Abstract
Employing a structure-based strategy, we have designed a new class of potent small-molecule inhibitors of the anti-apoptotic proteins Bcl-2 and Bcl-xL. An initial lead compound with a new scaffold was designed based upon the crystal structure of Bcl-xL and FDA-approved drugs and was found to have an affinity of 100 μM to both Bcl-2 and Bcl-xL. Linking this weak lead to another weak-affinity fragment derived from Abbott's ABT-737 led to an improvement of the binding affinity by a factor of >10,000. Further optimization ultimately yielded compounds with subnanomolar binding affinities to both Bcl-2 and Bcl-xL and potent cellular activity. The best compound (21) binds to Bcl-xL and Bcl-2 with Ki < 1 nM, inhibits cell growth in the H146 and H1417 small-cell lung cancer cell lines with IC50 values of 60–90 nM and induces robust cell death in the H146 cancer cell line at 30–100 nM.
Introduction
Resistance to apoptosis is a hallmark of human cancer1 and targeting key apoptosis regulators with the goal of promoting apoptosis is an exciting therapeutic strategy for cancer treatment.2, 3
The Bcl-2 protein family is a class of key apoptosis regulators and consists of both anti-apoptotic proteins, including Bcl-2, Bcl-xL, and Mcl-1, and pro-apoptotic proteins, such as BID, BIM, BAD, BAK, BAX and NOXA.4 The anti-apoptotic Bcl-2 and Bcl-xL proteins are overexpressed in many different types of human tumor samples and cancer cell lines and this overexpression confers resistance of cancer cells to current cancer treatments.5, 6 The anti-apoptotic proteins inhibit apoptosis via heterodimerization with pro-apoptotic Bcl-2 family proteins.5, 6 Despite their structural similarities, these anti-death Bcl-2 proteins confer a certain binding specificity on pro-death Bcl-2 proteins.5, 6 For example, while Bcl-2 and Bcl-xL bind to BIM and BAD proteins with high affinities, they have very weak affinities for NOXA. In contrast, Mcl-1 binds to BIM and NOXA with high affinities but has a very weak affinity to BAD. These data suggest that the pro-apoptotic proteins have non-redundant roles in the regulation of apoptosis.
It has been proposed that potent, non-peptide, small-molecules designed to block the protein-protein interactions between anti- and pro-apoptotic Bcl-2 members can antagonize the anti-death function of pro-apoptotic Bcl-2 proteins, and this in turn can overcome the apoptosis resistance of cancer cells mediated by the overexpression of these anti-apoptotic Bcl-2 proteins.5, 6 Design of potent, non-peptide, cell-permeable small-molecule inhibitors with the ability to block the protein-protein interactions involving the Bcl-2 family of proteins has been intensely pursued in the past decade as a novel cancer therapeutic strategy, and a number of laboratories have reported the design and characterization of non-peptide, small-molecule inhibitors.7–12
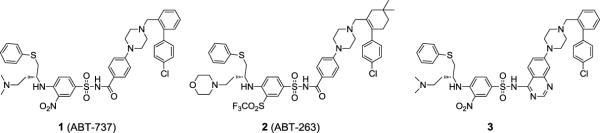
Among all the reported Bcl-2/Bcl-xL inhibitors, compound 1 (ABT-737, Figure 1) is arguably the most potent compound.13 Compound 1 binds to Bcl-2, Bcl-xL and Bcl-w with very high affinities (Ki <1 nM) and also shows a very high specificity over Mcl-1 and A1.13 Its analogue, 2 (ABT-263, Figure 1) has been advanced into Phase I/II clinical trials for the treatment of human cancer.14, 15 Recently, another class of potent Bcl-2/Bcl-xL inhibitors, exemplified by compound 3 (Figure 1), was designed starting from the chemical structure of compound 1.16 In this paper, we report our structure-based design of highly potent and specific small-molecule inhibitors of Bcl-2/Bcl-xL, started from a novel chemical scaffold designed based upon FDA-approved drugs and the crystal structures of Bcl-xL complexed with its inhibitors.
Figure 1.
Chemical structures of previously reported potent and specific Bcl-2/Bcl-xL inhibitors.
Results and Discussion
Structure-based Design of a New Chemical Scaffold to Target Bcl-xL
The crystal structure of Bcl-xL complexed with the BAD BH3 peptide17 reveals that the peptide interacts with two large binding pockets in Bcl-xL, shown in Figure 2. Site 1 is a deep, well-defined binding pocket while Site 2 is more exposed to solvents. We decided to focus on Site 1 for the design of initial lead compounds with novel chemical scaffolds.
Figure 2.
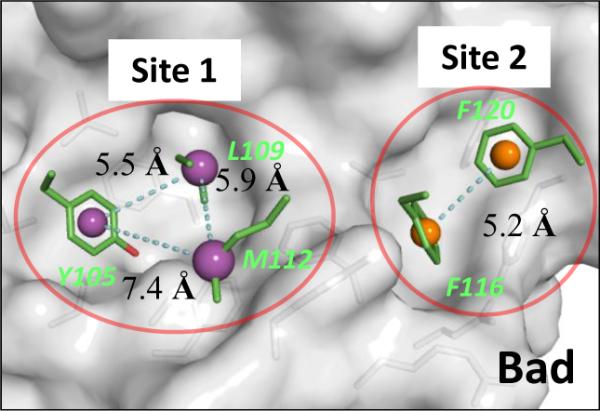
Crystal structure of Bcl-xL with five key residues of BAD BH3 peptide at the binding site. Centroids of hydrophobic pharmacophores are shown in spheres. The pharmacophore model based on three residues at Site 1 binding pocket (purple spheres in red circle) was used in pharmocophore search.
Site 1 of Bcl-xL interacts with Y105, L109, and M112, three hydrophobic residues of the BAD BH3 peptide. The distances between the centers of the mass of the side chains of any two of these three residues are between 5.5 and 7.4 Å (Figure 2). These three closely clustered hydrophobic residues in the BAD BH3 peptide offer a 3D pharmacophore template which we used to search for new scaffolds. A pharmacophore model was constructed using these three hydrophobic residues and the structural information, which consists of two aromatic rings and one hydrophobic group. The distance between the centers of the two aromatic rings was defined as 5 ± 1 Å and the distance between the center of each of the aromatic rings, and the center of mass of the hydrophobic group was set to 6 ± 1 Å. We were particularly interested in identifying scaffolds with good pharmacological and toxicological properties and accordingly, a pharmacophore search was made in a three-dimensional database of 1,410 FDA approved drugs constructed in our laboratory. Eleven compounds were identified and were grouped into three classes based on their scaffolds (Figure 3). Our initial efforts focused on the second class of compounds, which all contain the bis-aryl substituted five-membered heterocyclic template and include the well-known drugs Lipitor and Celecoxib (Figure 3).
Figure 3.
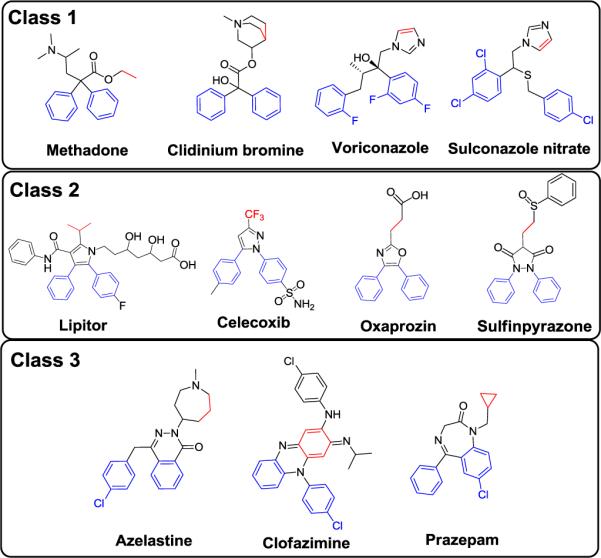
Three classes of scaffolds identified based on the pharmacophores in the Bad BH3 peptide. The three dimensional pharmocophore model is based on two aromatic rings (blue color) and one hydrophobic group (red color). The distance between the aromatic ring was set to 5 ± 1 Å and the distances between the aromatic ring to the hydrophobic group were set to 6 ± 1 Å.
To explore the possible binding models of Lipitor and Celexocib with Bcl-xL, we performed computational docking of both drugs to Bcl-xL using the crystal structure adopted by Bcl-xL in its complex with the BAD BH3 peptide. As can be seen in Figures 4A and 4B, the hydrophobic groups of Lipitor mimic Y105, L109 of the BAD BH3 peptide in its interaction with Bcl-xL, whereas the two phenyl groups of Celecoxib mimic the interaction between L109, M112 of the BAD BH3 peptide with Bcl-xL. Figures 4A and 4B suggested that the bis-aryl substituted five-membered heterocycle scaffold could mimic two of three critical hydrophobic residues in the BAD BH3 peptide.
Figure 4.
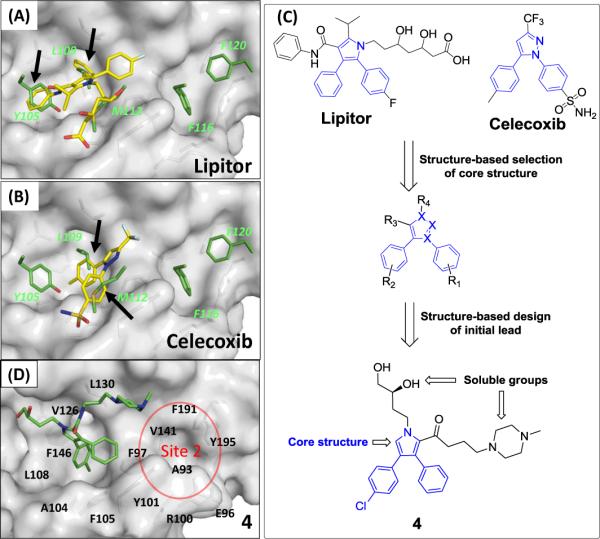
Structure-based design of a new scaffold as the initial lead compound 4 and its crystal structure in complex with Bcl-xL. (A), (B) Rank 1 pose of Lipitor and Celecoxib with Bcl-xL using the Bad BH3 peptide bound Bcl-xL structure (PDB ID: 2BZW). (C) Identification of the core scaffold from the FDA approved drugs database for a new class of Bcl-2/Bcl-XL inhibitors. (D) Co-crystal structure of 4 in complex with Bcl-xL (1.7 Å).
Based upon these analyses, compound 4 (Figure 4C), containing a 3,4-diphenyl-1H-pyrrole-2-carboxamide scaffold, was designed. Since it was critical to determine the crystal structure of our initial lead compound complexed with Bcl-xL for subsequent optimization efforts, we have decided to attach two soluble groups in compound 4 to facilitate crystallographic studies. Docking studies suggested that compound 4 can effectively interact with the deep hydrophobic pocket in Site 1.
Compound 4 was synthesized and evaluated for its binding to Bcl-xL and Bcl-2 proteins in fluorescence-polarization (FP) assays. Compound 4 has Ki values of 78 μM to Bcl-2 and 138 μM to Bcl-xL (Table 1). Although in a typical drug discovery program such weak affinities would seem to disqualify it as a useful lead compound, it does possess an attractive drug-like core structure and excellent aqueous solubility, and was proven to be an excellent starting point in our design of new and potent Bcl-2/Bcl-xL inhibitors.
Table 1.
Binding affinities of our designed compounds to Bcl-2 and Bcl-XL proteins in our FP-based assays and inhibition of cell growth in two cancer cell lines.
| CPDS | Binding Affinities | Cell Growth Inhibition IC50 ± SD (μM) | |||||
|---|---|---|---|---|---|---|---|
| Bcl-2 (FP) | Bcl-XL(FP) | Mcl-1 | |||||
| IC50 ± SD (nM) | Ki ± SD (nM) | IC50 ± SD (nM) | Ki ± SD (nM) | IC50 ± SD | H146 | H1417 | |
| 4 | 213 ± 16 (μM) | 78.0 ± 5.9 (μM) | 453 ± 25 (μM) | 138 ± 7.6 (μM) | > 100 (μM) | > 10 | > 10 |
| 5 | > 100 (μM) | 238 ± 14 (μM) | 75.0 ± 4.2 (μM) | > 100 (μM) | > 10 | > 10 | |
| 6 | 8.7 ± 1.9 | 2.0 ± 0.5 | 6.1 ± 1.5 | < 1 | > 10 (μM) | > 10 | > 10 |
| 7 | 1.3 ± 0.7 | < 0.6 | 6.2 ± 2.2 | < 1 | > 10 (μM) | 2.0 ± 1.1 | 1.8 ± 0.07 |
| 8 | 1.4 ± 0.8 | < 0.6 | 4.8 ± 1.1 | < 1 | > 10 (μM) | > 10 | > 10 |
| 9 | 1.9 ± 0.6 | ~ 0.6 | 5.7 ± 0.7 | < 1 | > 10 (μM) | > 10 | > 10 |
| 10 | 6.6 ± 2.2 | 1.5 ± 0.3 | 12.6 ± 3.1 | 1.7 ± 0.9 | > 10 (μM) | > 10 | > 10 |
| 11 | 60.6 ± 23.1 | 15.4 ± 6.0 | 44.1 ± 5.5 | 11.3 ± 0.6 | > 10 (μM) | > 10 | > 10 |
| 12 | 33.9 ± 3.8 | 8.5 ± 1.0 | 7.6 ± 1.6 | < 1 | > 10 (μM) | 2.5 ± 0.8 | 3.3 ± 1.4 |
| 13 | 85.3 ± 34.8 | 21.8 ± 9.0 | 88.3 ± 14.4 | 24.7 ± 4.0 | > 10 (μM) | > 10 | > 10 |
| 1 | 2 ± 0.2 | < 0.6 | 6 ± 2 | < 1 | > 1 (μM) | 0.097 ± 0.030 | 0.13 ± 0.05 |
| 14 | > 100 | 547 | 166 | > 100 (μM) | >10 | >10 | |
| 15 | 534 ±105 | 138 ± 27 | 19 ± 0.3 | 5.7 ± 0.1 | > 100 (μM) | >10,000 | 7.5 ±0.7 |
To provide a solid structural basis for our subsequent structure-based optimization of 4, we determined its crystal structure, at a resolution of 1.7 Å, in a complex with Bcl-xL (Figure 4D). This shows that 4 indeed binds to the large, deep hydrophobic pocket in Site 1 of Bcl-xL, supporting both our modeling prediction and our design premise.
Structure-based design of potent Bcl-2/Bcl-xL inhibitors
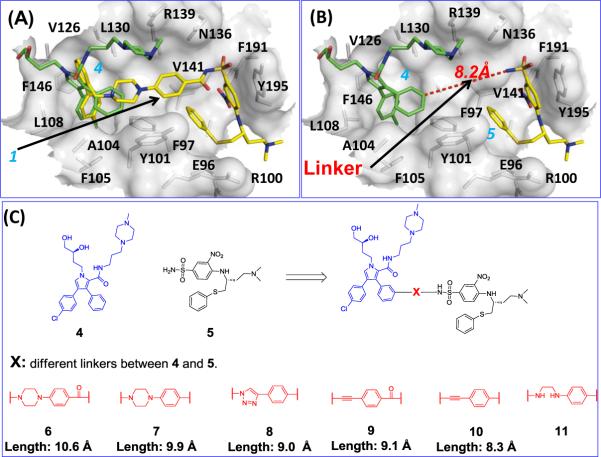
For a compound to achieve a high binding affinity to Bcl-xL, it may be necessary to occupy both Sites 1 and 2 in the protein.13 Since 4 occupies only Site 1, a second fragment, capable of occupying Site 2, is needed. We used the crystal structure of 1 complexed with Bcl-xL18 to identify a fragment that could be accommodated in Site 2 (red circle in Figure 4D).
