PubMed] [Google Scholar]
- 11.Wu C., King K., Kuo C., Fang J., Wu Y., Ho M. Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL protease. Chem. Biol. 2006;13:261–268. doi: 10.1016/j.chembiol.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ratia K., Saikatendu K., Santarsiero B., Barretto N., Baker S., Stevens R., Mesecar A. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl Acad. Sci. USA. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh A.K., Takayama J., Aubin Y., Ratia K., Chaudhuri R., Baez Y. Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors for the severe acute respiratory syndrome-coronavirus papain-like protease. J. Med. Chem. 2009;52:5228–5240. doi: 10.1021/jm900611t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh A.K., Takayama J., Rao K.V., Ratia K., Chaudhuri R., Mulhearn D.C. Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein–ligand X-ray structure and biological evaluation. J. Med. Chem. 2010;53:4968–4979. doi: 10.1021/jm1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S. A new noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl Acad. Sci. USA. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldsmith C., Tatti K., Ksiazek T., Rollin P., Comer J., Lee W. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004;10:320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snijder E., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J., van der Meulen J., Koerten H., Mommaas A. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong X.G., Wilkins J.W. Hyper molecular dynamics with a local bias potential. Phys. Rev. B: Condens. Matter Mater. Phys. 1999;59:54–57. [Google Scholar]
- 19.Marcelin R. Physicochemical kinetics. Ann. Phys. 1915;3:120–184. [Google Scholar]
- 20.Rahman J.A., Tully J.C. Puddle-skimming: an efficient sampling of multidimensional configuration space. J. Chem. Phys. 2002;116:8750–8760. [Google Scholar]
- 21.Steiner M.M., Genilloud P.A., Wilkins J.W. Simple bias potential for boosting molecular dynamics with the hyperdynamics scheme. Phys. Rev. B: Condens. Matter Mater. Phys. 1998;57:10236–10239. [Google Scholar]
- 22.Voter A.F. Hyperdynamics: accelerated molecular dynamics of infrequent events. Phys. Rev. Lett. 1997;78:3908–3911. [Google Scholar]
- 23.Wang J.-C., Pal S., Fichthorn K.A. Accelerated molecular dynamics of rare events using the local boost method. Phys. Rev. B: Condens. Matter Mater. Phys. 2001;63-085403:1–9. [Google Scholar]
- 24.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A., Baker S. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z., Wang Y., Ratia K., Mesecar A., Wilkinson K., Baker S. Proteolytic processing and deubiquitinating activity of papain-like proteases of HCoV-NL63. J. Virol. 2007;81:6007–6018. doi: 10.1128/JVI.02747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sulea T., Lindner H., Purisima E., Ménard R. Binding site-based classification of coronaviral papain-like proteases. Proteins. 2006;62:760–775. doi: 10.1002/prot.20802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartigan J.A., Wong M.A. Algorithm AS 136: a k-means clustering algorithm. J. R. Stat. Soc., Ser. C, Appl. Stat. 1979;28:100–108. [Google Scholar]
- 28.Case D.A., Cheatham T.E., III, Darden T., Gohlke H., Luo R., Merz K.M., Jr. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J., Cieplak P., Kollman P.A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 30.Miyamoto S., Kollman P.A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 31.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 32.Crowley M., Darden T., Cheatham T., Deerfield D. Adventures in improving the scaling and accuracy of a parallel molecular dynamics program. J. Supercomput. 1997;11:255–278. [Google Scholar]
- 33.Darden T., York D., Pedersen L. Particle mesh Ewald: an N∗log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089. [Google Scholar]
- 34.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577. [Google Scholar]
- 35.Sagui C., Darden T.A. AIP; Melville, NY: 1999. Simulation and Theory of Electrostatic Interactions in Solution. [Google Scholar]
- 36.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 37.Grubmuller H. Predicting slow structural transitions in macromolecular systems: conformational flooding. Phys. Rev. E. Stat. Phys., Plasmas Fluids Relat. Interdiscip. Topics. 1995;52:2893–2906. doi: 10.1103/physreve.52.2893. [DOI] [PubMed] [Google Scholar]
- 38.Hamelberg D., McCammon J.A. Mechanistic insight into the role of transition-state stabilization in cyclophilin A. J. Am. Chem. Soc. 2009;131:147–152. doi: 10.1021/ja806146g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamelberg D., Mongan J., McCammon J.A. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J. Chem. Phys. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 40.Lee H., Shieh C., Gorbalenya A., Koonin E., La Monica N., Tuler J. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schiller J., Kanjanahaluethai A., Baker S. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology. 1998;242:288–302. doi: 10.1006/viro.1997.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chouljenko V., Lin X., Storz J., Kousoulas K., Gorbalenya A. Comparison of genomic and predicted amino acid sequences of respiratory and enteric bovine coronaviruses isolated from the same animal with fatal shipping pneumonia. J. Gen. Virol. 2001;82:2927–2933. doi: 10.1099/0022-1317-82-12-2927. [DOI] [PubMed] [Google Scholar]
- 43.Lau S., Woo P., Yip C., Tse H., Tsoi H., Cheng V. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J. Clin. Microbiol. 2006;44:2063–2071. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woo P., Lau S., Chu C., Chan K., Tsoi H., Huang Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ortega J., Escors D., Laude H., Enjuanes L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 2002;76:11518–11529. doi: 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herold J., Siddell S., Gorbalenya A. A human RNA viral cysteine proteinase that depends upon a unique Zn2+-binding finger connecting the two domains of a papain-like fold. J. Biol. Chem. 1999;274:14918–14925. doi: 10.1074/jbc.274.21.14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thiel V., Herold J., Schelle B., Siddell S. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 2001;82:1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- 48.Vijgen L., Keyaerts E., Moes E., Thoelen I., Wollants E., Lemey P. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harcourt B., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K., Smith C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:1312–1360. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu D., Tibbles K., Cavanagh D., Brown T., Brierley I. Identification, expression, and processing of an 87-kDa polypeptide encoded by ORF 1a of the coronavirus infectious bronchitis virus. Virology. 1995;208:48–57. doi: 10.1006/viro.1995.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vijaykrishna D., Smith G., Zhang J., Peiris J., Chen H., Guan Y. Evolutionary insights into the ecology of coronaviruses. J. Virol. 2007;81:4012–4020. doi: 10.1128/JVI.02605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang X., Zhang J., Zhang S., Wang P., Fan X., Li L. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006;80:7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woo P., Wang M., Lau S., Xu H., Poon R., Guo R. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 2007;81:1574–1585. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindner H., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Ménard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sulea T., Lindner H., Purisima E., Menard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease. J. Virol. 2005;79:4550–4551. doi: 10.1128/JVI.79.7.4550-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fiser A., Do R., Sali A. Modeling of loops in protein structures. Protein Sci. 2003;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sali A., Blundell T. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 58.PyMOL. (2011). PyMOL (http://www.pymol.org/).
- 59.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Tripos. Benchware, 3D explorer, Tripos International, 1699 South Hanley Rd., St. Louis, Missouri 63144, USA.
- 61.Jones G., Willett P., Glen R.C., Leach A.R., Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material