Like all living organisms, plants must respond to many external stimuli. Mitogen-activated protein kinases (MAPKs) mediate signal transduction of stress, cell cycle, and growth control in all eukaryotes. MAPKs perform their function as part of protein kinase modules, which in addition to other components are composed of MAPKs, MAPKKs (MAPK kinases), and MAPKKKs (MAPKK kinases) (for reviews, see refs. 1 and 2). MAPKs are serine/threonine protein kinases with a two-lobed structure. The active site is found at the domain interface and contains the MAPK-specific TXY (threonine-X-tyrosine) motif that is targeted by MAPKKs, dual-specificity protein kinases that activate MAPKs by phosphorylation of both the threonine and tyrosine residue of the TXY motif. MAPKKs are activated themselves by phosphorylation of two conserved serine or threonine residues (S/TXXXS/T) by MAPKKKs. According to their divergent structures MAPKKKs can be classified into different subfamilies. MAPKKKs contain different regulatory motifs, including pleckstrin homology domains, proline-rich sequences involved in Src homology 3 binding, zinc finger motifs, leucine zippers, and binding sites for G proteins. MAPKKKs can be activated by a wide range of stimuli and by different mechanisms, such as phosphorylation and interaction with G proteins or receptors. Whereas MAPKKKs can feed into multiple MAPK pathways and also can target other protein kinase cascades (Fig. 1), MAPKKs usually have a restricted substrate specificity, functioning mainly in a single cascade. Different kinases are assembled into distinct modules by scaffold proteins. Scaffold proteins are important for preventing cross-talk between different cascades and allow a given kinase to function in more than one module without affecting the specificity of the response. Although scaffold proteins have so far only been characterized from yeast and mammals (3), several types also exist in plants (unpublished results). Upon activation of a MAPK pathway, MAPKs often induce expression of specific sets of genes. This expression mostly occurs through translocation of the active MAPK to the nucleus where the MAPK phosphorylates and thereby regulates the activity of specific transcription factors. However, MAPKs also can be targeted to other cellular locations where they phosphorylate structural regulators such as cytoskeleton-associated proteins or regulatory enzymes, including other protein kinases, phosphatases, and phospholipases.
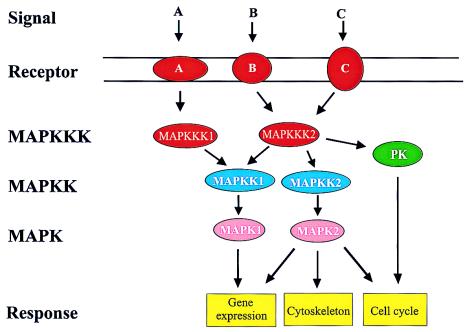
Figure 1.
Schematic illustration of the basic functioning of MAPK pathways. Receptors sense specific extracellular stimuli (denoted by A, B, and C) and activate MAPKKKs. This step usually involves other factors, such as G proteins or protein kinases. Activation of MAPKKK results in phosphorylation and activation of MAPKK. Some MAPKKKs are also capable of activating unrelated protein kinases (PK) of other pathways. MAPKKs activate MAPKs. MAPKs target different substrates, including transcription factors, cytoskeletal proteins, or other kinases and phosphatases, which are then directly or indirectly responsible for the responses. Arrows indicate activation of targets.
Whereas yeast has six MAPKs, 13 are known in mammals and more than 20 in plants (2). Various MAPKKs and MAPKKKs also have been isolated in plants, but so far no MAPK module has been conclusively established in plants. A report in this issue of PNAS by Kovtun et al. (4) demonstrates for the first time the functional existence of a MAPK module in plants. The authors show that ANP1 and NPK1, orthologous MAPKKKs from Arabidopsis and tobacco, respectively, mediate H2O2 signal transduction by activating the two Arabidopsis stress MAPKs, AtMPK3, and AtMPK6. H2O2 is a well-known product of oxidative stress playing multiple roles in plant physiology. H2O2 belongs to the class of reactive oxygen species (ROS) that are produced in photosynthetic tissues, mitochondria, and also in the cytosol under certain stress conditions, such as cold, drought, and salt stress, or pathogen attack (5, 6). ROS include singlet oxygen, superoxide, H2O2, and hydroxyl radicals and can damage proteins, membrane lipids, and other cellular components. Upon pathogen attack, ROS can help to induce cell death in infected cells or serve as a signal in activating defense responses in distant uninfected cells. The balance between the detrimental and beneficial role of ROS is determined by the local concentration of ROS and the state of the cellular detoxification system, which includes the levels of various antioxidants such as anthocyanins, glutathione, and carotenoids, and a panel of enzymes, including superoxide dismutases, ascorbate peroxidases, and catalases (6).
