Abstract
There is little understanding of the impact of tumor-associated neutrophils (TAN) on adaptive immunity to tumors. In this study, we report the results of an investigation of the pathobiological basis for the prognostic significance of neutrophil elastase (NE), a serine protease found in neutrophil granules, in a model of cyclin E-overexpressing breast cancer. We established that NE was expressed by TAN within breast cancer tissues but not by breast cancer cells. NE modulated killing of breast cancer cells by cytotoxic T lymphocytes (CTL) specific for cyclin E-derived HLA-A2 restricted peptide (ILLDWLMEV). Breast cancer cells exhibited striking antigen-specific uptake of NE from the microenvironment that was independent of NE enzymatic activity. Further, NE uptake increased expression of low molecular weight forms of cyclin E and enhanced susceptibility to peptide-specific CTL lysis, suggesting that cyclin E peptides are naturally presented on breast cancer cells. Taken together, our findings reveal a previously unknown mechanism of antitumor adaptive immunity that links cancer cell uptake of an inflammatory mediator to an effective cytolytic response against an important breast cancer antigen.
Keywords: neutrophil elastase, cyclin E, breast cancer, innate immunity, adaptive immunity
Introduction
Neutrophil elastase (NE) is a serine protease normally expressed in neutrophil primary granules. It plays a role in antimicrobial defenses and inflammation, and is aberrantly expressed in myeloid leukemia (1–3). Although NE is primarily restricted to hematopoietic cells of the myeloid lineage, it has been shown in breast cancer tissue extracts where it was prognostic (4–6). Foekens et al. demonstrated that high levels of NE detected by ELISA in primary breast tumors were associated with poor metastasis-free, disease-free (DFS), and overall survival (OS) (5). These results were corroborated by Yamashita et al. who determined that NE concentration correlated with DFS (4, 6). The prognostic value of NE has been attributed to its ability to degrade extracellular matrix thereby promoting invasion and metastasis (7, 8). The source of NE in breast tumors is unknown and has been attributed to endogenous production by breast cancer cells (9, 10).
Cyclin E (CCNE), an important cell cycle regulator, has also been shown to be prognostic in breast cancer. Overexpression of CCNE causes tumorigenesis by promoting the G1 to S phase transition, increasing CCNE-associated kinase activity, and causing genomic instability (11–14). Keyomarsi et al. demonstrated that CCNE levels were more powerful determinants of DFS and OS than commonly used clinicopathologic prognostic factors including tumor size, nodal status, clinical stage, and estrogen receptor expression (15). In tumors, the principal mode of CCNE deregulation is at the protein level. Some breast cancer cell lines and human breast cancers express tumor specific low molecular weight (LMW) isoforms that are more active than full-length CCNE and are resistant to cyclin dependent kinase inhibitors (12, 16–19). Importantly, NE was shown to cleave CCNE into its LMW isoforms suggesting that generation of LMW CCNE may be another mechanism linking NE expression and poor prognosis in breast cancer (18, 20).
The CCNE LMW isoforms have been described in other tumors including leukemia (21). We have investigated CCNE as a leukemia-associated antigen and identified the human leukocyte antigen (HLA)-A2-restricted CCNE-derived peptide CCNE144-152 (ILLDWLMEV) as a candidate target for immunotherapy. Importantly, the sequence for CCNE144-152 is contained in full-length CCNE and the LMW isoforms. CCNE144-152-specific cytototxic T lymphocytes (CCNE-CTL) were shown to specifically lyse leukemia cells overexpressing CCNE and its LMW isoforms (21). Because CCNE is aberrantly expressed in breast cancer, we hypothesized that it may represent a target for immunotherapy in breast cancer as well.
Neutrophils and other myeloid cells are present in the tumor microenvironment, and because it has been demonstrated that lung cancer cells can take up NE (22), we postulated that breast cancer cells may take up NE. Since NE has been shown to cleave full-length CCNE, we further hypothesized that NE uptake may lead to increased cleavage of CCNE to its LMW isoforms. The LMW isoforms lack the portion of the full-length protein’s amino terminus that contains the nuclear localization sequence, therefore, LMW CCNE isoforms have altered subcellular localization, accumulating in the cytoplasm where they may be preferentially processed and presented as antigens complexed with HLA-I molecules (23, 24). This in turn could increase susceptibility to lysis by CCNE-CTL. In this report, we show that breast cancer cells lack endogenous NE expression but can take up NE at concentrations similar to that encountered in the tumor microenvironment due to the presence of activated tumor-associated neutrophils (TAN). NE uptake resulted in increased LMW CCNE expression and susceptibility of breast cancer cells to specific lysis by CCNE-CTL. Taken together, these data provide strong evidence for a previously undescribed mechanism linking innate immunity and an adaptive immune response against a novel breast cancer antigen.