Superposition of the crystal structure of 4 and the crystal structure of 1 in its complex with Bcl-xL showed that the core structure in 4 and the p-chlorobiphenyl fragment in 1 (Figure 5A) both occupy Site 1. It is also evident that fragment 5 occupies Site 2 in Bcl-xL (Figure 5A and 5B). Therefore, we reasoned that linking compounds 4 and 5 could yield new compounds with high affinity to Bcl-xL.
Figure 5.
Computational structure-based design of a new class of Bcl-2/Bcl-xL inhibitors by linking two fragments with weak binding affinities. (A) Superposition of the crystal structure of 4 (green) onto the crystal structure of 1 (yellow) in complex with Bcl-xL. (B) Measurement of the distance (8.2 Å) between 4 and 5. The 1 bound Bcl-xL structure was used in the surface representation. (C) Chemical structures of 5 and the new designed Bcl-2/Bcl-xL inhibitors with different linkers.
Compound 5 was synthesized using the published method19 and found to have very weak affinities (IC50 > 100 μM) to Bcl-xL and Bcl-2 proteins in our FP-based assays. These data further showed that interacting with either Site 1 or Site 2 is insufficient to yield compounds with high affinities to Bcl-xL and Bcl-2, and occupying both sites are needed to achieve high affinities to Bcl-xL and Bcl-2. Therefore, we decided to link 4 and 5 together using a proper linker for the design of potent inhibitors of Bcl-xL and Bcl-2.
Analysis of the modeled structure of 5 complexed with Bcl-xL and the crystal structures of 1 and 4 complexed with Bcl-xL suggested that the meta position of the unsubstituted phenyl ring in 4 and the sulfonamido nitrogen atom of 5 are sites at which 4 and 5 could be linked together for the design of potent Bcl-xL inhibitors (Figure 5B). The distance between this ring in 4 and the sulfonamido nitrogen atom of 5 is 8.2 Å (Figure 5B).
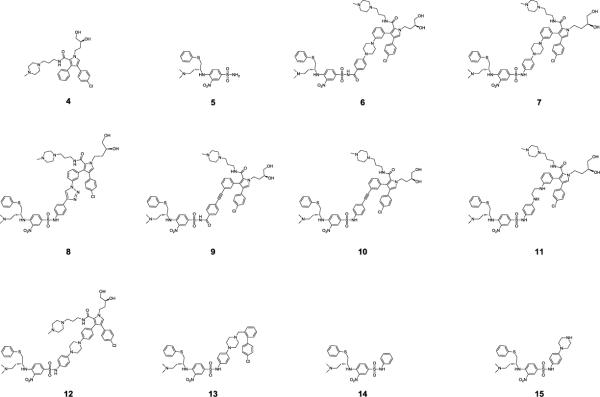
Compound 6, designed to make use of the linker in 1 (Figure 5C and Figure 6), which is 10.6 Å in length, was found to have Ki = 2.0 nM to Bcl-2 and Ki < 1 nM to Bcl-xL, and is thus >10,000-times more potent than either 4 or 5. The dramatically improved binding affinities of 6 over those of 4 and 5 supported our design strategy.
Figure 6.
Chemical structures of designed Bcl-2 and Bcl-xL inhibitors.
Compounds 1 and 6 have similar affinities to Bcl-xL, but 6 is less potent than 1 to Bcl-2 (Table 1). To further optimize the binding affinities of 6, we next tested linkers with various lengths, flexibility, orientation and chemical properties (Figure 5C and Figure 6). Modeling suggested that removal of the carbonyl group in the N-acylsulfonamide in 6 may lead to further improvement in the binding affinity (Figure S1 in SI). Removal of this carbonyl group from 6 gives 7, in which the linker has been shortened from 10.6 Å to 9.9 Å. The affinity of 7 to both Bcl-2 and Bcl-xL (Ki < 1 nM) is very similar to that of 1. Replacement of the piperazine in 7 by a triazole aromatic ring shortens the linker to 9.0 Å, giving 8, which also binds to both Bcl-2 and Bcl-xL with Ki < 1 nM. Replacement of the piperazine ring in the linker in 6 with a rigid ethynyl group leads to 9, which has a linker length of 9.1 Å and binds to both Bcl-2 and Bcl-xL with Ki < 1 nM. Removal of the carbonyl group from the N-acylsulfonamide group in 9 leads to 10, which has a linker length of 8.3 Å and binds to Bcl-2 and Bcl-xL with Ki = 1.5 nM and 1.7 nM, respectively.
Hence, compounds 7, 8 and 9 bind to Bcl-2 and Bcl-xL with Ki <1 nM and are as potent as 1 to both proteins. The binding data show that tethering 4 and 5, two weak Bcl-2/Bcl-xL inhibitors, with linkers approximately 9.0 Å in length, produce highly potent Bcl-2/Bcl-xL inhibitors, whose affinities to Bcl-2/Bcl-xL are four orders of magnitude better than those of 4 or 5. When the linker is longer (10.6 Å in 6) or shorter (8.3 Å in 10), the resulting compounds are less potent. To investigate the influence of the flexibility of the linker, we synthesized 11, with a more flexible linker than that in 7, and found that 11 is at least 10-times less potent than 7.
To examine binding specificity, we evaluated the binding of these compounds to Mcl-1. Similar to 1, compounds 6, 7, 8, 9, 10 and 11 all were found to bind to Mcl-1 with IC50 > 10 μM, thus showing very high specificity over Mcl-1 (Table 1).
Changing the attachment position of the linker from the meta position to the para position of the phenyl ring in 7 results in 12 (Figure 6), which is >10-times less potent than 7 in binding to Bcl-2, but is equipotent with 7 in its binding to Bcl-xL (Table 1).
We also synthesized 13 (Figure 6), an analogue of 1 lacking the benzamido carbonyl group. This compound binds to Bcl-2 and Bcl-xL with Ki values of 18 nM and 17 nM, respectively, an affinity >10-times weaker than that of 1. Hence, the carbonyl group in 1 evidently contributes significantly to its binding affinities with Bcl-2 and Bcl-xL. By contrast, removal of the corresponding carbonyl group in 6 enhances the binding affinity to Bcl-2 by 6 times, while having no effect on the binding to Bcl-xL.
Compounds 14 and 15, fragments of 7 (Figure 6), were synthesized and their binding to Bcl-2 and Bcl-xL was examined. Both 14 and 15 bind to Bcl-2 and Bcl-xL with very weak affinities (Table 1).
As a single agent, 1 was shown to be very effective in inhibition of cancer cell growth in cancer cell lines such as small-cell lung cancer cell lines H146 and H1417, with high levels of Bcl-2/Bcl-xL, but low levels of Mcl-1. Since 7 and several other compounds bind to Bcl-2 and Bcl-xL with very high affinities, show high specificity over Mcl-1 and have the same binding profiles as 1, we evaluated their ability, in comparison to 1, to inhibit cell growth in the H146 and H1417 cancer cell lines with the results shown in Table 1.
Consistent with their weak binding affinities, four compounds (4, 5, 14 and 15) have poor activities (IC50 > 10 μM) in inhibition of cell growth in these two cancer cell lines. Although compounds 6, 8, 9, 10 and 11 all have high affinities to both Bcl-2 and Bcl-xL, they also have weak cellular activities (IC50 > 10 μM), suggesting that these compounds may suffer from poor cell permeability. Removal of the carbonyl group from the linker in 6 to give 7, significantly improves both the binding affinities to Bcl-2 and Bcl-xL and the cellular activity. Compound 7 inhibits cell growth in these two cancer cell lines and has IC50 values of approximately 2.0 μM against both cancer cell lines. In contrast, removal of the corresponding carbonyl group in 1, which results in compound 13, is detrimental to binding affinities to both Bcl-2 and Bcl-xL, as well as to the cell growth inhibitory activities. The binding and cellular data thus identify 7 as a promising lead compound for further optimization and, accordingly, we focused next on 7 to further investigate the structure-activity relationships (SAR) for this class of compounds.
Structure-Activity relationships of compound 7
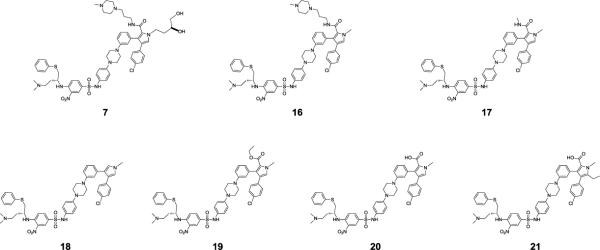
Modeling suggested that the dihydroxybutyl side chain in 7 lacks any specific interactions with Bcl-xL (Figure S1B in SI). Removal of this dihydroxybutyl side chain yields 16 (Figure 7), which binds to Bcl-2 and Bcl-xL with Ki < 1 nM. It inhibits cell growth in the H146 and H1417 cancer cell lines with IC50 values of 0.43 μM and 0.65 μM, respectively, and is thus 3–5 times more potent than 7 (Table 2). The data thus show that truncation of the dihydroxybutyl side chain in 7 to a methyl group is accompanied by retention of the high binding affinities to Bcl-2/Bcl-xL and significant improvement in its cellular activities in both the H146 and H1417 cancer cell lines.
Figure 7.
Chemical structures of 7 and its analogues.
Table 2.
Binding affinities of our designed compounds to Bcl-2 and Bcl-XL proteins in FP-based assays and inhibition of cell growth in two small-cell lung cancer cell lines.
| CPDS | Binding Affinities | Cell Growth Inhibition IC50 ± SD (μM) | |||||
|---|---|---|---|---|---|---|---|
| Bcl-2 (FP) | Bcl-XL(FP) | Mcl-1 | |||||
| IC50 ± SD (nM) | Ki ± SD (nM) | IC50 ± SD (nM) | Ki ± SD (nM) | IC50 ± SD | H146 | H1417 | |
| 7 | 1.3 ± 0.7 | < 0.6 | 6.2 ± 2.2 | < 1 | > 10 (μM) | 2.0 ± 1.1 | 1.8 ± 0.07 |
| 16 | 0.6 ± 0.2 | < 0.6 | 4.9 ± 1.2 | < 1 | > 10 (μM) | 0.43 ± 0.25 | 0.65 ± 0.51 |
| 17 | 5.3 ± 0.6 | 1.2 ± 0.2 | 6.3 ± 0.4 | < 1 | > 2 (μM) | 0.40 ± 0.31 | 0.65 ± 0.15 |
| 18 | 82.6 ± 19.5 | 21.1 ± 5.0 | 34.4 ± 3.5 | 8.3 ± 1.0 | > 2 (μM) | > 10 | > 10 |
| 19 | 45.8 ± 26.1 | 11.6 ± 6.8 | 12.6 ± 5.4 | 1.7 ± 0.6 | > 2 (μM) | 4.9 ± 3.2 | 7.3 ± 2.8 |
| 20 | 1.7 ± 1.0 | < 0.6 | 3.6 ± 1.1 | < 1 | > 2 (μM) | 0.34 ± 0.01 | 0.552 ± 0.089 |
| 21 | 4.4 | 0.28 | 8.9 | < 1 | > 2 (μM) | 0.061 ± 0.009 | 0.090 ± 0.003 |
| 1 | 2 ± 0.2 | < 0.6 | 6 ± 2 | < 1 | > 1 (μM) | 0.097 ± 0.03 | 0.13 ± 0.05 |
The N-(3-(4-methylpiperazin-1-yl)propyl)amide side chain in 16 was used to improve the aqueous solubility. We investigated the effect of modifications of this side chain in 16 on binding to Bcl-2/Bcl-xL. Compound 17, in which the N-(3-(4-methylpiperazin-1-yl)propyl)amide side chain in 16 was truncated to an N-methyl carbamoyl group (Figure 7), binds to Bcl-2 with Ki = 1.2 nM and to BclxL with Ki < 1 nM. Compounds 16 and 17 thus have similar potencies in inhibition of cell growth against both the H146 and H1417 cell lines. Compound 18, obtained by removal of the N-methyl carbamoyl group from 17, has a much weaker binding affinity to both Bcl-2 and Bcl-xL than 16 and 17. It also has very weak cellular activity in both H146 and H1417 cancer cell lines with IC50 values >10 μM (Table 2). These data show that the amide group in 16 and 17 plays a role in binding to Bcl-2 and Bcl-xL and in inhibition of cell growth.
Replacement of the N-methyl carbamoyl substituent on the pyrrole ring in 17 by a carbethoxyl results in 19 (Figure 7), which binds to Bcl-2 10 times less potently than 17, but 19 is only slightly less potent than 17 in binding to Bcl-xL. It has IC50 values of 4.9 μM and 7.3 μM in inhibition of cell growth in H146 and H1417 cancer cell lines, respectively, and is thus 10 times less potent than 17 (Table 2).
Replacement of the N-methyl carbamoyl substituent on the pyrrole ring of 17 by a carboxyl substituent generates 20, which binds to Bcl-2 and Bcl-xL with high affinities, with Ki < 1 nM, and has IC50 values of 0.34 μM and 0.55 μM in inhibition of cell growth against the H146 and H1417 cancer cell lines, respectively.
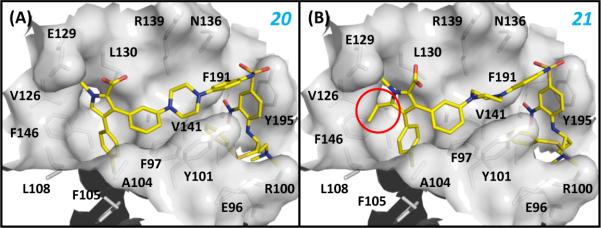
In an effort to further improve the binding affinities and especially the cellular activity of 20, we performed docking simulations of 20 based on the crystal structures of 4 and 1 complexed with Bcl-xL. The predicted binding model for 20 (Figure 8A) suggested that there is an unoccupied small hydrophobic pocket close to the 5-position of the pyrrole core in 20. Accordingly, we designed and synthesized 21, in which an additional ethyl group was introduced to the 5-position of the pyrrole ring in 20 (Figure 7 and Figure 8B). Compound 21 binds to both Bcl-2 and Bcl-xL with very high affinities (Ki< 1 nM), actually exceeding the lower limits of the assays, and it has an improved cell-growth inhibitory activity against the H146 and H1417 cancer cell lines, with IC50 values of 61 nM and 90 nM, respectively. Hence, 21 is as potent as 1, both in binding to Bcl-2 and Bcl-xL and in inhibition of cell growth against both the H146 and H1417 cancer cell lines.
Figure 8.
Binding models between (A) 20, (B) 21 and Bcl-XL. The Bcl-XL protein from the crystal structure between 1 and Bcl-XL was used in the docking simulations. The highest ranked poses of both compounds were selected as the binding models. The added ethyl group to 20 was denoted by the red circle. Residues of Bcl-XL at the binding site were labeled.
Further evaluation of compounds 20 and 21
We next evaluated the ability of 20 and 21 to induce cell death in the H146 cell line (Figure 9A). Both compounds effectively induce cell death in a dose-dependent manner in H146 cells as determined in a trypan blue assay, 21 being more potent than 20. Compound 21 induces substantial cell death at 30–100 nM and >70% of cell death in 24 h at 300 nM.
Figure 9.