By now, it is well established that not only ROS but also stress MAPKs are induced upon fungal, bacterial, and viral attack (7). Because MAPK activation precedes ROS production in most systems, it is possible that the MAPK is involved in the onset of ROS production. Recent evidence indicates, however, that ROS production is induced by an independent mechanism of the activation of the stress MAPKs (8). The work of Kovtun et al. (4) shows that H2O2 triggers the expression of several stress-induced reporter genes, and constitutively active ANP1 can mimic the response in the absence of oxidative stress. These data put the stress MAPKs into the context of mediating the oxidative stress response itself. Interestingly, although H2O2, cold, and abscisic acid seem to activate the stress MAPK AtMPK3, coexpression of ANP1 only enhances the effect of H2O2. These results suggest that the stress MAPK pathway is activated by multiple MAPKKKs and that ANP1 mainly is involved in oxidative stress signaling (Fig. 2A). ANP1 also has a negative effect on auxin-induced gene expression, suggesting that the MAPK pathway acts positively on specific stress-responsive genes, but negatively on auxin signaling. At present, it is not clear whether these effects are directly mediated by the stress MAPKs AtMPK3 and AtMPK6. Because MAPKKKs can stimulate multiple MAPK cascades and also unrelated pathways (1, 2), it is possible that other factors also may be involved in the regulation of the stress- and auxin-responsive genes (Fig. 2A).
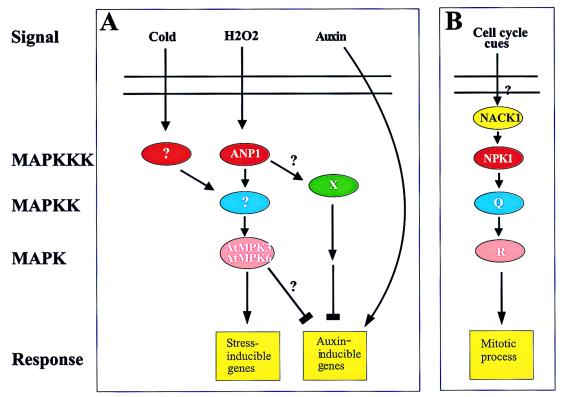
Figure 2.
Schematic illustration of the possible functions of the Arabidopsis ANP1 and tobacco NPK1 MAPK pathways. (A) Cold and H2O2 activate the AtMPK3 and AtMPK6 pathway through either an unknown MAPKKK or ANP1, respectively (4). ANP1 activation results in induction of stress-inducible genes and inhibition of auxin-inducible genes. Auxin stimulates auxin-inducible gene expression by an independent pathway. (B) Mitotic regulation of the cell cycle is mediated by the NPK1 pathway, which also consists of the MAPKK Q and MAPK R (13). NACK1, a kinesin-like protein, is an upstream activator of NPK1 (13).
Several lines of evidence suggest that ANP1 and NPK1 may play a role in the cell cycle. ANP1 and NPK1 transcripts are most abundant in proliferating tissues (9, 10), and NPK1 shows a cell cycle M phase-dependent transcript and protein pattern in tobacco cells (11). Moreover, in a functional interaction screen in yeast, the NACK1 gene was isolated and encodes a kinesin-like protein (11). Kinesins are microtubule-associated motor proteins and have been shown to be associated with MAPKs at kinetochores in mammalian cells (12). NACK1 cannot only activate NPK1 in yeast cells, but also shows a similar expression pattern as NPK1 in tobacco cells. Because NPK1 and NACK1 localize to the central region of the mitotic spindle and the phragmoplast, it was suggested that NPK1 and NACK1 might be involved in mitotic regulation, such as phragmoplast or cell plate formation (11) (Fig. 2B). Although the isolation of the downstream MAPKK and MAPK was reported, which were called Q and R, respectively (13), it is unclear whether R is related to any of the stress Arabidopsis MAPKs or constitutes another pathway.