Methods
Patients, Cells and Cell Lines
Peripheral blood samples were obtained through an institutional IRB-approved protocol. MCF-7, MDA-MB-231, T47D, and MDA-MB-453 breast cancer cells, U-937, Jurkat (JKT), HL-60 and T2 cell lines were obtained from American Type Culture Collection (Manassas, VA). HER-18 was a gift from Dr. Mien-Chie Hung (MD Anderson Cancer Center). Cell lines were validated by STR DNA fingerprinting using the AmpF/STR Identifiler kit according to manufacturer instructions (Applied Biosystems, Carlsbad, CA). Breast cancer cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS), 100U/mL penicillin, and 100μg/mg streptomycin. Media for HER-18 cells was supplemented with 0.5mg/ml G418. U-937, JKT, T2 and HL-60 cell lines were cultured in RPMI-1640 (RPMI) with 10% FBS, 100U/mL penicillin, and 100μg/mg streptomycin. All cells were maintained in 5% CO2 at 37°C.
Western Blot Analysis
Whole cells lysates were generated in RIPA buffer containing protease inhibitors (Santa Cruz Biotechnology, Santa Cruz, CA). Lysates were run on 10% SDS-page gels then transferred to polyvinylidene fluoride membranes. After blocking, blots were probed with antibodies targeting NE (Santa Cruz Biotechnology) or cyclin E (Santa Cruz Biotechnology).
RNA Extraction and Amplification, cDNA Synthesis and Reverse Transcription Polymerase Chain Reaction
Breast cancer cells were isolated from fresh frozen tumor samples (Origene, Rockville, MD) by laser capture microdissection (LCM) using an Arcturus PixCell laser capture microscope with an IR diode laser (Life Technologies, Applied Biosystems, Carlsbad, CA).. Total RNA was extracted and purified using the Arcturus PicoPure RNA Isolation Kit (Life Technologies, Applied Biosystems). RNA integrity and quantity were evaluated by spectrophotometry (Nano Drop ND-1000 Spectrophotometer, Thermo Scientific, Wilmington, DE). Prior to PCR, RNA was amplified using the Arcturus RiboAmp RNA Amplification Kit (Life Technologies, Applied Biosystems) to generate aRNA. cDNA was synthesized from 1μg of aRNA using the Roche Transcriptor First Strand cDNA Synthesis kit (Roche Applied Science, Indianapolis, IN). For cultured cell lines, total cellular RNA was extracted and isolated using RNA STAT-60 RNA extraction reagent (Amsbio, Lake Forest, CA). cDNA was synthesized as described above.
RT-PCR reactions were performed on an iCycler iQ™ thermal cycler (Bio-Rad Laboratories, Hercules, CA). Primer sequences used included: NE (forward primer 5′-CACGGAGGGGCAGAGACC-3′, reverse primer 5′-TATTGTGCCAGATGCTGGAG-3′), mammaglobin (forward primer 5′-AGCACTGCTACGCAGGCTCT-3′, reverse primer 5′-ATAAGAAAGAGAAGGTGTGG-3′), and GAPDH, an endogenous control, (forward primer 5′-TAGACGGGAAGCTCACTGGC-3′, reverse primer 5′-AGGTCCACCACCCTGTTGCT-3′) (oligonucleotides from Sigma Aldrich, St. Louis, MO).
Immunohistochemistry
Following LCM, remaining tumor tissue was fixed in formalin and paraffin-embedded for immunohistochemistry. Tissue sections were de-paraffinized and re-hydrated. Non-specific binding was blocked after which sections were incubated with primary anti-NE mAb (1:200) (Clone NP-57, Dako, Carpinteria, CA). Slides were incubated with secondary anti-mouse IgG-biotin antibody (1:200) (Vectastain Elite ABC Kit; Vector laboratories, CA) then with the avidin-biotin peroxidase complex (1:100) (Vectastain Elite ABC Kit) after which visualization was performed with chromagen 3, 3′-diaminobenzidine (DAB, Dako). Sections of normal tonsil tissue with neutrophils were used as positive controls. Omission of the primary antibodies were used as negative staining control.
