Abstract
The hypothesis that codon usage regulates gene expression at the level of translation is tested. Codon usage of Escherichia coli and phage lambda is compared by correspondence analysis, and the basis of this hypothesis is examined by connecting codon and tRNA distributions to polypeptide elongation kinetics. Both approaches indicate that if codon usage was random tRNA limitation would only affect the rarest tRNA species. General discrimination against their cognate codons indicates that polypeptide elongation rates are maintained constant. Thus, differences in expression of E. coli genes are not a consequence of their variable codon usage. The preference of codons recognized by the most abundant tRNAs in E. coli genes encoding abundant proteins is explained by a constraint on the cost of proof-reading.
Full text
PDF


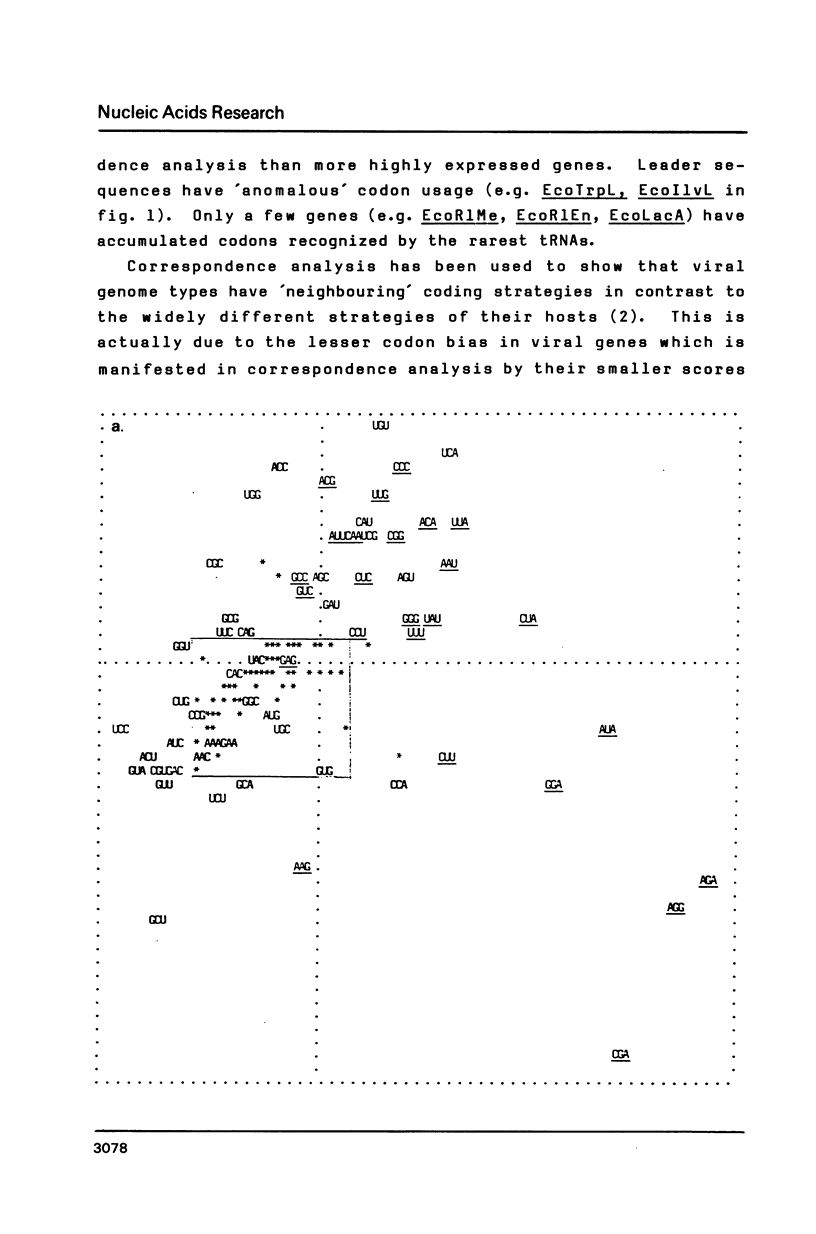
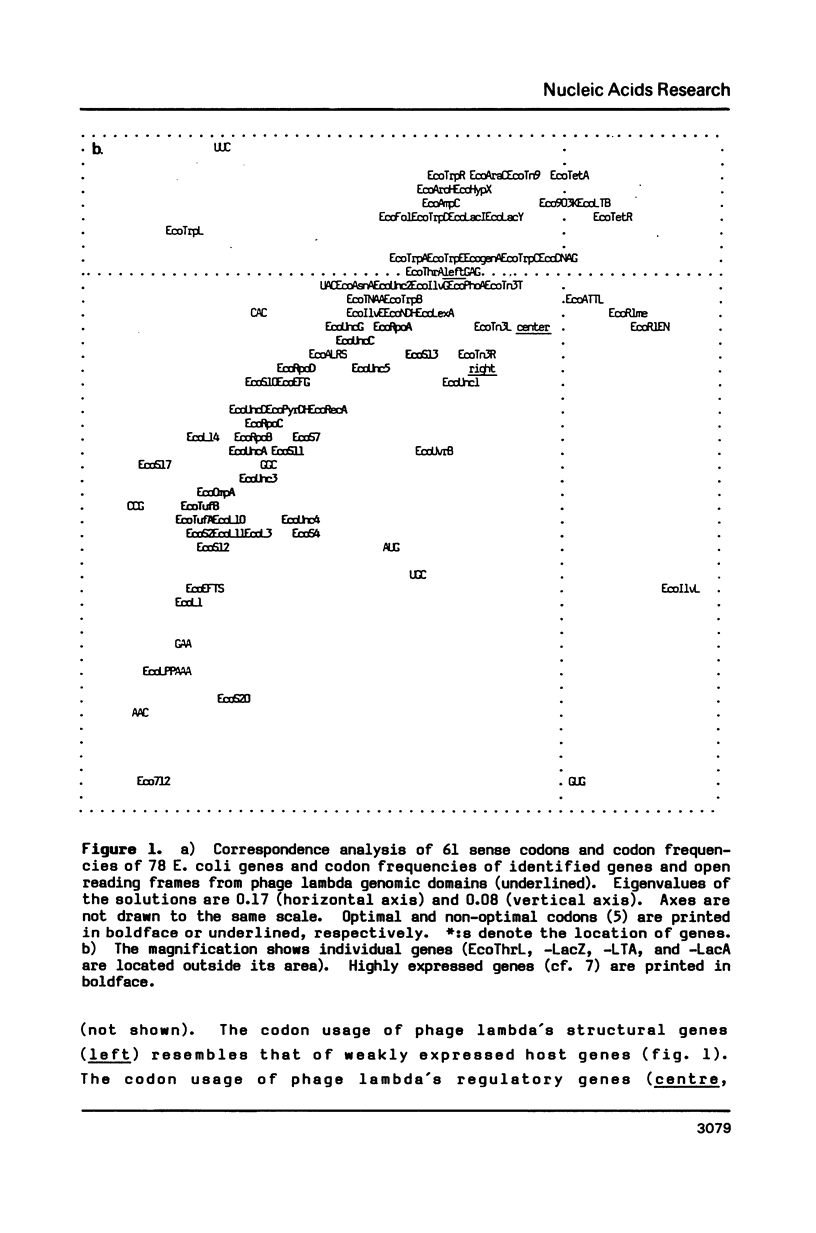
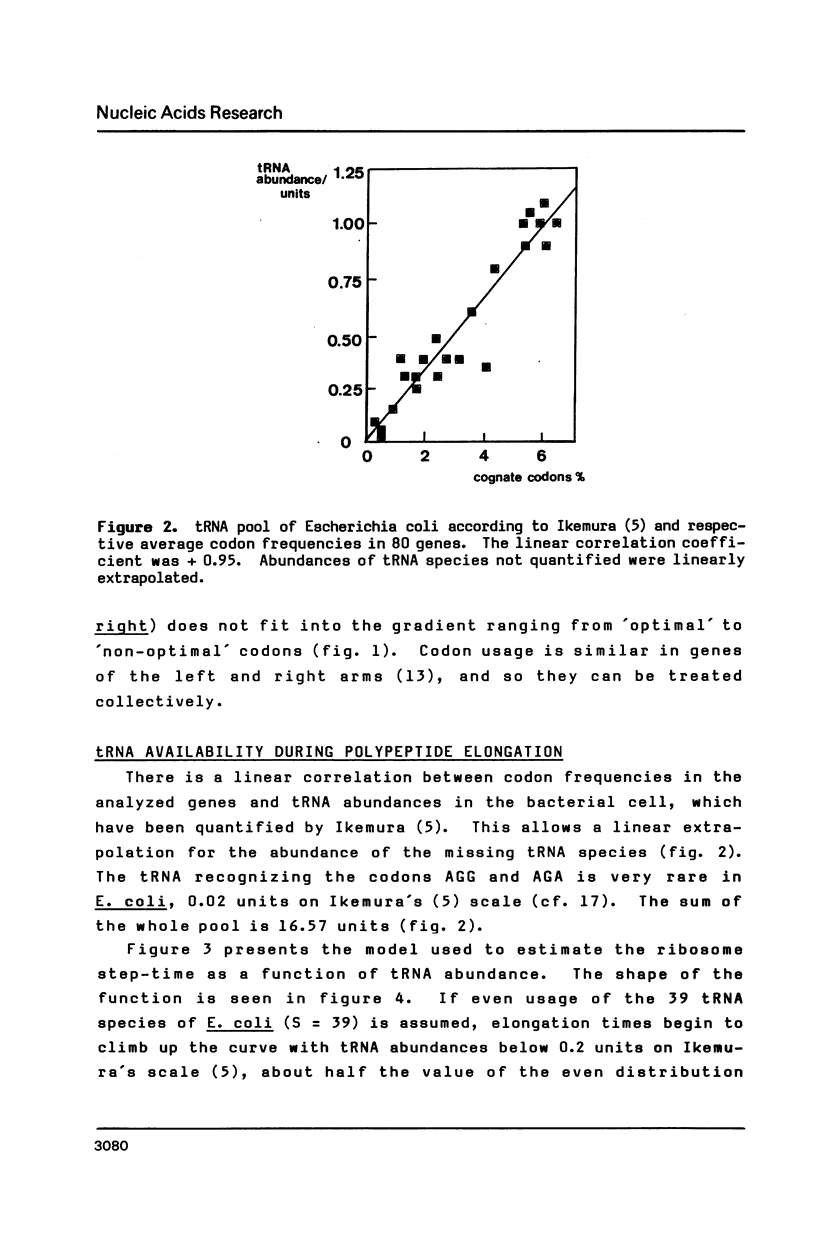

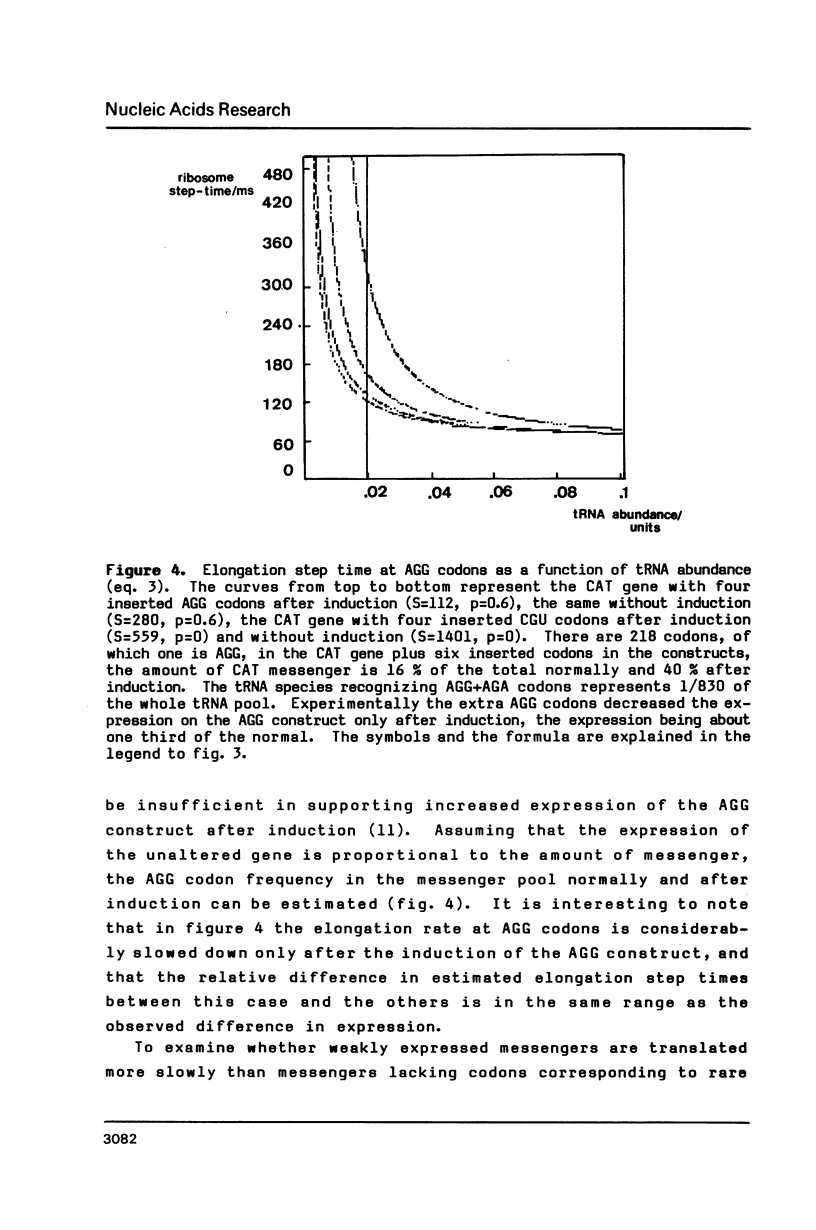





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersson S. G., Buckingham R. H., Kurland C. G. Does codon composition influence ribosome function? EMBO J. 1984 Jan;3(1):91–94. doi: 10.1002/j.1460-2075.1984.tb01766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Beatty J. T., Adams C. W., von Gabain A., Cohen S. N. Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell. 1985 Jan;40(1):171–181. doi: 10.1016/0092-8674(85)90320-4. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Blake R. D., Hinds P. W. Analysis of the codon bias in E. coli sequences. J Biomol Struct Dyn. 1984 Dec;2(3):593–606. doi: 10.1080/07391102.1984.10507593. [DOI] [PubMed] [Google Scholar]
- Bonekamp F., Andersen H. D., Christensen T., Jensen K. F. Codon-defined ribosomal pausing in Escherichia coli detected by using the pyrE attenuator to probe the coupling between transcription and translation. Nucleic Acids Res. 1985 Jun 11;13(11):4113–4123. doi: 10.1093/nar/13.11.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzone A. J. Stringent control and protein synthesis in bacteria. Biochimie. 1980;62(10):647–664. doi: 10.1016/s0300-9084(80)80022-8. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Grantham R. Polypeptide elongation and tRNA cycling in Escherichia coli: a dynamic approach. FEBS Lett. 1980 Jun 30;115(2):151–155. doi: 10.1016/0014-5793(80)81155-0. [DOI] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M. Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res. 1980 May 10;8(9):1893–1912. doi: 10.1093/nar/8.9.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Jacobzone M., Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981 Jan 10;9(1):r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H. J., de Henau S., Crothers D. M. On