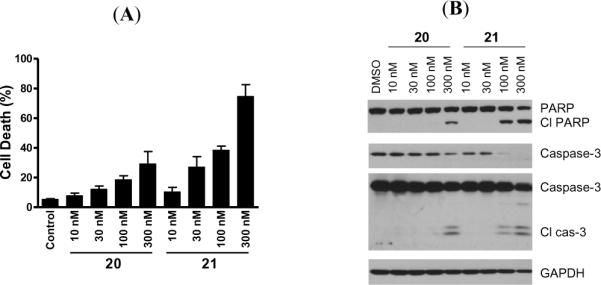
(A). Cell death induction by 20 and 21 in the H146 cancer cell line. Cells were treated for 24 h and cell death was analyzed by trypan blue assay. (B). Induction of cleavage of PARP and caspase-3 by 20 and 21 in the H146 cell line. Cells were treated for 24 h and caspase-3 and PARP were probed by western blotting.
We further tested 20 and 21 in the H146 cell line for their ability to induce cleavage of PARP and caspase-3, two biochemical markers of apoptosis (Figure 9B), and found that 21 is more potent than 20 and can effectively induce cleavage of PARP and caspase-3 in 24 h at concentrations as low as 100 nM. These data are consistent with their activities in the cell growth assay.
Synthesis
The synthesis of the initial lead compound (4) is outlined in Scheme 1. Briefly, benzaldehyde, 4-chlorophenyl cyanide and K2CO3 were heated in methanol to generate 2-(4-chlorophenyl)-3-phenylacrylonitrile that was used in a 2+3 cycloaddition reaction with ethyl isocyanoacetate to produce pyrrole 22.20 Alkylation of 22 with (S)-4-(2-iodoethyl)-2,2-dimethyl-1,3-dioxolane in the presence of K2CO3 at 60°C in DMF gave 23, hydrolysis of which afforded the corresponding acid, which was coupled to 1-(3-aminopropyl)-4-methylpiperazine. Removal of the acetal protecting group with HCl produced 4.
Scheme 1.
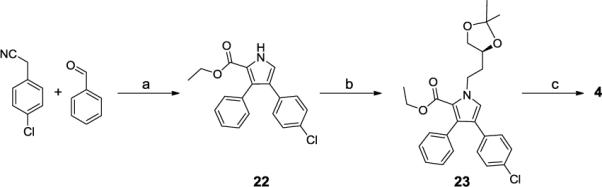
Synthesis of compound compound 4
Reagents and conditions: a) i. K2CO3, MeOH, reflux; ii. CNCH2COOEt, t-BuOK; b) (S)-4-(2-iodoethyl)-2,2-dimethyl-1,3-dioxolane, K2CO3; c) i. KOH, H2O/THF/MeOH; ii. 3-(4-methylpiperazin-1-yl)propan-1-amine, EDCI, HOBt, DIEA, DCM; iii. 4 M HCI in dioxane, MeOH.
The synthesis of compound 6 is shown in Scheme 2. Compounds 24, 25 and 26 were synthesized employing the same strategy as in Scheme 1 and intermediate 27 was prepared as described.19 Compound 6 was formed by palladium-catalyzed amination 21 of 26 with 27 in the presence of Pd(dba)2 and tri-tert-butylphosphine, followed by acetal-deprotection.
Scheme 2.
Synthesis of compound 6
Reagents and conditions: a) i. K2CO3, MeOH, reflux; ii. CNCH2COOEt, t-BuOK; b) (S)-4-(2-iodoethyl)-2,2-dimethyl-1,3-dioxolane, K2CO3; c) i. KOH, H2O/THF/MeOH; ii. 3-(4-methylpiperazin-1-yl)propan-1-amine, EDCI, HOBt, DIEA, DCM; d) i. 27, Pd(dba)2, tri-tert-butylphosphine, sodium tert-butoxide, DMF, toluen; ii. 4 M HCI in dioxane, MeOH, 10 min
Compounds 7, 13, 14 and 15 were prepared as shown in Scheme 3. Commercially available 4-fluoro-3-nitrobenzene-1-sulfonyl chloride was treated with the corresponding aniline in pyridine at 0°C to produce the sulfonamides 28 and 29. (R)-N1,N1-dimethyl-4-(phenylthio)butane-1,3-diamine was used to displace the fluorine in 28 and the Boc group was removed to give compound 15. Using the same strategy, 14 was synthesized from 29. Palladium-catalyzed amination was again employed to couple 15 and 26, and the resulting compound was deprotected to generate compound 7. Reductive amination of 15 with 4'-chloro-[1, 1'-biphenyl]-2-carbaldehyde was employed in the presence of sodium triacetoxyborohydride and 1,2-dichloroethane to give 13.
Scheme 3.
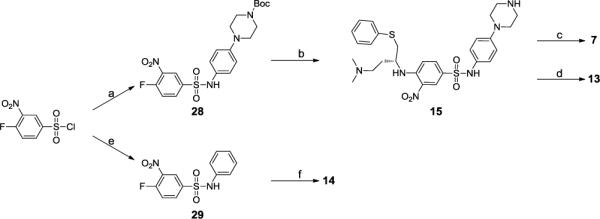
Synthesis of compounds 7,13,14 and 15
Reagents and conditions: a) ferf-butyl 4-(4-aminophenyl)piperazine-1-carboxylate, pyridine; b) i. (R)-N1,N1-dimethyl-4-(phenylthio)butane-i, 3-diamine, DIEA, DMF; ii. TFA, CH2CI2; c) i. 26, Pd(dba)2, tri-terf-butylphosphine, sodium terf-butoxide, DMF, toluen; ii. 4 M HCI in dioxane, MeOH, 10 min; d)4'-chloro-[1,1'-biphenyl]-2-carbaldehyde, Na(OAc)3BH, CICH2CH2CI; e) Aniline, pyridine; f) (R)-N1, N1-dimethyl-4-(phenylthio)butane-1,3-diamine, DIEA, DMF.
Compounds 9 and 10, in which the linker is an ethynyl group, were synthesized as shown in Scheme 4. Compound 5 was synthesized using the published procedure.19 Compound 5 was then coupled with 4-ethynylbenzoic acid in the presence of EDCI to give compound 30. Pd(0)-catalyzed coupling22 of the iodide 26 with the terminal alkyne 30 followed by deprotection of the acetal with HCl yielded compound 9. Compound 10 was synthesized using a procedure similar to compound 9.
Scheme 4.
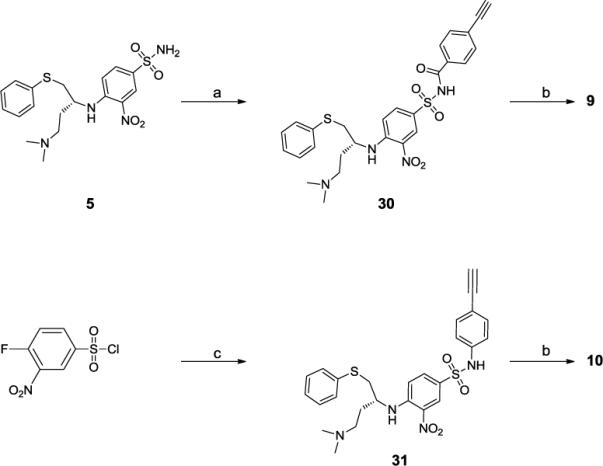
Synthesis of compounds 9 and 10
a) 4-ethynylbenzoic acid, EDCI, DMAP, CH2CI2, b) i. 26, Pd(PPh3)4, Cul, Et3N, DMF; ii. 4 M HCI in dioxane, MeOH, 10 min; c) i. 4-ethynylaniline, Pyridine, ii. (R)-N1,N1-dimethyl-4-(phenylthio)butane-1,3-diamine, DIPEA, DMF;
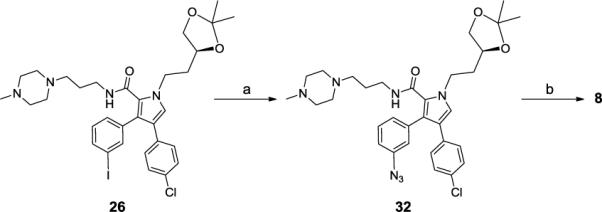
Scheme 5 shows synthesis of compound 8 with a triazole ring as the linker. CuI/L-proline-catalyzed coupling reaction23 of aryl iodide 26 with sodium azide was carried out at 70°C in DMSO to produce 32. This aryl azide 32 and alkyne 31 were joined by a CuI-catalyzed Huisgen cycloaddition24 in a mixture of water and t-butanol in the presence of CuSO4.5H2O and (+)-sodium L-ascorbate. The acetal group in the resulting triazole was removed with HCl to produce compound 8.
Scheme 5.
Synthesis of compound 8
a) NaN3, Cul, L-proline, NaOH, DMSO, 70 °C; b) i. 31, CuSO4-5H2O, Sodium L-ascorbate, H2O/t-BuOH; ii. 4 M HCI in dioxane, MeOH, 10 min
The linear synthetic strategy shown in scheme 6 was employed to prepare compounds 12 and 11. Compounds 33 and 34 were synthesized using the strategy described in Scheme 1. Using L-proline as a promotor, Ullmann-type C-N bond formation reaction25 of 34 with 1-(p-nitrophenyl)piperazine provided intermediate 35, and subsequent hydrolysis of the ethyl ester and coupling with 1-(3-aminopropyl)-4-methylpiperazine in the presence of EDCI gave 36. Hydrogenation of the nitro group in 36 gave the aniline, which was treated with 4-fluoro-3-nitrobenzene-1-sulfonyl chloride in pyridine. Subsequent displacement of the fluoro group with (R)-N1,N1-dimethyl-4-(phenylthio)butane-1,3-diamine and acetal-deprotection with HCl produced 12. Compound 11 was synthesized similarly.
Scheme 6.
Synthesis of compounds 12 and 11.
Reagents and conditions: a) i. K2CO3, MeOH, reflux; ii. CNCH2COOEt, t-BuOK; b) (S)-4-(2-iodoethyl)-2,2-dimethyl-1,3-dioxolane, K2CO3; c) 1-(4-nitrophenyl)piperazine, Cul, L-Proline, K2CO3, 80 °C, 2h; d) i. KOH, H2O/THF/MeOH; ii. 3-(4-methylpiperazin-1-yl)propan-1-amine, EDCI, HOBt, DIEA, DCM; e) i. H2, Pd/C; ii. 4-fluoro-3-nitrobenzene-1-sulfonyl chloride, pyridine; iii. (R)-N1,N1-dimethyl-4-(phenylthio)butane-1,3-diamine, DIPEA, DMF; iv. 4 M HCI in dioxane, MeOH; f) N1-(4-nitrophenyl)ethane-1,2-diamine, Cul, L-Proline, K2CO3, 80 °C, overnight;
The syntheses of compounds 16, 17, 18, 19 and 20 are outlined in Scheme 7. Methylation with MeI and subsequent Ullmann-type C-N bond formation reaction of 24 yielded compound 39. Compound 20 was prepared from 39 by a strategy similar to that used for 12. The amides 16 and 17 were produced by coupling the acid 20 to the corresponding amines using EDCI and HOBt. Treatment of 20 with TFA afforded the decarboxylated compound 18. The ester 19 was synthesized by condensation of ethanol and the acid 20 with N,N'-diisopropylcarbodiimide in the presence of 4-(dimethylamino)-pyridine.
Scheme 7.
Synthesis of compounds 20,16,17,18 and 19
Reagents and conditions: a) Mel, K2CO3; b) 1-(4-nitrophenyl)piperazine, Cul, L-Proline, K2CO3, 80 °C, 2h; c) i. KOH, H2O/THF/MeOH, reflux; ii. H2, Pd/C; iii. 4-fluoro-3-nitrobenzene-1-sulfonyl chloride, pyridine; iv. (R)-N1,N1-dimethyl-4-(phenyltriio)butane-1,3-diamine, DIPEA, DMF; d) 3-(4-methylpiperazin-1-yl)propan-1-amine, EDCI, HOBt, DIEA, DCM; e) methyl amine, EDCI, HOBt, DIEA, DCM, f) TFA, CH2CI2; g) EtOH, N,N'-Diisopropylcarbodiimide, 4-(dimethylamino)pyridine, THF.
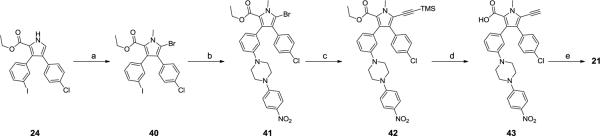
Compound 21 was prepared as shown in Scheme 8. Bromination of 24 with NBS in DMF and subsequent methylation afforded 40, which was coupled to 1-(p-nitrophenyl)piperazine to give the intermediate 41. Sonogashira coupling of 41 with ethynyltrimethylsilane in the presence of CuI and Pd(PPh3)4 yielded 42. Hydrolysis of the ethyl ester and simultaneous removal of the trimethylsilane protecting group in 42 afforded 43. Reduction of the nitro and ethynyl groups in 43 with H2 and Pd/C as the catalyst gave a product which, subjected to the same strategy as described in Scheme 7 yielded compound 21.
Scheme 8.
Synthesis of compound 21
Reagents and conditions: a) i. NBS, DMF; ii. Mel, K2CO3; b) 1-(4-nitrophenyl)piperazine, Cul, L-Proline, K2CO3, 80 °C, 2h; c) Ethynyltrimethylsilane, Pd(PPh3)4, Cul, Et3N, DMF, 85 °C; d) KOH, Dioxane, EtOH, H2O, reflux, 2h; e) i. H2, Pd/C; ii. 4-fluoro-3-nitrobenzene-1-sulfonyl chloride, pyridine; iii. (R)-N1,N1-dimethyl-4-(phenylthio)butane-1,3-diamine, DIPEA, DMF.
Summary
Employing a computational structure-based design strategy, we have designed compound 4, which contains a novel and druglike scaffold and binds to one well-defined binding pocket in Bcl-xL. The binding model of 4 with Bcl-xL was determined experimentally by x-ray crystallography. Based upon the crystal structure of 4 and of 1 (ABT-737) in complex with Bcl-xL, we employed 4 and 5, a large fragment of 1, for the design of new small-molecule inhibitors that occupy two separate binding pockets in Bcl-xL. Although both 4 and 5 bind to Bcl-2 and Bcl-xL with very weak affinities, linking them together with appropriate linkers have yielded compounds with very high affinities (Ki < 1 nM) to both Bcl-2 and Bcl-xL. Optimization of the linker between them resulted in 7, which not only binds to Bcl-2 and Bcl-xL with high affinities (Ki < 1 nM), but also potently inhibits cell growth in two small-cell lung cancer cell lines, which are sensitive to potent and specific Bcl-2/Bcl-xL inhibitors. Our results demonstrate that linking two molecules with very weak affinities to Bcl-2/Bcl-xL with appropriate linkers can result in highly potent Bcl-2/Bcl-xL inhibitors. The nature of the linkers plays a key role in achieving high binding affinities to Bcl-2/Bcl-xL and potent cell growth inhibitory activity against cancer cells. Further structure-activity relationship studies of 7 yielded a very promising lead compound, 21, which binds to Bcl-2 and Bcl-xL with Ki values < 1 nM, exceeding the limits of the binding assay. Compound 21 achieves IC50 values of 60 and 90 nM against the H146 and H1417 cancer cell lines and induces robust cell death in the H146 cancer cell line at 30 nM. It is now the subject of further optimization, the results of which will be reported in future publications.
EXPERIMENTAL SECTION
General Chemistry Information
Unless otherwise stated, all reactions were performed under nitrogen atmosphere in dry solvents under anhydrous conditions. Reagents were used as supplied without further purification unless otherwise noted. NMR spectra were acquired at a proton frequency of 300 MHz and chemical shifts are reported in parts per million (ppm) relative to an internal standard. The final products were purified by a C18 reverse phase semi-preparative HPLC column with solvent A (0.1% of TFA in water) and solvent B (0.1% of TFA in CH3CN) as eluents.