Confocal microscopy and flow cytometry analysis
To evaluate uptake of soluble NE, cells were maintained in low serum (0.5%) media supplemented with NE prepared from whole blood and purified to >95% (Athens Research and Technology, Athens, GA). Cathepsin G (Athens Research and Technology) was prepared in an identical fashion therefore used as a control to demonstrate specificity of uptake. After culture in media supplemented with NE over a range of concentrations, viability was assessed at one, four or 24 hours by trypan blue exclusion assay or by staining with SYTOX blue dead cell stain (Invitrogen). NE activity was determined using a fluorescent substrate assay (Enzcheck Protease Assay, Invitrogen) according to the manufacturer’s instructions. Dose and time course experiments were performed. Briefly, 2×105 cells were maintained in 6-well plates in media supplemented with various concentrations of NE at 37°. At designated timepoints, cells were harvested, permeabilized and stained with the following antibodies: Alexa-647- or -488-conjugated anti-NE (clone NP57; Santa Cruz), FITC-conjugated anti-EEA-1 (BD Biosciences, San Jose, CA), or FITC -conjugated anti-LAMP-2 (eBioscience, San Diego, CA). Direct conjugation of anti-NE antibody was performed using Alexa-647 and 488 conjugation kits (Invitrogen, Carlsbad, CA). Aqua live/dead stain (Invitrogen) was used to assess cell viability. Flow cytometry was performed using the Cytomation CyAn flow cytometer (Beckman Coulter, Brea, CA). Data were analyzed using FlowJo software (Tree Star Inc, Ashland, OR). Confocal imaging was performed using a Leica Microsystems SP2 SE confocal microscope (Buffalo Grove, IL). To evaluate uptake of cell-associated NE, neutrophils were isolated from healthy donors by double ficoll after which they were irradiated and co-cultured with MDA-MB-231 cells at a 3:1 ratio for four hours.
Peptide-specific CTL lines and cell-mediated cytotoxicity assay
Healthy donor HLA-A2+ peripheral blood mononuclear cells (PBMC) were stimulated with CCNE144-152-peptide, as previously described (25). Briefly, T2 cells were incubated with 20μg/mL of CCNE for 90 minutes at 37°C then irradiated and cultured with freshly isolated PBMC at a 1:1 ratio. On days 7, 14, and 21, re-stimulation with CCNE-pulsed T2 cells was performed, and the following day 20IU/mL of recombinant human interleukin-2 (Biosource International) was added. On day 25, CTLs were harvested and used in cytotoxicity assays as previously described (25). Target cells, including T2 cells ± CCNE peptide and HLA-A2+ breast cancer cells, were stained with 10μg/mL of Calcein-AM (Sigma Aldrich), washed and plated in a 60-well Terasaki tray (2×103 cells/10ul/well). Effector cells (CCNE-CTL) were resuspended in 10μl at increasing effector to target dilutions and added to target cells. After 4 hours, trypan blue was added as a quenching agent. Flourescence was measured (FLx800 Microplate fluorescence reader, Bio-Tek Instruments, Winooski, VT) and the percentage of cell lysis was calculated as follows: % cytotoxicity= (1−(Eexperimental−EMedia)/(EControl−Emedia))*100, where E is fluorescence emission and control group is targets alone.
Staining for CCNE-CTL in breast cancer patients
PBMC from HLA-A2+ breast cancer patients and healthy donors were stained with aqua live/dead stain (Invitrogen) and the following antibodies; CD8 APC-H7 (BD Biosciences), CD3 PE Cy7 (BD Biosciences), CD4 pacific orange (Invitrogen), CCNE-APC conjugated tetramer and the following pacific blue conjugated lineage antibodies: CD14 (BD Biosciences), CD16 (BD Biosciences) and CD19 (Biolegend). Data was acquired on a Canto flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star Inc). The frequency of CCNE-CTLs was determined as the percentage of cells that were alive, lineage-, CD4-, CD3+, CD8+ and CCNE-tetramer+.
Statistical Analysis
GraphPad Prism 5.0 software was used to perform statistical analyses and P-values <0.05 were considered significant.
Results
Breast cancer cells do not produce endogenous elastase
Since breast cancer cells are not derived from myeloid hematopoietic progenitors, the source of NE in breast tumors is not fully understood. To investigate this, we evaluated cultured breast cancer cell lines for the presence of NE at the protein and mRNA level. We did not detect NE protein (Fig. 1A) or mRNA transcripts (Fig. 1B) in any of five cell lines investigated. To evaluate whether the lack of expression was limited to cell lines, laser capture microdissection was used to isolate breast cancer cells from primary tumors. Following RNA extraction, RT-PCR confirmed the lack of NE mRNA in all breast cancer specimens evaluated (Fig. 1C). Immunohistochemistry performed on breast tumor tissue showed NE in TAN within the tumor microenvironment, but not in breast cancer cells (Fig. 1D). Taken together, these data are consistent with our hypothesis that TAN present in the microenvironment are the primary source of NE in breast tumors.