the physical basis for ambiguity in genetic coding interactions. Proc Natl Acad Sci U S A. 1978 Feb;75(2):610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982 Jul 15;158(4):573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Konigsberg W., Godson G. N. Evidence for use of rare codons in the dnaG gene and other regulatory genes of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Feb;80(3):687–691. doi: 10.1073/pnas.80.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland C. G., Ehrenberg M. Optimization of translation accuracy. Prog Nucleic Acid Res Mol Biol. 1984;31:191–219. doi: 10.1016/s0079-6603(08)60378-5. [DOI] [PubMed] [Google Scholar]
- McCarthy J. E., Schairer H. U., Sebald W. Translational initiation frequency of atp genes from Escherichia coli: identification of an intercistronic sequence that enhances translation. EMBO J. 1985 Feb;4(2):519–526. doi: 10.1002/j.1460-2075.1985.tb03659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan A. D., Staden R., Boswell D. R. A method for measuring the non-random bias of a codon usage table. Nucleic Acids Res. 1984 Dec 21;12(24):9567–9575. doi: 10.1093/nar/12.24.9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J., Pollard J. W., Friesen J. D., Stanners C. P. Stuttering: high-level mistranslation in animal and bacterial cells. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1091–1095. doi: 10.1073/pnas.75.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984 Dec 1;3(12):2895–2898. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postle K., Nguyen T. T., Bertrand K. P. Nucleotide sequence of the repressor gene of the TN10 tetracycline resistance determinant. Nucleic Acids Res. 1984 Jun 25;12(12):4849–4863. doi: 10.1093/nar/12.12.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M., Lilley R., Little S., Emtage J. S., Yarranton G., Stephens P., Millican A., Eaton M., Humphreys G. Codon usage can affect efficiency of translation of genes in Escherichia coli. Nucleic Acids Res. 1984 Sep 11;12(17):6663–6671. doi: 10.1093/nar/12.17.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982 Dec 25;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the pyruvate dehydrogenase component. Eur J Biochem. 1983 Jun 1;133(1):155–162. doi: 10.1111/j.1432-1033.1983.tb07441.x. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Gay N. J. The unc operon. Nucleotide sequence, regulation and structure of ATP-synthase. Biochim Biophys Acta. 1984 Sep 6;768(2):164–200. doi: 10.1016/0304-4173(84)90003-x. [DOI] [PubMed] [Google Scholar]
- Yudelevich A. Specific cleavage of an Escherichia coli leucine transfer RNA following bacteriophage T4 infection. J Mol Biol. 1971 Aug 28;60(1):21–29. doi: 10.1016/0022-2836(71)90444-x. [DOI] [PubMed] [Google Scholar]