Ethyl 4-(4-chlorophenyl)-3-phenyl-1H-pyrrole-2-carboxylate (22)
A mixture of benzaldehyde (1.06 g, 10 mmol), 4-chlorophenyl cyanide (1.52 g, 10 mmol), and K2CO3 (1.66 g, 12 mmol) was refluxed overnight in MeOH (15 mL) under N2. The mixture was cooled, poured into H2O (15 mL), and stirred for 20 min. The precipitate was collected by filtration, washed with H2O (2 × 15 mL) and petroleum ether (2 × 15 mL), and air-dried to give 2-(4-chlorophenyl)-3-phenylacrylonitrile. A solution of this compound and ethyl isocyanoacetate (1.13 g, 10 mmol) in THF (20 mL) was added dropwise to a stirred solution of potassium t-butoxide (1.34g, 12 mmol) in DMF (15 mL) at 0°C under N2. After stirring at 0°C for 1 h, the reaction mixture was diluted with H2O (20 mL) and extracted with EtOAc (2 × 20 mL). The combined organic fractions were washed with water (20 mL) and then with brine (20 mL). After removal of the solvent under vacuum, the residue was purified by flash chromatography on silica gel to afford 22 (1.7 g, 52% over two steps).1H NMR (300 MHz, CCl3D), δ 9.62 (br. 1H), 7.32~7.30 (m, 5H), 7.18 (d, J=8.3, 2H), 7.11 (d, J=2.9, 1H), 7.05 (d, J=8.3, 2H), 4.22 (q, J=7.1, 2H), 1.16 (t, J=7.1, 3H); 13C NMR (75 MHz, CCl3D), δ 161.4, 134.2, 133.1, 131.9, 130.8, 129.5, 129.2, 128.4, 127.6, 127.0, 125.5, 120.4, 120.2, 60.4, 14.0;
(S)-ethyl 4-(4-chlorophenyl)-1-(2-(2,2-dimethyl-1,3-dioxolan-4-yl)ethyl)-3-phenyl-1H-pyrrole-2-carboxylate (23)
Compound 22 (1.7g 5.2 mmol), (S)-4-(2-iodoethyl)-2,2-dimethyl-1,3-dioxolane (2.0 g, 7.8 mmol), and K2CO3 (2.2 g, 15.6 mmol) in DMF (15 mL) were heated to 60°C for 8 hours. The reaction was cooled, diluted with water (20 mL), and extracted into EtOAc (30 mL, 2 × 20 mL). The combined organic fractions were washed with water (4 × 10 mL) then with brine (10 mL) and dried over Na2SO4. After evaporation of the solvent, the residue was purified by flash chromatography on silica gel to provide 23 (2.2 g, 92% yield). 1H NMR (300 MHz, CCl3D), δ 7.30~7.28 (m, 3H), 7.21~7.18 (m, 2H), 7.13 (d, J=8.5, 2H), 7.07 (s, 1H), 6.99 (d, J=8.5, 2H), 4.65~4.57 (m, 1H), 4.47~4.38 (m, 1H), 4.17~4.01 (m, 4H), 3.61 (t, J=7.3, 1H), 2.23~2.15 (m, 1H), 2.09~1.98 (m, 1H), 1.48 (s, 3H), 1.39 (s, 3H), 0.93 (t, J=7.1, 3H); 13C NMR (75 MHz, CCl3D), δ 161.5, 135.8, 133.0, 131.6, 131.4, 130.6, 129.2, 128.3, 127.6, 126.7, 126.3, 123.0, 119.9, 73.2, 69.1, 59.8, 46.8, 35.5, 27.1, 25.7, 13.6;
(S)-4-(4-chlorophenyl)-1-(3,4-dihydroxybutyl)-N-(3-(4-methylpiperazin-1-yl)propyl)-3-phenyl-1H-pyrrole-2-carboxamide (4)
Potassium hydroxide (0.2 g, 3.6 mmol) was added to a solution of 23 (0.54 g, 1.2mmol) in a mixture of THF/methanol/water (1:1:1, 10 ml) and the solution was refluxed until no starting material could be detected by TLC. After cooling the reaction was neutralized with 1M HCl and extracted with EtOAc. The EtOAc solution was washed with brine, dried over Na2SO4 and concentrated in vacuum to produce the crude acid which was used in the next step without purification. A solution of this acid, 1-(3-aminopropyl)-4-methylpiperazine (0.23 g. 1.4 mmol), EDCI (0.35 g 1.8 mmol), HOBt (0.23 g, 1.8 mmol) and N,N-diisopropylethylamine (0.42 mL, 2.4 mmol) in DCM (10 mL) was stirred for 8 h and then concentrated. The residue was resolved in MeOH (10 mL) and treated with 1 mL HCl solution (4N in dioxane) for 10 min and then the solvent was removed under vacuum. The residue was then purified by HPLC to provide 4 (0.5 g, 79%).1H NMR (300 MHz, CD3OD), δ 7.24~7.20 (m, 3H), 7.09~6.89 (m, 5H), 6.88 (d, J=8.5, 2H), 4.28~4.16 (m, 2H), 3.50~3.35 (m, 11H), 3.12~3.04 (m, 2H), 2.85 (s, 3H), 2.82~2.75 (m, 2H), 1.95~1.89 (m, 1H), 1.68~1.63 (m, 3H); 13C NMR (75 MHz, CD3OD), δ 165.6, 136.2, 134.9, 132.6, 131.8, 130.5, 129.7, 129.2, 128.4, 126.6, 126.1, 124.9, 123.6, 70.2, 67.3, 55.5, 51.6, 49.9, 46.2, 43.4, 37.3, 36.4, 25.1; ESI MS: m/z 525.8 (M + H)+.
Ethyl 4-(4-chlorophenyl)-3-(3-iodophenyl)-1H-pyrrole-2-carboxylate (24)
Compound 24 was prepared by a procedure similar to that used for compound 22. The yield was 49% in two steps. 1H NMR (300 MHz, CCl3D), δ 9.48 (s, 1H), 7.74 (s, 1H), 7.64 (d, J=7.8, 1H), 7.20 (d, J=8.4, 2H), 7.14 (d, J=7.7, 1H), 7.10 (d, J=2.9, 1H), 7.05~7.00 (m, 3H), 4.22 (q, J=7.1, 2H), 1.20 (t, J=7.1, 3H); 13C NMR (75 MHz, CCl3D), δ 161.2, 139.7, 136.4, 135.9, 132.6, 132.2, 130.0, 129.5, 129.3, 128.5, 127.1, 125.5, 120.42, 120.39, 93.3, 60.6, 14.1;
(S)-Ethyl 4-(4-chlorophenyl)-1-(2-(2,2-dimethyl-1,3-dioxolan-4-yl)ethyl)-3-(3-iodophenyl)-1H-pyrrole-2-carboxylate (25)
Compound 25 was prepared in 87% yield from 24 using a procedure similar to that used with compound 23. 1H NMR (300 MHz, CCl3D), δ 7.67 (s, 1H), 7.62 (d, J=7.8, 1H), 7.16 (d, J=8.5, 2H), 7.10~6.97 (m, 5H), 4.63~4.56 (m, 1H), 4.46~4.39 (m, 1H), 4.15~4.03 (m, 4H), 3.60 (t, J=6.7, 1H), 2.21~2.14 (m, 1H), 2.07~1.99 (m, 1H), 1.47 (s, 3H), 1.38 (s, 3H), 1.00 (t, J=7.1, 3H); 13C NMR (75 MHz, CCl3D), δ 161.2, 139.7, 138.1, 135.6, 132.6, 131.9, 129.7, 129.4, 129.3, 128.4, 126.5, 123.0, 119.9, 93.3, 73.1, 69.1, 60.0, 46.8, 35.5, 27.1, 25.7, 13.8; ESI MS: m/z 580.1 (M + H)+.
(S)-4-(4-Chlorophenyl)-1-(2-(2,2-dimethyl-1,3-dioxolan-4-yl)ethyl)-3-(3-iodophenyl)-N-(3-(4-methylpiperazin-1-yl)propyl)-1H-pyrrole-2-carboxamide (26)
Potassium hydroxide (0.71 g, 12.6 mmol) was added to a solution of 25 (2.44 g, 4.2 mmol) in a mixture of THF/MeOH/H2O (1:1:1, 30 ml) and the solution was refluxed until no starting material was detectable by TLC. After cooling the reaction was neutralized with 1M HCl and the compound was extracted with EtOAc. The EtOAc solution was washed with brine, dried over Na2SO4 and concentrated in vacuum to produce the crude acid which was used directly in the next step without purification. A solution of this acid, 1-(3-aminopropyl)-4-methylpiperazine (0.86 g, 5.5 mmol), EDCI (1.5g 6.3 mmol), HOBt (1.0 g, 6.3 mol) and N,N-diisopropylethylamine (1.46 mL, 8.4 mmol) in DCM (15 mL) was stirred for 8 h and then concentrated. The residue was purified by flash chromatography on silica gel to afford 26 (2.26g, 78% in two steps). 1H NMR (300 MHz, CCl3D), δ 7.66~7.64 (m, 2H), 7.16~7.13 (m, 3H), 7.05 (t, J=7.8, 1H), 6.95~6.93 (m, 3H), 5.59 (t, J=5.1, 1H), 4.47~4.43 (m, 1H), 4.38~4.31 (m, 1H), 4.11~4.00 (m, 2H), 3.55 (t, J=7.1, 1H), 3.26~3.19 (m, 2H), 2.35~1.97 (m, 15H), 1.49~1.43 (m, 5H), 1.34 (s, 3H); 13C NMR (75 MHz, CCl3D), δ 161.5, 139.3, 137.0, 136.5, 132.8, 131.8, 130.4, 129.9, 129.2, 128.4, 124.3, 124.1, 123.1, 122.1, 94.6, 73.2, 69.1, 55.8, 55.0, 53.0, 46.2, 46.0, 38.0, 35.7, 27.0, 26.1, 25.7; ESI MS: m/z 691.6 (M + H)+.
4-(4-Chlorophenyl)-1-((S)-3,4-dihydroxybutyl)-3-(3-(4-(4-(((4-(((S)-4-(dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitrophenyl)sulfonyl)carbamoyl)phenyl)piperazin-1-yl)phenyl)-N-(3-(4-methylpiperazin-1-yl)propyl)-1H-pyrrole-2-carboxamide (6)
Pd(dba)2 (3.5 mg, 0.006 mmol), tri-tert-butylphosphine (1M in toluene, 4.8 μL), and sodium tert-butoxide (18 mg, 0.18 mmol) were added to a stirred slurry of 26 (83mg, 0.12 mmol) and 27 (88mg, 0.14 mmol) in a mixture of toluene/DMF (1:1, 4 mL) at room temperature under N2. The mixture was heated to 70°C and monitored by thin-layer chromatography. After complete consumption of starting materials, the reaction mixture was filtered through celite and concentrated. The residue was resolved in MeOH (5 mL) and treated with 0.2 mL HCl solution (4M in dioxane) for 10 minutes and then the solvent was removed in vacuum. Purification of the residue by HPLC afforded 6 (39 mg, 29%). 1H NMR (300 MHz, CD3OD), δ 8.66 (d, J=2.2, 1H), 7.93 (dd, J=2.2, 9.2, 1H), 7.75 (d, J=9.0, 2H), 7.29~6.95 (m, 15H), 6.81 (s, 1H), 6.74 (d, J=7.4, 1H), 4.38~4.27 (m, 2H), 4.16~4.13 (m, 1H), 3.54~3.32 (m, 11H), 3.24~3.08 (m, 14H), 2.84 (s, 6H), 2.82 (s, 3H), 2.61 (t, J=7.1, 2H), 2.27~2.14 (m, 2H), 2.03~2.00 (m, 1H), 1.78~1.75 (m, 1H), 1.66~1.61 (m, 2H); 13C NMR (75 MHz, CD3OD), δ 167.2, 165.4, 155.8, 152.1, 148.5, 137.2, 136.2, 135.6, 135.0, 132.5, 132.2, 131.5, 130.6, 130.4, 130.1, 129.7, 127.8, 127.5, 126.8, 126.0, 124.9, 123.7, 123.5, 121.7, 119.7, 116.5, 115.7, 114.8, 70.2, 67.3, 55.9, 55.4, 53.0, 52.4, 50.7, 50.2, 48.0, 46.2, 43.6, 43.5, 39.3, 37.6, 36.4, 30.1, 25.9; ESI MS: m/z 1135.6 (M + H)+.
t-Butyl 4-(4-(4-fluoro-3-nitrophenylsulfonamido)phenyl)piperazine-1-carboxylate (28)
4-Fluoro-3-nitrobenzene-1-sulfonyl chloride (312 mg, 1.3mmol) was added to t-butyl 4-(4-aminophenyl)piperazine-1-carboxylate (360 mg, 1.3mmol) in pyridine (10 mL) at 0°C. The mixture was stirred at 0°C for 30 min and then concentrated under vacuum. The residue was purified by flash chromatography on silica gel to afford 28 (474 mg, 76%). 1H NMR (300 MHz, CCl3D), δ 8.43 (dd, J=2.2, 6.8, 1H), 7.95~7.90 (m, 1H), 7.37 (t, J=9.3, 1H), 7.01~6.98 (m, 3H), 6.81 (d, J=8.9, 2H), 3.59~3.56 (m, 4H), 3.13~3.10 (m, 4H), 1.49 (s, 9H); ESI MS: m/z 480.9 (M + H)+.
(R)-4-((4-(Dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitro-N-(4-(piperazin-1-yl)phenyl)benzenesulfonamide 15
DIEA (70 μL, 0.4mmol) was added to a solution of 28 (100 mg, 0.21 mmol) and (R)-N1,N1-dimethyl-4-phenylthio)butane-1,3-diamine (47 mg, 0.21mmol) in DMF. The solution was stirred overnight then concentrated. The residue was resolved in MeOH (5 mL) and treated with HCl solution (4M in dioxane, 4mL), which was stirred for 5 h and then concentrated. This residue was purified by HPLC to afford 15 (170 mg, 81% over two steps). 1H NMR (300 MHz, CD3OD), δ 8.24 (d, J=2.2 Hz, 1H), 7.56 (dd, J=2.2, 9.1, 1H), 7.13~7.10 (m, 2H), 7.04~6.87 (m, 8H), 4.09~4.07 (m, 1H), 3.37~3.27 (m, 10H), 3.21~3.13 (m, 2H), 2.82 (s, 6H), 2.24~2.14 (m, 2H); 13C NMR (75 MHz, CD3OD), δ 149.3, 148.0, 136.2, 134.4, 132.1, 132.0, 131.6, 130.1, 128.0, 127.8, 127.5, 124.4, 118.8, 116.3, 55.9, 52.4, 47.9, 44.7, 43.5, 39.5, 30.1; ESI MS: m/z 585.7 (M + H)+.
4-(4-Chlorophenyl)-1-((S)-3,4-dihydroxybutyl)-3-(3-(4-(4-(4-(((R)-4-(dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitrophenylsulfonamido)phenyl)piperazin-1-yl)phenyl)-N-(3-(4-methylpiperazin-1-yl)propyl)-1H-pyrrole-2-carboxamide (7)
Compound 7 was prepared in 24% yield from compounds 15 and 26 using a similar procedure as that used to prepare BM-977. 1H NMR (300 MHz, CD3OD), δ 8.30 (d, J=2.2, 1H), 7.59 (dd, J=2.3, 9.2, 1H), 7.28 (t, J=7.9, 1H), 7.18~6.98 (m, 15H), 6.91 (d, J=9.4, 1H), 6.83 (s, 1H), 6.78 (d, J=7.5, 1H), 4.38~4.28 (m, 2H), 4.09~4.08 (m, 1H), 3.55~3.32 (m, 11H), 3.21~3.15 (m, 14H), 2.84 (s, 3H), 2.83 (s, 6H), 2.70~2.65 (m, 2H), 2.25~1.99 (m, 3H), 1.78~1.64 (m, 3H); 13C NMR (75 MHz, CD3OD), δ 165.4, 151.7, 147.9, 137.2, 136.2, 135.0, 134.5, 132.6, 132.2, 131.6, 130.7, 130.5, 130.2, 129.3, 129.2, 128.0, 127.9, 127.7, 126.7, 126.1, 124.8, 124.3, 124.1, 123.5, 120.0, 118.8, 116.8, 116.2, 115.8, 70.2, 67.2, 55.9, 55.4, 53.1, 52.4, 51.0, 50.7, 50.5, 46.2, 43.7, 43.5, 39.6, 37.7, 36.4, 30.1, 25.9; ESI MS: m/z 1107.7 (M + H)+.