The available data indicate that ANP1 and NPK1 play roles in H2O2, auxin, and cell cycle signaling. How is it possible that a specific MAPKKK can perform such diverse functions? One possible answer comes from the finding that a given MAPKKK can function in different cascades. This was clearly shown to be the case for STE11, a yeast MAPKKK that is part of both the FUS3 mating response and the HOG1 high osmolarity glycerol pathway (14). Because MAPKKKs cannot only activate multiple MAPKKs but also other unrelated protein kinases (15), part of the response of ANP1 might be executed by pathways distinct from MAPK cascades (Fig. 2A). In this context, it also should be noted that deletion of the regulatory domain of MAPKKKs not only leads to constitutive activation but also to a loss of signaling specificity of the kinases (11, 16). These considerations make it clear that more research is required to clarify the functioning of ANP1 and NPK1. Whatever the exact mechanism of the action of ANP1 and NPK1 is, expression of constitutively active NPK1 in tobacco resulted in enhanced tolerance to abiotic stresses but apparently had no detrimental effect on growth or development (4). Although an improvement of stress tolerance has been achieved in several cases by overexpression of either stress target genes or transcription factors, most of the time negative side effects were observed on crop growth or yield (17, 18). Therefore, the work of Kovtun et al. (4) has great economic potential to become a powerful new strategy in the production of stress-resistant crops, especially because the approach is not restricted to NPK1, but can be applied to any signaling pathway for which a kinase can be genetically engineered to become constitutively active.
Acknowledgments
This work was supported by the Austrian Science Foundation (Grants P13535-GEN and P12188-GEN) and the European Union Training and Mobility of Researchers program.
Footnotes
See companion article on page 2940.
References
- 1.Widmann C, Gibson S, Jarpe M B, Johnson G L. Phys Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 2.Ligterink, W. & Hirt, H. (2000) Int. Rev. Cytol., in press. [DOI] [PubMed]
- 3.Whitmarsh A J, Davis R J. Trends Biochem Sci. 1998;23:481–485. doi: 10.1016/s0968-0004(98)01309-7. [DOI] [PubMed] [Google Scholar]
- 4.Kovtun Y, Chiu W-L, Tena G, Sheen J. Proc Natl Acad Sci USA. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamb C, Dixon R A. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 6.Noctor G, Foyer C H. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S, Klessig D F. In: MAP Kinases in Plant Signal Transduction. Hirt H, editor. Heidelberg: Springer; 2000. pp. 65–84. [Google Scholar]
- 8.Romeis T, Piedras P, Zhang S, Klessig D F, Hirt H, Jones J D G. Plant Cell. 1999;11:273–287. doi: 10.1105/tpc.11.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishihama R, Banno H, Kawahara E, Irie K, Machida Y. Plant J. 1997;12:39–48. doi: 10.1046/j.1365-313x.1997.12010039.x. [DOI] [PubMed] [Google Scholar]
- 10.Nakashima M, Hirano K, Nakashima S, Banno H, Nishihama R, Machida Y. Plant Cell Physiol. 1998;39:690–700. doi: 10.1093/oxfordjournals.pcp.a029423. [DOI] [PubMed] [Google Scholar]
- 11.Machida Y, Nakashima M, Morikiyo K, Banno H, Ishikawa M, Soyano T, Nishihama R. J Plant Res. 1998;111:243–246. [Google Scholar]
- 12.Zecevic M, Catling A D, Eblen S T, Renzi L, Hittle J C, Yen T J, Gorbsky G J, Weber M J. J Cell Biol. 1998;142:1547–1558. doi: 10.1083/jcb.142.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nisihama R, Machida Y. In: MAP Kinases in Plant Signal Transduction. Hirt H, editor. Heidelberg: Springer; 2000. pp. 119–130. [Google Scholar]
- 14.Gustin M C, Albertyn J, Alexander M, Davenport K. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee F S, Hagler J, Chen Z J, Maniatis T. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- 16.Ichimura K, Mizoguchi T, Irie K, Morris P, Giraudat J, Matsumoto K, Shinozaki K. Biochem Biophys Res Commun. 1998;253:532–543. doi: 10.1006/bbrc.1998.9796. [DOI] [PubMed] [Google Scholar]
- 17.Bohnert H J, Sheveleva E. Curr Opin Plant Biol. 1998;1:267–274. doi: 10.1016/s1369-5266(98)80115-5. [DOI] [PubMed] [Google Scholar]
- 18.Holmberg N, Bulow L. Trends Plant Sci. 1998;3:61–66. [Google Scholar]