(R)-N-(4-(4-((4'-Chloro-[1,1'-biphenyl]-2-yl)methyl)piperazin-1-yl)phenyl)-4-((4-(dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitrobenzenesulfonamide (13)
Compound 15 (58.5 mg, 0.1 mmol) and 4'-chloro-[1, 1'-biphenyl]-2-carbaldehyde (21.7 mg, 0.1 mmol) were mixed in 1,2-dichloroethane (5 mL) and then treated with sodium triacetoxyborohydride (30 mg, 0.14 mmol). The mixture was stirred at room temperature under a N2 atmosphere for 24 h until the reactants were consumed. Then the reaction mixture was quenched by adding 1N NaOH, and the product was extracted with CH2Cl2. The CH2Cl2 extract was washed with brine and dried over MgSO4. The solvent was evaporated and purified by HPLC to give 13 (61 mg, 78%). 1H NMR (300 MHz, CD3OD), δ 8.24 (s, 1H), 7.70~7.68 (m, 1H), 7.58~7.47 (m, 5H), 7.39~7.31 (m, 3H), 7.12 (d, J=7.0, 2H), 7.02~6.89 (m, 6H), 8.18 (d, J=8.7, 2H), 4.40 (s, 2H), 4.08 (m. 1H), 3.38~3.31 (m, 3H), 3.21~3.08 (m, 9H), 2.84 (s, 6H), 2.25~2.15 (m, 2H); 13C NMR (75 MHz, CD3OD), δ 148.6, 147.9, 144.4, 139.7, 136.2, 135.3, 134.4, 132.6, 132.3, 132.2, 132.1, 131.6, 131.4, 130.2, 130.1, 130.0, 128.0, 127.8, 127.6, 127.5, 124.3, 118.6, 116.2, 58.0, 55.9, 52.8, 52.4, 47.5, 43.5, 39.6, 30.1; ESI MS: m/z 785.8 (M + H)+;
4-Fluoro-3-nitro-N-phenylbenzenesulfonamide (29)
Compound 29 was prepared in 80% yield using a procedure similar to that used to prepare compound 28. 1H NMR (300 MHz, CCl3D), δ 8.51 (dd, J=2.3, 6.8, 1H), 8.02~7.97 (m, 1H), 7.42~7.30 (m, 3H), 7.14~7.11 (m, 2H), 6.92 (s, 1H);
(R)-4-((4-(Dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitro-N-phenylbenzenesulfonamide (14)
Compound 14 was prepared from 29 using a similar procedure as that used for compound 15 in 83% yield. 1H NMR (300 MHz, CD3OD), δ 8.38 (d, J=2.2 Hz, 1H), 8.15 (d, J=9.3, 1H), 7.62 (dd, J=2.1, 9.1, 1H), 7.28~7.23 (m, 2H), 7.18~6.97 (m, 8H), 6.90, (d, J=9.3, 1H), 4.11~4.07 (m, 1H), 3.40~3.33 (m, 1H), 3.23~3.09 (m, 3H), 2.85 (s, 6H), 2.31~2.11 (m, 2H); 13C NMR (75 MHz, CD3OD), δ 147.9, 139.0, 136.1, 134.4, 132.3, 131.5, 130.3, 128.0, 127.9, 127.7, 125.8, 121.8, 116.1, 55.9, 52.3, 43.5, 39.4, 30.1; ESI MS: m/z 501.7 (M + H)+.
(R)-N-((4-((4-(Dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitrophenyl)sulfonyl)-4-ethynylbenzamide (30)
A suspension of 5 (400 mg, 0.94 mmol), 4-ethynylbenzoic acid (138 mg, 0.94 mmol), DMAP (230 m g, 1.88 mmol) and EDCI (362 g, 1.88 mmol) in CH2Cl2 (10 mL) was stirred at room temperature for 8 h. The reaction mixture was washed with saturated NH4Cl (3 × 8 mL) and concentrated. The crude residue was purified by HPLC to provide 30 (365 mg, 70%). 1H NMR (300 MHz, CD3OD), δ 8.66 (s, 1H), 8.30 (d, J=9.2, 1H), 7.89 (d, J=8.0, 2H), 7.52 (d, J=8.0, 2H), 7.21~7.19 (m, 2H), 7.05~6.89 (m, 4H), 4.16~4.15 (m, 1H), 3.80 (s, 1H), 3.40~3.34 (m, 1H), 3.24~3.13 (m, 3H), 2.86 (s, 6H), 2.30~2.14 (m, 2H); 13C NMR (75 MHz, CD3OD), δ 165.5, 147.2, 134.8, 134.1, 132.0, 131.6, 130.7, 130.1, 128.7, 128.5, 128.0, 127.4, 126.5, 125.4, 114.6, 81.8, 81.0, 54.5, 51.1, 42.2, 38.0, 28.7; ESI MS: m/z 553.8 (M + H)+.
4-(4-Chlorophenyl)-1-((S)-3,4-dihydroxybutyl)-3-(3-((4-(((4-(((R)-4-(dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitrophenyl)sulfonyl)carbamoyl)phenyl)ethynyl)phenyl)-N-(3-(4-methylpiperazin-1-yl)propyl)-1H-pyrrole-2-carboxamide (9)
CuI (6.9 mg,, 0.036 mmol) and Pd(PPh3)4 (13.9 mg, 0.012 mmol) were added to a solution of 26 (83 mg, 0.12 mmol), 30 (66mg, 0.12mmol) and Et3N (50μL, 0.36mmol) in DMF (5 mL) at room temperature under nitrogen. The mixture was stirred at 40 °C for 3 h and then filtered through Celite, washed with 50 mL of CH2Cl2, and concentrated under reduced pressure. The residue was resolved in MeOH (5 mL) and treated with 0.2 mL HCl solution (4M in dioxane) for 10 minutes and then the solvent was removed in vacuum. Purification of the residue by HPLC afforded 9 (97 mg, 75%). 1H NMR (300 MHz, CD3OD), δ 8.70 (d, J=1.9, 1H), 7.94 (d, J=7.4, 1H), 7.83 (d, J=8.3, 2H), 7.58 (d, J=8.2, 2H), 7.48 (d, J=7.7, 1H), 7.38~7.34 (m, 2H), 7.24~7.01 (m, 12H), 4.37~4.29 (m, 2H), 4.19~4.17 (m, 1H), 3.56~3.39 (m, 8H), 3.27~3.13 (m, 9H), 2.87~2.76 (m, 11H), 2.31~2.22 (m, 2H), 2.04~2.01 (m, 1H), 1.75~1.70 (m, 3H); 13C NMR (75 MHz, CD3OD), δ 166.9, 165.4, 148.6, 136.8, 136.2, 135.5, 134.8, 134.7, 132.9, 132.8, 132.6, 132.2, 131.5, 130.6, 130.1, 129.9, 129.5, 129.4, 129.3, 127.9, 126.9, 126.8, 125.0, 124.7, 123.9, 123.5, 93.4, 89.6, 70.2, 67.3, 55.9, 55.5, 52.6, 52.5, 50.4, 46.1, 43.6, 39.4, 37.7, 36.4, 30.1, 25.7; ESI MS: m/z 1075.8 (M + H)+.
(R)-4-((4-(Dimethylamino)-1-(phenylthio)butan-2-yl)amino)-N-(4-ethynylphenyl)-3-nitrobenzenesulfonamide (31)
Compound 31 was prepared using a similar procedure to that used for compound 30 in 80% yield. 1H NMR (300 MHz, CD3OD), δ 8.43 (s, 1H), 7.64 (d, J=8.8, 1H), 7.38 (d, J=8.2, 2H), 7.21~7.14 (m, 4H), 7.10~6.93 (m, 4H), 4.13~4.12 (m, 1H), 3.44~3.37 (m, 2H), 3.26~3.14 (m, 3H), 2.88 (s, 6H), 2.30~2.21 (m, 2H); 13C NMR (75 MHz, CD3OD), δ 146.7, 138.0, 134.7, 132.9, 132.7, 130.9, 130.1, 128.7, 126.6, 126.5, 125.9, 119.6, 118.3, 114.9, 82.4, 77.3, 54.5, 51.0, 42.1, 38.0, 28.7; ESI MS: m/z 526.0 (M + H)+.
4-(4-chlorophenyl)-1-((S)-3,4-dihydroxybutyl)-3-(3-((4-(4-(((R)-4-(dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitrophenylsulfonamido)phenyl)ethynyl)phenyl)-N-(3-(4-methylpiperazin-1-yl)propyl)-1H-pyrrole-2-carboxamide (10)
Compound 10 was prepared from 31 using a similar procedure as that for 9 in 79% yield. 1H NMR (300 MHz, CD3OD), δ 8.39 (d, J=2.2, 1H), 7.63 (dd, J=2.2, 9.0, 1H), 7.40~7.37 (m, 3H), 7.32~7.27 (m, 2H), 7.15~7.10 (m, 8H), 7.01~6.89 (m, 6H), 4.37~4.21 (m, 2H), 4.09~4.06 (m, 1H), 3.53~3.49 (m, 1H), 3.45~3.43 (m, 2H), 3.33~3.31 (m, 5H), 3.24~3.08 (m, 9H), 2.82 (s, 9H), 2.72~2.68 (m, 2H), 2.24~1.98 (m, 3H), 1.79~1.63 (m, 3H); 13C NMR (75 MHz, CD3OD), δ 165.4, 148.1, 139.4, 136.6, 136.2, 134.8, 134.4, 134.3, 133.7, 132.7, 132.2, 132.0, 131.5, 131.2, 130.5, 130.1, 129.8, 129.3, 128.0, 127.9, 127.4, 126.7, 125.2, 124.7, 124.6, 123.5, 121.1, 120.1, 116.3, 90.2, 90.0, 70.2, 67.3, 55.9, 55.4, 52.7, 52.3, 50.5, 46.1, 43.6, 43.5, 39.4, 37.6, 36.4, 30.1, 25.7; ESI MS: m/z 1048.4 (M + H)+.
(S)-3-(3-Azidophenyl)-4-(4-chlorophenyl)-1-(2-(2,2-dimethyl-1,3-dioxolan-4-yl)ethyl)-N-(3-(4-methylpiperazin-1-yl)propyl)-1H-pyrrole-2-carboxamide (32)
A mixture of 26 (300 mg, 0.43 mmol), sodium azide (42mg, 0.65 mmol), CuI (8.3mg, 0.043 mmol), L-proline (10mg, 0.087 mmol) and NaOH (3.5mg, 0.087 mmol) in DMSO (4 mL) in a sealed tube was heated to 70°C under N2. After the reaction was completed, the cooled mixture was partitioned between CH2Cl2 and H2O. The organic layer was separated, and the aqueous layer was extracted twice with CH2Cl2. The combined organic layers were washed with brine, dried over Mg2SO4, and concentrated in vacuo. The residue was purified by flash chromatography on silica gel to afford 32 (134mg, 51%).1H NMR (300 MHz, CCl3D), δ 7.35~7.29 (m, 1H), 7.11 (d, J=8.3, 2H), 6.98 (d, J=7.7, 2H), 6.93~6.87 (m, 4H), 5.54 (br. 1H), 4.47~4.43 (m, 1H), 4.38~4.29 (m, 1H), 4.10~4.08 (m, 1H), 4.03~3.00 (m, 1H), 3.53 (t, J=7.3, 1H), 3.21~3.15 (m, 2H), 2.36~1.96 (m, 15H), 1.42~1.40 (m, 5H), 1.32 (s, 3H); 13C NMR (75 MHz, CCl3D), δ 161.5, 140.6, 136.8, 132.8, 131.8, 130.2, 129.1, 128.4, 127.2, 124.1, 123.8, 122.2, 121.2, 118.1, 108.9, 73.2, 69.1, 55.7, 54.9, 52.7, 46.3, 45.8, 37.8, 35.7, 27.0, 26.1, 25.6; ESI MS: m/z 606.8 (M + H)+.
4-(4-Chlorophenyl)-1-((S)-3,4-dihydroxybutyl)-3-(3-(4-(4-(4-(((R)-4-(dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitrophenylsulfonamido)phenyl)-1H-1,2,3-triazol-1-yl)phenyl)-N-(3-(4-methylpiperazin-1-yl)propyl)-1H-pyrrole-2-carboxamide (8)
Compound 32 (90 mg, 0.15 mmol) and compound 31 (78 mg, 0.15 mmol) were suspended in a 1:2 mixture of H2O and t-BuOH (9 mL) and then a solution of CuSO4.5H2O (3.7 mg, 0.015 mmol) and (+)-sodium L-ascorbate (8.8 mg, 0.045 mmol) in H2O (1 mL) was added. The resulting mixture was stirred at room temperature overnight and then was diluted with water (10 ml) and extracted with CH2Cl2 (3 × 20 mL). The combined organic layer was washed with brine, dried over Na2SO4, and concentrated under reduced pressure. The residue was resolved in MeOH (5 mL) and treated with HCl solution (4M in dioxane, 0.2 mL) for 10 min and then the solvent was removed in vacuum. Purification of the residue by HPLC afforded 8 (97 mg, 75%). 1H NMR (300 MHz, CD3OD), δ 8.67 (s, 1H), 8.38 (d, J=2.2, 1H), 7.78~7.70 (m, 4H), 7.62 (dd, J=2.2, 9.2, 1H), 7.47 (t, J=7.0, 1H), 7.24~7.21 (m, 3H), 7.12~7.01 (m, 7H), 6.94~6.88 (m, 4H), 4.35~4.28 (m, 2H), 4.07~4.06 (m, 1H), 3.54~3.34 (m, 9H), 3.20~3.11 (m, 8H), 2.84~2.80 (m, 11H), 2.19~2.15 (m, 3H), 1.71~1.66 (m, 3H); 13C NMR (75 MHz, CD3OD), δ 165.4, 148.9, 148.1, 139.3, 138.3, 138.0, 136.1, 134.7, 134.3, 132.9, 132.4, 132.2, 131.5, 131.0, 130.8, 130.0, 129.4, 128.1, 127.8, 127.7, 127.4, 127.1, 124.8, 124.5, 123.6, 123.4, 121.9, 120.1, 119.9, 116.4, 70.2, 67.3, 55.9, 55.5, 52.4, 50.3, 46.1, 43.5, 39.4, 37.7, 36.4, 30.1, 25.7; ; ESI MS: m/z 1091.4 (M + H)+.
Ethyl 4-(4-chlorophenyl)-3-(4-iodophenyl)-1H-pyrrole-2-carboxylate
Compound 33 was prepared using a similar procedure as was used to prepare 24 in 58% yield in two steps. 1H NMR (300 MHz, CCl3D), δ 9.52 (br. 1H), 7.56 (d, J=8.2, 2H), 7.20 (d, J=8.4, 2H), 7.09 (d, J=3.0, 1H), 7.04~7.01 (m, 4H), 4.23 (q, J=7.1, 2H), 1.20 (t, J=7.1, 3H); 13C NMR (75 MHz, CCl3D), δ 161.0, 136.8, 133.7, 132.7, 132.2, 129.6, 128.5, 127.9, 125.5, 120.4, 120.2, 92.9, 60.5, 14.1.
(S)-Ethyl 4-(4-chlorophenyl)-1-(2-(2,2-dimethyl-1,3-dioxolan-4-yl)ethyl)-3-(4-iodophenyl)-1H-pyrrole-2-carboxylate (34)
Compound 34 was prepared from 33 using a procedure similar to that used for 25 in 90% yield. 1H NMR (300 MHz, CCl3D), δ 7.62 (d, J=8.4, 2H), 7.16 (d, J=8.6, 2H), 7.05 (s, 1H), 6.98~6.93 (m, 4H), 4.62~4.55 (m, 1H), 4.46~4.36 (m, 1H), 4.15~4.03 (m, 4H), 3.63~3.57 (m, 1H), 2.21~2.12 (m, 1H), 2.08~1.94 (m, 1H), 1.47 (s, 3H), 1.38 (s, 3H), 0.99 (t, J=7.1, 3H); 13C NMR (75 MHz, CCl3D), δ 161.2, 136.7, 135.4, 132.6, 131.9, 130.0, 129.3, 128.4, 126.4, 123.0, 119.9, 109.1, 92.3, 73.1, 69.0, 60.0, 46.8, 35.5, 27.0, 25.6, 13.7.
(S)-Ethyl 4-(4-chlorophenyl)-1-(2-(2,2-dimethyl-1,3-dioxolan-4-yl)ethyl)-3-(4-(4-(4-nitrophenyl)piperazin-1-yl)phenyl)-1H-pyrrole-2-carboxylate (35)
Compound 34 (380 mg, 0.66 mmol), 1-(4-nitrophenyl)piperazine (271 mg, 1.31 mmol), CuI (12 mg, 0.066 mmol), L-proline (15 mg, 0.13 mmol) and K2CO3 (181 mg, 1.31 mmol) were dissolved in 5 mL of DMSO. This mixture was heated to 80 °C for 2 h under nitrogen. After the solution was cooled, saturated ammonium chloride solution was added and extracted with CH2Cl2. The combined organic layers were washed with brine, dried over sodium sulfate and concentrated. Purification of the residue by flash chromatography on silica gel afforded 35 (354 mg, 82%). 1H NMR (300 MHz, CCl3D), δ 8.15 (d, J=8.9, 2H), 7.15~7.11 (m, 4H), 7.05~7.00 (m, 3H), 6.90~6.87 (m, 4H), 4.63~4.54 (m, 1H), 4.45~4.36 (m, 1H), 4.12~4.04 (m, 4H), 3.62~3.57 (m, 5H), 3.39~3.37 (m, 4H), 2.21~2.15 (m, 1H), 2.09~2.00 (m, 1H), 1.47 (s, 3H), 1.38 (s, 3H), 1.02 (t, J=7.1, 3H); 13C NMR (75 MHz, CCl3D), δ 161.5, 154.7, 149.4, 138.6, 133.2, 131.5, 131.1, 129.3, 128.3, 127.5, 126.4, 126.0, 123.1, 120.0, 115.3, 112.7. 109.1, 73.2, 69.1, 59.8, 48.9, 47.1, 46.9, 35.6, 27.0, 25.6, 13.8; ESI MS: m/z 659.8 (M + H)+.
(S)-4-(4-Chlorophenyl)-1-(2-(2,2-dimethyl-1,3-dioxolan-4-yl)ethyl)-N-(3-(4-methyl-piperazin-1-yl)propyl)-3-(4-(4-(4-nitrophenyl)piperazin-1-yl)phenyl)-1H-pyrrole-2-carboxamide (36)
Compound 36 was prepared from 35 in 85% yield in two steps using a procedure similar to that used for 26. 1H NMR (300 MHz, CCl3D), δ 8.14 (d, J=8.9, 2H), 7.17~7.10 (m, 4H), 6.89~6.86 (m, 7H), 5.61 (t, J=5.3, 1H), 4.58~4.49 (m, 1H), 4.44~4.35 (m, 1H), 4.13~4.09 (m, 1H), 4.05~4.01 (m, 1H), 3.62~3.53 (m, 5H), 3.41~3.40 (m, 4H), 3.18 (q, J=6.2, 2H), 2.35~1.98 (m, 15H), 1.48~1.40 (m, 5H), 1.35 (s, 3H); 13C NMR (75 MHz, CCl3D), δ 161.8, 154.5, 149.8, 138.7, 133.4, 131.7, 131.4, 129.1, 128.3, 126.0, 125.8, 125.0, 124.2, 123.5, 122.3, 116.0, 112.8, 108.9, 73.4, 69.1, 55.8, 55.0, 53.0, 48.2, 47.0, 46.5, 45.9, 37.5, 35.7, 27.0, 26.4, 25.7; ESI MS: m/z 770.4 (M + H)+.
4-(4-Chlorophenyl)-1-((S)-3,4-dihydroxybutyl)-3-(4-(4-(4-(4-(((R)-4-(dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitrophenylsulfonamido)phenyl)piperazin-1-yl)phenyl)-N-(3-(4-methylpiperazin-1-yl)propyl)-1H-pyrrole-2-carboxamide (12)
10% Pd-C (20 mg) was added to a solution of compound 36 (124 mg, 0.16 mmol) in CH2Cl2 (3 mL) and MeOH (3 mL). The solution was stirred under 1 atm of H2 at room temperature for 0.5 h before it was filtered through celite and concentrated. The resulting aniline was used in the next step without purification. To this aniline in pyridine (5 mL), 4-fluoro-3-nitrobenzene-1-sulfonyl chloride (39 mg, 0.16 mmol) was added at 0°C. The mixture was stirred at 0°C for 30 min. The pyridine was removed under vacuum and the residue was purified by flash chromatography on silica gel to give (S)-4-(4-chlorophenyl)-1-(2-(2,2-dimethyl-1,3-dioxolan-4-yl)ethyl)-3-(4-((2-((4-(4-fluoro-3-nitrophenylsulfonamido)phenyl)amino)ethyl)amino)-phenyl)-N-(3-(4-methylpiperazin-1-yl)propyl)-1H-pyrrole-2-carboxamide. DIEA (56 μL, 0.32 mmol) was added to a solution of this sulfonamide and (R)-N1,N1-dimethyl-4-phenylthio)butane-1,3-diamine (39 mg, 0.16 mmol) in DMF. The solution was stirred overnight and concentrated. The residue was dissolved in MeOH (5 mL) and treated with HCl solution (4M in dioxane, 0.2 mL) for 10 min and then the solvent was removed in vacuum. Purification of the residue by HPLC afforded 12 (94 mg, 51% in four steps). 1H NMR (300 MHz, CD3OD), δ 8.36 (s, 1H), 7.66 (d, J=9.1, 1H), 7.22~7.04 (m, 18H), 6.97 (d, J=9.2, 1H), 4.43~4.31 (m, 2H), 4.15~4.13 (m, 1H), 3.59~3.37 (m, 17H), 3.26~3.21 (m, 8H), 2.90~2.82 (m, 11H), 2.30~2.05 (m, 3H), 1.83~1.74 (m, 3H); 13C NMR (75 MHz, CD3OD), δ 165.6, 147.9, 136.2, 135.1, 134.4, 132.7, 132.5, 132.3, 131.6, 130.5, 130.1, 129.2, 128.0, 127.9, 127.7, 126.3, 125.9, 125.1, 124.2, 123.7, 119.2, 117.9, 116.2, 70.3, 67.3, 55.9, 55.5, 52.5, 52.4, 51.4, 50.6, 50.4, 46.3, 43.6, 43.5, 39.5, 37.5, 36.5, 30.1, 25.7; ESI MS: m/z 1107.7 (M + H)+.
(S)-Ethyl 4-(4-chlorophenyl)-1-(2-(2,2-dimethyl-1,3-dioxolan-4-yl)ethyl)-3-(3-((2-((4-nitrophenyl)amino)ethyl)amino)phenyl)-1H-pyrrole-2-carboxylate (37)
Compound 37 was prepared in 78% yield from 26 using a procedure similar to that used for 35. 1H NMR (300 MHz, CCl3D), δ 8.07 (d, J=8.6, 2H), 7.15~7.13 (m, 3H), 7.05~7.02 (m, 3H), 6.66 (d, J=7.5, 1H), 6.59 (d, J=8.5, 1H), 6.52 (d, J=8.8, 2H), 6.46 (s, 1H), 4.58~4.56 (m, 1H), 4.45~4.35 (m, 1H), 4.11~3.99 (m, 4H), 3.59 (t, J=6.9, 1H), 3.37 (br., 4H), 2.18~2.16 (m, 1H), 2.06~2.01 (m, 1H), 1.47 (s, 3H), 1.38 (s, 3H), 1.01 (t, J=7.1, 3H); 13C NMR (75 MHz, CCl3D), δ 161.5, 153.3, 146.9, 138.1, 136.9, 133.1, 131.6, 129.2, 128.6, 128.2, 126.4, 126.2, 122.8, 121.1, 120.0, 115.3, 111.9, 109.1, 73.2, 69.1, 59.8, 46.8, 43.0, 42.5, 35.5, 27.0, 25.6, 13.7; ESI MS: m/z 633.4 (M + H)+.
4-(4-Chlorophenyl)-1-((S)-3,4-dihydroxybutyl)-3-(3-((2-((4-(4-(((R)-4-(dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitrophenylsulfonamido)phenyl)amino)ethyl)amino)phenyl)-N-(3-(4-methylpiperazin-1-yl)propyl)-1H-pyrrole-2-carboxamide (11)
Compound 11 was prepared from 37 in 40% yield over six steps using a procedure similar to that used for compound 12. 1H NMR (300 MHz, CD3OD), δ 8.35 (d, J=1.9, 1H), 7.65 (d, J=7.4, 1H), 7.33~6.94 (m, 14H), 6.78~6.61 (m, 5H), 4.45~4.31 (m, 2H), 4.15~4.12 (m, 1H), 3.70~3.39 (m, 8H), 3.28~3.17 (m, 13H), 2.90~2.88 (m, 9H), 2.69~2.64 (m, 2H), 2.30~2.05 (m, 3H), 1.85~1.82 (m, 1H), 1.70~1.68 (m, 2H); 13C NMR (75 MHz, CD3OD) 165.2, 147.9, 137.6, 136.2, 135.0, 134.5, 132.5, 132.3, 131.6, 130.9, 130.3, 130.1, 129.2, 128.02, 127.96, 127.7, 125.9, 125.1, 125.0, 123.4, 116.2, 70.2, 67.3, 55.9, 55.4, 53.0, 52.3, 50.7, 46.4, 43.7, 43.5, 39.6, 37.6, 36.4, 30.1, 25.8; ESI MS: m/z 1081.6 (M + H)+.
Ethyl 4-(4-chlorophenyl)-3-(3-iodophenyl)-1-methyl-1H-pyrrole-2-carboxylate (38)
K2CO3 (1.83 g, 13.3 mmol) and MeI (1.89 g, 13.3 mmol) were added to a solution of 24 (3.0 g, 6.6 mmol) in DMF (40 mL). The reaction mixture was stirred for 4 h under nitrogen at room temperature. Then the reaction was diluted with water (40 mL), and extracted with EtOAc (2 × 60 mL). The combined organics were washed with water (60 mL) and brine (60 mL) and then dried over Na2SO4. After evaporation of the solvent, the residue was purified by flash chromatography on silica gel to provide 38 (2.88 g, 93%). 1H NMR (300 MHz, CCl3D), δ 7.64 (t, J=1.6, 1H), 7.60 (dt, J=1.6, 7.8, 1H), 7.15~7.12 (m, 2H), 7.07~7.04 (m, 1H), 7.00~6.91 (m, 4H), 4.06 (q, J=7.1, 2H), 3.97 (s, 3H), 1.00 (t, J=7.1, 3H); 13C NMR (75 MHz, CCl3D), δ 161.6, 139.8, 138.2, 135.7, 132.8, 132.0, 129.42, 129.39, 129.0, 128.5, 127.0, 122.9, 121.0, 93.4, 60.1, 37.8, 14.0;
Ethyl 4-(4-chlorophenyl)-1-methyl-3-(3-(4-(4-nitrophenyl)piperazin-1-yl)phenyl)-1H-pyrrole-2-carboxylate (39)
Compound 39 was prepared in 83% yield by a procedure similar to that used for compound 35.1H NMR (300 MHz, CCl3D), δ 8.12 (d, J=9.1, 2H), 7.26~7.21 (m, 1H), 7.13 (d, J=8.3, 2H), 7.03 (d, J=8.3, 2H), 6.96 (s, 1H), 6.90~6.79 (m, 5H), 4.09 (q, J=7.1, 2H), 3.99 (s, 3H), 3.54~3.51 (m, 4H), 3.27~3.24 (m, 4H), 1.01 (t, J=7.1, 3H); 13C NMR (75 MHz, CCl3D), δ 161.7, 154.7, 150.1, 138.5, 136.7, 133.2, 131.5, 131.0, 129.2, 128.4, 128.2, 126.6, 125.9, 123.1, 122.7, 121.0, 119.0, 114.9, 112.7, 59.8, 49.0, 46.9, 37.6, 13.8; ESI MS: m/z 545.8 (M + H)+.
(R)-4-(4-Chlorophenyl)-3-(3-(4-(4-(4-((4-(dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitrophenylsulfonamido)phenyl)piperazin-1-yl)phenyl)-1-methyl-1H-pyrrole-2-carboxylic acid (20)
Compound 20 was prepared from 38 in 49% yield in four steps using a procedure similar to that used for 12. 1H NMR (300 MHz, CD3OD), δ 8.29 (d, J=2.2, 1H), 7.58 (dd, J=2.3, 9.2, 1H), 7.27 (t, J=7.8, 1H), 7.15~6.89 (m, 18H), 4.08~4.05 (m, 1H), 3.93 (s, 3H), 3.67~3.30 (m, 9H), 3.20~3.14 (m, 3H), 2.82 (s, 6H), 2.23~2.14 (m, 2H); 13C NMR (75 MHz, CD3OD), δ 164.3, 147.9, 138.9, 136.2, 134.8, 134.4, 132.8, 132.6, 132.2, 131.7, 131.6, 130.6, 130.1, 130.0, 129.2, 128.7, 128.3, 128.0, 127.9, 127.6, 124.1, 124.0, 122.4, 122.0, 119.1, 117.8, 116.2, 55.9, 52.8, 52.4, 50.6, 43.5, 39.5, 38.1, 30.1; ESI MS: m/z 895.1 (M + H)+.
(R)-4-(4-Chlorophenyl)-3-(3-(4-(4-(4-((4-(dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitrophenylsulfonamido)phenyl)piperazin-1-yl)phenyl)-1-methyl-N-(3-(4-methylpiperazin-1-yl)propyl)-1H-pyrrole-2-carboxamide (16)
A mixture of 20 (76 mg, 0.085mmol), 1-(3-aminopropyl)-4-methylpiperazine (20 mg, 0.127 mmol), EDCI (24.5 mg, 0.127 mmol), HOBt (16.4 mg, 0.127 mol) and DIEA (44 μL, 0.25 mmol) in DCM (5 mL) was stirred for 8 h and then concentrated. The residue was purified by HPLC to provide 16 (74 mg, 84%).1H NMR (300 MHz, CD3OD), δ 8.30 (d, J=2.2, 1H), 7.59 (dd, J=2.2, 9.1, 1H), 7.28 (t, J=7.9, 1H), 7.16~6.89 (m, 16H), 6.82~6.77 (m, 2H), 4.09~4.06 (m, 1H), 3.82 (s, 3H), 3.33~3.28 (m, 6H), 3.25~3.09 (m, 16H), 2.83 (s, 9H), 2.62~2.57 (m, 2H), 2.24~2.14 (m, 2H), 1.65~1.61 (m, 2H); 13C NMR (75 MHz, CD3OD), δ 165.1, 151.6, 147.9, 137.4, 136.2, 135.0, 134.4, 132.5, 132.2, 131.6, 130.7, 130.4, 130.1, 129.2, 128.0, 127.9, 127.6, 126.7, 126.4, 125.8, 124.2, 123.3, 120.1, 118.9, 116.9, 116.2, 55.9, 55.4, 52.9. 52.4, 51.1, 50.6, 50.5, 43.6, 43.5, 39.5, 37.5, 36.3, 30.1, 25.9; ESI MS: m/z 1033.7 (M + H)+.
(R)-4-(4-Chlorophenyl)-3-(3-(4-(4-(4-((4-(dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitrophenylsulfonamido)phenyl)piperazin-1-yl)phenyl)-N,1-dimethyl-1H-pyrrole-2-carboxamide (17)
A mixture of 20 (42 mg, 0.047 mmol), 2M methylamine in THF (47 μM, 0.094 mmol), EDCI (18.0 mg, 0.094mmol), HOBt (12.1mg, 0.094mol) and DIEA (25 μL, 0.14 mmol) in DCM (3 mL) was stirred for 8 h and then concentrated. The residue was purified by HPLC to provide 17 (36.6 mg, 86%).1H NMR (300 MHz, CD3OD), δ 8.32 (d, J=1.2, 1H), 7.58 (d, J=9.1, 1H), 7.28 (t, J=7.9, 1H), 7.17~6.99 (m, 15H), 6.90 (d, J=9.3, 1H), 6.86 (s, 1H), 6.80 (d, J=7.5, 1H), 4.09~4.07 (m, 1H), 3.80 (s, 3H), 3.45~3.33 (m, 9H), 3.21~3.08 (m, 3H), 2.84 (s, 6H), 3.05 (s, 3H), 2.25~2.10 (m, 2H) ; 13C NMR (75 MHz, CD3OD), δ 165.4, 149.9, 148.0, 147.2, 137.4, 136.2, 135.1, 134.4, 133.6, 132.6, 132.3, 131.6, 130.7, 130.6, 130.1, 129.2, 128.0, 127.9, 127.6, 126.9, 125.9, 125.8, 125.6, 124.0, 123.1, 120.8, 119.5, 117.4, 116.2, 55.9, 52.4, 51.4, 43.5, 39.6, 36.2, 30.1, 26.4; ESI MS: m/z 907.6 (M + H)+.
(R)-N-(4-(4-(3-(4-(4-Chlorophenyl)-1-methyl-1H-pyrrol-3-yl)phenyl)piperazin-1-yl)phenyl)-4-((4-(dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitrobenzenesulfonamide (18))
TFA (0.5 mL) was added to a solution of 20 (37mg, 0.041 mmol) in DCM (2 mL). The solution was stirred for 15 minutes and then evaporated. The residue was purified by HPLC to afford compound 18 (28 mg, 80%). 1H NMR (300 MHz, CD3OD), δ 8.30 (d, J=2.2, 1H), 7.59 (dd, J=2.2, 9.2, 1H), 7.25 (m, 1H), 7.18~6.89 (m, 17H), 6.80 (dd, J=2.3, 9.7, 2H), 4.10~4.06 (m, 1H), 3.67 (s, 3H), 3.35~3.30 (m, 9H), 3.20~3.14 (m, 3H), 2.82 (s, 6H), 2.23~2.14 (m, 2H); 13C NMR (75 MHz, CD3OD), δ 148.0, 147.9, 139.3, 136.3, 136.2, 134.4, 133.0, 132.34, 132.25, 131.6, 131.0, 130.8, 130.1, 129.3, 128.0, 127.9, 127.6, 126.0, 124.1, 123.6, 123.4, 123.2, 123.1, 123.0, 119.7, 119.2, 116.8, 116.2, 55.9, 53.0, 52.4, 50.5, 43.5, 39.5, 36.4, 30.1; ESI MS: m/z 851.6 (M + H)+.
(R)-Ethyl 4-(4-chlorophenyl)-3-(3-(4-(4-(4-((4-(dimethylamino)-1-(phenylthio)butan-2-yl)-amino)-3-nitrophenylsulfonamido)phenyl)piperazin-1-yl)phenyl)-1-methyl-1H-pyrrole-2-carboxylate (19)
A mixture of 20 (37 mg, 0.041 mmol), EtOH (1 mL), N,N'-diisopropylcarbodiimide (16 mg, 0.124 eq), 4-(dimethylamino)pyridine (1.0 mg, 0.008 mmol) in THF (3 mL) was stirred for 8 h then concentrated. The residue was purified by HPLC to provide 19 (29 mg, 76%). 1H NMR (300 MHz, CD3OD), δ 8.31 (d. J=2.1, 1H), 7.60 (dd, J=2.1, 9.1, 1H), 7.31 (t, J=7.8, 1H), 7.17~6.90 (m, 18H), 4.11~4.07 (m, 1H), 3.99 (q, J=7.1, 2H), 3.93 (s, 3H), 3.39~3.31 (m, 9H), 3.21~3.14 (m, 3H), 2.83 (s, 6H), 2.27~2.11 (m, 2H), 0.93 (t, J=7.1, 3H); 13C NMR (75 MHz, CD3OD), δ 162.9, 148.0, 139.1, 136.2, 134.6, 134.4, 133.3, 132.7, 132.3, 131.6, 130.5, 130.1, 130.0, 129.2, 128.6, 128.0, 127.9, 127.6, 124.0, 123.9, 122.0, 121.9, 119.4, 117.7, 116.2, 60.9, 55.9, 52.44, 52.37, 51.0, 43.5, 39.5, 37.8, 30.1, 14.2; ESI MS: m/z 922.8 (M + H)+.
Ethyl 5-bromo-4-(4-chlorophenyl)-3-(3-iodophenyl)-1-methyl-1H-pyrrole-2-carboxylate (40)
N-Bromosuccinimide (0.59 g, 3.3 mmol) was added in portions to a solution of 24 (1.5 g, 3.3 mmol) in DMF (20 mL) and the result mixture was stirred at room temperature for 2 h. The reaction mixture was diluted with H2O (30 mL) and extracted into EtOAc (2 × 30 mL). The combined organics were washed with water (30 mL) and brine (30 mL), and dried over Na2SO4. After removal of the solvent under vacuum, a solution of the residue in DMF (20 mL) was added to K2CO3 (0.92 g, 6.6 mmol) and MeI (0.94 g, 6.6 mmol). This reaction mixture was stirred for 4 h under nitrogen at room temperature. The reaction was diluted with water (20 mL), and extracted into EtOAc (2 × 30 mL). The combined organics were washed with water (30 mL) and then brine (30 mL) and dried over Na2SO4. After evaporation of the solvent, the residue was purified by flash chromatography on silica gel to provide 40 (1.3 g, 71% yield over two steps). 1H NMR (300 MHz, CCl3D), δ 7.61 (s, 1H), 7.56 (d, J=7.5, 1H), 7.21 (d, J=8.4, 2H), 7.05 (d, J=8.4, 2H), 6.99~6.89 (m, 2H), 4.09 (q, J=7.1, 2H), 4.05 (s, 3H), 1.01 (t, J=7.1, 3H); 13C NMR (75 MHz, CCl3D), δ 160.9, 139.7, 137.2, 135.7, 132.8, 131.7, 131.5, 130.0, 129.7, 129.1, 128.2, 123.6, 121.7, 111.7, 93.1, 60.3, 35.6, 13.8;
Ethyl 5-bromo-4-(4-chlorophenyl)-1-methyl-3-(3-(4-(4-nitrophenyl)piperazin-1-yl)phenyl)-1H-pyrrole-2-carboxylate (41)
Compound 41 was prepared in 74% yield by a procedure similar to that used for compound 35. 1H NMR (300 MHz, CCl3D), δ 8.15 (d, J=9.3, 2H), 7.21~7.14 (m, 3H), 7.07 (d, J=8.5, 2H), 6.87~6.81 (m, 3H), 6.72 (d, J=7.6, 1H), 6.63 (s, 1H), 4.09 (q, J=7.1, 2H), 4.04 (s, 3H), 3.53~3.49 (m, 4H), 3.20~3.17 (m, 4H), 0.99 (t, J=7.1, 3H); 13C NMR (75 MHz, CCl3D), δ 161.20, 154.7, 149.8, 138.6, 135.6, 132.5, 132.1, 131.9, 131.7, 128.2, 128.1, 125.9, 123.6, 123.1, 121.7, 119.1, 114.9, 112.7, 111.3, 60.1, 48.9, 46.9, 35.5, 13.8; ESI MS: m/z 623.3 (M + H)+.
Ethyl 4-(4-chlorophenyl)-1-methyl-3-(3-(4-(4-nitrophenyl)piperazin-1-yl)phenyl)-5-((trimethylsilyl)ethynyl)-1H-pyrrole-2-carboxylate (42)
CuI (37 mg, 0.19 mmol) and Pd(PPh3)4 (111 mg, 0.096 mmol) were added to a solution of 41 (600 mg, 0.96 mmol), ethynyltrimethylsilane (473 mg, 4.8 mmol) and Et3N (0.4 mL, 2.9 mmol) in DMF (20 mL) in a sealed tube at room temperature under nitrogen. After stirring at 80°C for 6 h, the reaction mixture was cooled and filtered through Celite, washed with 50 mL of CH2Cl2, and concentrated under reduced pressure. Purification of the residue by flash chromatography on silica gel afforded 42 (160 mg, 26%). 1H NMR (300 MHz, CCl3D), δ 8.15 (d, J=9.3, 2H), 7.23~7.14 (m, 5H), 6.87~6.84 (m, 3H), 6.74 (d, J=7.6, 1H), 6.69 (s, 1H), 4.09 (q, J=7.1, 2H), 4.06 (s, 3H), 3.54~3.51 (m, 4H), 3.24~3.21 (m, 4H), 0.99 (t, J=7.1, 3H), 0.26 (s, 9H); 13C NMR (75 MHz, CCl3D), δ 161.2, 154.7, 150.0, 138.7, 135.9, 132.1, 130.9, 130.4, 128.4, 127.8, 127.5, 125.9, 123.2, 121.7, 119.7, 119.1, 114.9, 112.7, 103.9, 95.3, 60.1, 49.0, 46.9, 34.9, 13.7, −0.26; ESI MS: m/z 641.9(M + H)+.
4-(4-chlorophenyl)-5-ethynyl-1-methyl-3-(3-(4-(4-nitrophenyl)piperazin-1-yl)phenyl)-1H-pyrrole-2-carboxylic acid (43)
KOH (56 mg, 1.0 mmol) was added to a solution of 42 (160 mg, 0.25 mmol) in a mixture of dioxane/EtOH/H2O (1:1:1, 10 ml) and the solution was refluxed for 2 h. After cooling, the reaction was neutralized with 1M HCl and extracted with EtOAc. The EtOAc solution was washed with brine, dried over Na2SO4 and concentrated in vacuum. Purification of the residue by flash chromatography on silica gel afforded 43 (116 mg, 86%). 1H NMR (300 MHz, CCl3D), δ 8.15 (d, J=9.1, 2H), 7.22~7.10 (m, 5H), 6.86~6.83 (m, 3H), 6.76~6.73 (m, 2H), 4.06 (s, 3H), 3.51~3.49 (m, 5H), 3.22~3.19 (m, 4H); 13C NMR (75 MHz, CCl3D), δ 154.7, 149.9, 138.7, 134.9, 132.5, 132.1, 131.6, 131.0, 128.6, 128.1, 126.0, 123.1, 120.1, 119.4, 115.2, 112.7, 86.1, 74.2, 48.8, 46.8, 35.5; ESI MS: m/z 541.8(M + H)+.
(R)-4-(4-Chlorophenyl)-3-(3-(4-(4-(4-((4-(dimethylamino)-1-(phenylthio)butan-2-yl)amino)-3-nitrophenylsulfonamido)phenyl)piperazin-1-yl)phenyl)-5-ethyl-1-methyl-1H-pyrrole-2-carboxylic acid (21)
10% Pd-C (15 mg) was added to a solution of compound 43 (82 mg, 0.15 mmol) in a mixture of CH2Cl2 (3 mL) and MeOH (3 mL). The mixture was stirred under 1 atm of H2 at room temperature for 0.5 h before being filtered through celite and concentrated. The resulting aniline was used in the next step without purification. To this, aniline in pyridine (5 mL), 4-fluoro-3-nitrobenzene-1-sulfonyl chloride (36 mg, 0.15 mmol) was added at 0°C. The mixture was stirred at 0°C for 30 minutes. The pyridine was removed under vacuum and the residue was purified by flash chromatography on silica gel to give 4-(4-chlorophenyl)-5-ethyl-3-(3-(4-(4-(4-fluoro-3-nitrophenylsulfonamido)phenyl)-piperazin-1-yl)phenyl)-1-methyl-1H-pyrrole-2-carboxylic acid. DIEA (53 μL, 0.30 mmol) was added to a solution of this sulfonamide and (R)-N1,N1-dimethyl-4-phenylthio)butane-1,3-diamine (34 mg, 0.15 mmol) in DMF. The reaction mixture was stirred overnight and concentrated. The residue was resolved in a mixture of H2O and CH3CN (a trace of HCl was added to assist with the solubility) and purified by HPLC with H2O and CH3CN (without TFA) as eluents afforded 21 (41 mg, 29 % in four steps). 1H NMR (300 MHz, CD3OD), δ 8.34 (d, J=1.9, 1H), 7.60 (dd, J=1.9, 9.1, 1H), 7.20~6.93 (m, 16H), 6.83~6.81 (m, 2H), 4.13~4.11 (m, 1H), 3.92 (s, 3H), 3.41~3.35 (m, 2H), 3.24~3.21 (m, 10H), 2.86 (s, 6H), 2.66 (q, J=7.5, 2H), 2.35~2.13 (m, 2H), 1.16 (t, J=7.5, 3H); 13C NMR (75 MHz, CD3OD), δ 164.8, 147.9, 140.3, 138.3, 136.2, 135.6, 134.5, 133.3, 133.1, 132.3, 131.6, 130.1, 129.2, 129.0, 128.0, 127.9, 127.7, 124.5, 123.0, 122.1, 120.3, 118.5, 116.8, 116.2, 56.0, 52.4, 52.1, 50.3, 43.5, 39.6, 33.6, 30.1, 18.9, 14.5; ESI MS: m/z 922.5 (M + H)+.
Fluorescence polarization based binding assays
Details of the expression and purification of Bcl-2, Bcl-xL and Mcl-1 proteins and determination of Kd values of fluorescent probes to proteins are provided in the SI. IC50 and Ki values to Bcl-2/Bcl-xL/Mcl-1 of our synthesized compounds and reference compounds were determined in competitive binding experiments, in which an inhibitor in serial dilutions was allowed to compete with a fixed concentration of a fluorescent probe for a fixed concentration of a protein. Mixtures of 5 μl of the tested compound in DMSO and 120 μl of pre-incubated protein/probe complex in the assay buffer were added to assay plates and incubated at room temperature for 2 h with gentle shaking. The final concentrations of the protein and probe were 1.5 nM and 1 nM for the Bcl-2 assay, 10 nM and 2 nM for the Bcl-xL assay, and 20 nM and 2 nM for the Mcl-1 assay, respectively. Controls containing protein/probe complex only (equivalent to 0% inhibition) or free probe only (equivalent to 100% inhibition), were included in each assay plate. FP values were measured as described above. IC50 values were determined by nonlinear regression fitting of the competition curves. The Ki value of a compound to a protein was calculated using the equation described previously26, based upon the measured IC50 value, the Kd value of the probe to the protein, and the concentration of the protein and probe in the competitive assays. Ki values were also calculated using an equation published in the literature.27 The values obtained from both equations were found to be in excellent agreement.
Molecular modeling
Crystal structures of Bcl-xL with 118 (PDB entry: 2YXJ) and 4 were used to model the binding poses of our designed compounds with Bcl-xL. In the binding models, the structure of Bcl-xL in complex with 4 was superimposed on that of Bcl-xL in complex with 1. The core scaffold of 4 then replaced the 4'-chloro-2-methyl-1,1'-biphenyl group of 1 to generate the initial binding model, which was then refined by a 1 ns molecular dynamics simulation. All the modifications of the ligands were performed using the Sybyl program.28
The charge and force field parameters of the compounds were obtained using the most recent Antechamber module in the Amber 10 program suite29, where the charge models were calculated from the Gaussian 98 program30 at the Hartree-Fock level using the 6-31G** basis sets. Protocols for the MD simulation are the following. The total charge of the system was neutralized by first adding counter ions. Then, the system was solvated in a 10 Å cubic box of water using the TIP3P water model31. 2000 steps of minimization of the system were performed where the protein and the modeled compound were constrained by a force constant of 50 kcal/mol/Å2. After minimization, a 20 ps simulation was used to gradually raise the temperature of the system to 298K while the whole system was constrained by a force constant of 10 kcal/mol/Å2. Another 40 ps of equilibrium run was used where only the backbone atoms of the protein and the ligand atoms were constrained by a force constant of 2 kcal/mol/Å2. A final production run of 1 ns was performed with no constraints on any atoms of the complex structure. When applying constraints, the initial complex structure was used as a reference structure. All the MD simulations were at NTP. The SHAKE algorithm32 was used to fix the bonds involving hydrogen. The PME method33 was used and the non-bonded cutoff distance was set at 10 Å. The time step was 2 fs, and the neighboring pairs list was updated in every 20 steps. Final conformations of Bcl-xL in complex with 6 and 7 are shown in Figure S1 in SI.
Cell growth assay
The effect of compounds on cell growth was evaluated by a WST-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt] assay (Dojindo Molecular Technologies, Gaithersburg, Maryland). Human small cell lung cancer cell lines H146 and H1417 were purchased from the American Type Culture Collection (ATCC) and were maintained in RPMI-1640 medium containing 10% FBS. Cells were seeded in 96-well flat bottom cell culture plates at a density of 1×104 cells/well with various concentrations of compounds and incubated for 4 days. At the end of incubation, WST-8 dye (20 μL) was added to each well and incubated for an additional 1–2 h, and then the absorbance was measured in a microplate reader (Molecular Devices) at 450 nm. The concentration of compounds that inhibited cell growth by 50% (IC50) was calculated by comparing absorbance in the untreated cells and the cells treated with the compounds using the GraphPad Prism software (GraphPad Software, La Jolla). At least three independent experiments were performed to obtain the standard deviation for each compound in each cell line.
Cell death assay
Cell death assays were performed using trypan blue staining. Cells were treated with the indicated compounds. At the end of treatment, cells were collected and stained with trypan blue. Cells that stained blue or the morphologically unhealthy cells were scored as dead cells. At least 100 cells were counted for each sample.
Western blotting
Cells were lysed using radioimmunoprecipitation assay lysis buffer (PBS containing 1% NP40, 0.5% Na-deoxycholate, and 0.1% SDS) supplemented with 1 μmol/L phenylmethylsulfonyl fluoride and 1 protease inhibitor cocktail tablet per 10 mL on ice for 20 min, and lysates were then cleared by centrifugation before protein concentration determination using the Bio-Rad protein assay kit according to the manufacturer's instructions. Proteins were electrophoresed onto 4–20% SDS-PAGE gels (Invitrogen) and transferred onto polyvinylidenedifluoride membranes. Following blocking in 5% milk, membranes were incubated with a specific primary antibody, washed, and incubated with horseradish peroxidase–linked secondary antibody (Amersham). The signals were visualized with the chemiluminescent horseradish peroxidase antibody detection reagent (Denville Scientific). Rabbit antibodies against PARP and caspase-3 were from Cell Signaling Technology and rabbit anti-GAPDH was from Santa Cruz Biotechnology.
Bcl-xL Crystallographic studies
Expression and Purification of Bcl-xLΔTM ΔLP is described in the SI. Prior to crystallization, Bcl-xL was incubated with a 5-fold molar excess of compound 4 in the presence of 4% DMSO for 1 hr at 4 °C and then concentrated to 7 mg/mL. Crystals of Bcl-xL:4 were grown by vapor diffusion in a sitting drop tray with 1.2M sodium citrate and 25 mM CHES pH 9.0 as the mother liquor. Crystals did not appear until after the well was opened, perturbed with a loop and resealed. In the experiment, the crystallization drops contained equal volumes of protein and well solution. Prior to data collection, crystals were cryoprotected in well solution with increasing amounts of glycerol to a final concentration of 20%, then flash frozen in liquid nitrogen.
X-ray data was collected at LS-CAT ID-21-F and G lines at the Advanced Photon Source at Argonne National Lab. Data were processed with HKL2000.34 The Bcl-xL:4 complex crystallized in the P42212 space group and diffracted to 1.7 Å resolution. The structure contained one molecule in the asymmetric unit. The structure of the complex was solved by molecular replacement with Phaser35 using a structure of Bcl-xL previously solved in our laboratory as a starting model. Iterative rounds of refinement and model building were completed using Buster36 and Coot37 respectively. The initial Fo-Fc electron density map revealed the presence of the compounds in the binding site of Bcl-xL. Only the three-ring core of 4 was visible in the Fo-Fc electron density map contoured at 3 sigma. The PRODRG server38 was used to create the starting coordinates and restraint files for the compounds. The current Rfree/Rwork for the Bcl-xL:4 structure is 0.2036/0.1854. All amino acids fall into the allowed regions of the Ramanchandran plot with 98% in the preferred regions. Data collection and refinement statistics are given in Table S1 in SI.
Supplementary Material
ACKNOWLEDGMENT
This research was supported in part by a grant from the National Cancer Institute, National Institutes of Health (U19CA113317). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817). Coordinates for Bcl-xL complexed with 4 were deposited into the Protein Data Bank with accession numbers 3SPF.
Footnotes
Supporting Information (SI) Available: Experimental procedures and spectral data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Ziegler DS, Kung AL. Therapeutic targeting of apoptosis pathways in cancer. Curr Opin Oncol. 2008;20(1):97–103. doi: 10.1097/CCO.0b013e3282f310f6. [DOI] [PubMed] [Google Scholar]
- 3.Reed JC. Apoptosis-based therapies. Nat Rev Drug Discov. 2002;1(2):111–121. doi: 10.1038/nrd726. [DOI] [PubMed] [Google Scholar]
- 4.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2(9):647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 5.Cory S, Adams JM. Killing cancer cells by flipping the Bcl-2/Bax switch. Cancer Cell. 2005;8(1):5–6. doi: 10.1016/j.ccr.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Labi V, Erlacher M, Kiessling S, Villunger A. BH3-only proteins in cell death initiation, malignant disease and anticancer therapy. Cell Death Differ. 2006;13(8):1325–1238. doi: 10.1038/sj.cdd.4401940. [DOI] [PubMed] [Google Scholar]
- 7.Lessene G, Czabotar PE, Colman PM. BCL-2 family antagonists for cancer therapy. Nat Rev Drug Discov. 2008;7(12):989–1000. doi: 10.1038/nrd2658. [DOI] [PubMed] [Google Scholar]
- 8.Di Micco S, Vitale R, Pellecchia M, Rega MF, Riva R, Basso A, Bifulco G. Identification of lead compounds as antagonists of protein Bcl-xL with a diversity-oriented multidisciplinary approach. J Med Chem. 2009;52(23):7856–7867. doi: 10.1021/jm9010687. [DOI] [PubMed] [Google Scholar]
- 9.Feng Y, Ding X, Chen T, Chen L, Liu F, Jia X, Luo X, Shen X, Chen K, Jiang H, Wang H, Liu H, Liu D. Design, synthesis, and interaction study of quinazoline-2(1H)-thione derivatives as novel potential Bcl-xL inhibitors. J Med Chem. 2010;53(9):3465–3479. doi: 10.1021/jm901004c. [DOI] [PubMed] [Google Scholar]
- 10.Wei J, Stebbins JL, Kitada S, Dash R, Placzek W, Rega MF, Wu B, Cellitti J, Zhai D, Yang L, Dahl R, Fisher PB, Reed JC, Pellecchia M. BI-97C1, an optically pure Apogossypol derivative as pan-active inhibitor of antiapoptotic B-cell lymphoma/leukemia-2 (Bcl-2) family proteins. J Med Chem. 2010;53(10):4166–4176. doi: 10.1021/jm1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei J, Kitada S, Stebbins JL, Placzek W, Zhai D, Wu B, Rega MF, Zhang Z, Cellitti J, Yang L, Dahl R, Reed JC, Pellecchia M. Synthesis and biological evaluation of Apogossypolone derivatives as pan-active inhibitors of antiapoptotic B-cell lymphoma/leukemia-2 (Bcl-2) family proteins. J Med Chem. 2010;53(22):8000–8011. doi: 10.1021/jm100746q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rega MF, Wu B, Wei J, Zhang Z, Cellitti JF, Pellecchia M. SAR by interligand nuclear overhauser effects (ILOEs) based discovery of acylsulfonamide compounds active against Bclx(L) and Mcl-1. J Med Chem. 2011;54(17):6000–6013. doi: 10.1021/jm200826s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM, O'Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435(7042):677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 14.Park CM, Bruncko M, Adickes J, Bauch J, Ding H, Kunzer A, Marsh KC, Nimmer P, Shoemaker AR, Song X, Tahir SK, Tse C, Wang X, Wendt MD, Yang X, Zhang H, Fesik SW, Rosenberg SH, Elmore SW. Discovery of an orally bioavailable small molecule inhibitor of prosurvival B-cell lymphoma 2 proteins. J Med Chem. 2008;51(21):6902–6915. doi: 10.1021/jm800669s. [DOI] [PubMed] [Google Scholar]
- 15.Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, Johnson EF, Marsh KC, Mitten MJ, Nimmer P, Roberts L, Tahir SK, Xiao Y, Yang X, Zhang H, Fesik S, Rosenberg SH, Elmore SW. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68(9):3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 16.Sleebs BE, Czabotar PE, Fairbrother WJ, Fairlie WD, Flygare JA, Huang DC, Kersten WJ, Koehler MF, Lessene G, Lowes K, Parisot JP, Smith BJ, Smith ML, Souers AJ, Street IP, Yang H, Baell JB. Quinazoline sulfonamides as dual binders of the proteins B-cell lymphoma 2 and B-cell lymphoma extra long with potent proapoptotic cell-based activity. J Med Chem. 2011;54(6):1914–1926. doi: 10.1021/jm101596e. [DOI] [PubMed] [Google Scholar]
- 17.Muchmore SW, Sattler M, Liang H, Meadows RP, Harlan JE, Yoon HS, Nettesheim D, Chang BS, Thompson CB, Wong SL, Ng SL, Fesik SW. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature. 1996;381(6580):335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 18.Lee EF, Czabotar PE, Smith BJ, Deshayes K, Zobel K, Colman PM, Fairlie WD. Crystal structure of ABT-737 complexed with Bcl-xL: implications for selectivity of antagonists of the Bcl-2 family. Cell Death Differ. 2007;14(9):1711–1713. doi: 10.1038/sj.cdd.4402178. [DOI] [PubMed] [Google Scholar]
- 19.Bruncko M, Oost TK, Belli BA, Ding H, Joseph MK, Kunzer A, Martineau D, McClellan WJ, Mitten M, Ng SC, Nimmer PM, Oltersdorf T, Park CM, Petros AM, Shoemaker AR, Song X, Wang X, Wendt MD, Zhang H, Fesik SW, Rosenberg SH, Elmore SW. Studies leading to potent, dual inhibitors of Bcl-2 and Bcl-xL. J Med Chem. 2007;50(4):641–662. doi: 10.1021/jm061152t. [DOI] [PubMed] [Google Scholar]
- 20.Bullington JL, Wolff RR, Jackson PF. Regioselective preparation of 2-substituted 3,4-diaryl pyrroles: a concise total synthesis of ningalin B. J Org Chem. 2002;67(26):9439–9442. doi: 10.1021/jo026445i. [DOI] [PubMed] [Google Scholar]
- 21.Hartwig JF, Kawatsura M, Hauck SI, Shaughnessy KH, Alcazar-Roman LM. Room-Temperature Palladium-Catalyzed Amination of Aryl Bromides and Chlorides and Extended Scope of Aromatic C-N Bond Formation with a Commercial Ligand. J Org Chem. 1999;64(15):5575–5580. doi: 10.1021/jo990408i. [DOI] [PubMed] [Google Scholar]
- 22.Chinchilla R, Najera C. The Sonogashira reaction: a booming methodology in synthetic organic chemistry. Chem Rev. 2007;107(3):874–922. doi: 10.1021/cr050992x. [DOI] [PubMed] [Google Scholar]
- 23.Zhu W, Ma D. Synthesis of aryl azides and vinyl azides via proline-promoted CuI-catalyzed coupling reactions. Chem Commun (Camb) 2004;(7):888–889. doi: 10.1039/b400878b. [DOI] [PubMed] [Google Scholar]
- 24.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41(14):2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Cai Q, Ma D. Amino acid promoted CuI-catalyzed C-N bond formation between aryl halides and amines or N-containing heterocycles. J Org Chem. 2005;70(13):5164–5173. doi: 10.1021/jo0504464. [DOI] [PubMed] [Google Scholar]
- 26.Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, Wang S. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004;332(2):261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 27.Huang X. Fluorescence polarization competition assay: the range of resolvable inhibitor potency is limited by the affinity of the fluorescent ligand. J Biomol Screen. 2003;8(1):34–38. doi: 10.1177/1087057102239666. [DOI] [PubMed] [Google Scholar]
- 28.Sybyl; a molecular modeling system; is supplied by Tripos, I. St. Louis, MO; 63144. [Google Scholar]
- 29.Case DA, Darden TA, T.E. Cheatham I, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA. AMBER 10. University of California; San Francisco: 2008. AMBER10. [Google Scholar]
- 30.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Jr., Montgomery JA, Stratmann RE, Burant JC, Dapprich S, Millam JM, Daniels AD, Kudin KN, Strain MC, Farkas O, Tomasi J, Barone V, Cossi M, Cammi R, Mennucci B, Pomelli C, Adamo C, Clifford S, Ochterski J, Petersson GA, Ayala PY, Cui Q, Morokuma K, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Cioslowski J, Ortiz JV, Baboul AG, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Gomperts R, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PM, Johnson B, Chen W, Wong MM, Andres JL, Gonzalez C, Head-Gordon M, Replogle ES, Pople JA. Gaussian 98, Revision A-11. Gaussian, Inc.; Pittsburgh PA: 2001. [Google Scholar]
- 31.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics. 1983;79(2):926–935. [Google Scholar]
- 32.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Journal of Computational Physics. 1977;23(3):327–341. [Google Scholar]
- 33.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J Chem Phys. 1993;98(12):10089–10092. [Google Scholar]
- 34.Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. In: Carter Charles W., Jr., editor. Methods in Enzymology. Volume 276. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 35.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart O, Vonrhein C, Womack T. BUSTER. version 2.9 Global Phasing Ltd; Cambridge, United Kingdom: 2010. [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.