Figure 1. Breast cancer cells do not express endogenous neutrophil elastase (NE).
(A) Lysates harvested from breast cancer cell lines cultured in 0.5% serum media were used in western blots showing no NE protein expression. The leukemia cell line U937 served as a positive control. (B) RNA extracted from cultured breast cancer cells was used to perform RT-PCR using primers to NE. The leukemia cell line HL60 and T cell line jurkat (JKT) served as positive and negative controls respectively for NE. No NE mRNA was detected in any of the cell lines evaluated. (C) RT-PCR evaluating for NE mRNA was performed using RNA extracted from breast cancer cells isolated from tumors using laser capture microdissection (LCM1, LCM2 and LCM3). Primers to mammoglobin (MGB-1) were used to confirm LCM was specific for breast cancer cells. Positive controls included MDA-MB-453 breast cancer cells for mammoglobin and HL60 for NE. No LCM specimens demonstrated NE mRNA. (D) A breast tumor was evaluated by hematoxylin and eosin staining to confirm tumor cells and inflammatory infiltrate. TAN present in the microenvironment stained positive for NE by immunohistochemistry.
Soluble and cell associated NE are taken up by breast cancer cells
We have previously shown that antigen presenting cells are capable of taking up soluble NE and that this uptake leads to cross presentation of PR1, a nonameric peptide derived from NE that has been extensively investigated in myelogenous leukemias (25–28). In addition, Houghton et al. demonstrated the ability of lung cancer cells to take up NE (22). We therefore investigated whether breast cancer cells can take up NE using MDA-MB-231 breast cancer cells cultured in NE-supplemented media. At 24 hours, flow cytometry was used to show NE uptake (3.4-fold increase in MFI vs. unpulsed). At that point, the extent of NE following uptake was 76% of the level of NE in HL-60, a promyelocytic leukemia cell line known to express endogenous NE (Fig. 2A). Cell viability was not affected by NE in the culture media (Supplementary Fig. 1). Experiments were repeated using additional breast cancer cell lines (HER18 and MDA-MB-453), which showed NE uptake at one, four and 24-hour time points (Supplementary Fig. 2). The extent of NE uptake varied among cell lines.
Figure 2. Soluble neutrophil elastase (NE) is taken up by breast cancer cells.
(A) MDA-MB-231 cells were maintained in media supplemented with NE (10μg/ml). After 24 hours, cells were analyzed for intracellular NE. NE uptake was significant when compared to unpulsed cells (T-test; P<0.01). NE expression in MDA-MB-231 cells was also compared with T2 cells amd HL60, a cell line known to express significant endogenous NE. (B) MDA-MB-231 cells were maintained in media supplemented with NE at various concentrations for 18 hours after which NE uptake was determined. Cells maintained in media supplemented with OVA and cathepsin G (CG), were used to evaluate for non-specific uptake. (C) Timing of NE uptake was determined using MDA-MB-231cells maintained in media supplemented with NE (10μg/ml). Cells were harvested at various time points and NE uptake determined. (D) To determine if NE uptake was enzyme dependent, NE was co-incubated with two different inhibitors, elafin (6kD) and alpha-1 antitrypsin (65kD), prior to addition to media. After 4 hours, cells were harvested and NE uptake determined. All experiments were performed in triplicate and mean MFI ± SD is shown. Comparisons were made using ANOVA with Tukey’s multiple comparison test. MFI = median fluorescence intensity.
To further study soluble NE uptake by MDA-MB-231 cells, concentration-response and time course experiments were performed. Concentration-response experiments showed concentration-dependent uptake of NE. Importantly, there was no uptake of the non-specific proteins OVA or cathepsin G, a second serine protease (Fig. 2B), suggesting NE uptake is antigen specific. Time course experiments showed that NE uptake was time-dependent and occurred as early as one minute after addition of soluble NE to the culture media (Fig. 2C). Confocal imaging confirmed early NE uptake by breast cancer cells and localization within distinct compartments, as shown by focal NE staining (Supplementary Fig. 3).
We next sought to investigate whether enzymatic activity of NE was required for uptake. NE was incubated with one of two NE inhibitors, α-1 antitrypsin (65kD) or elafin (6kD), prior to its addition to culture media. NE enzymatic activity inhibition by α-1 antitrypsin and elafin was confirmed (Supplementary Fig. 4). α-1 antitrypsin inhibited NE uptake while elafin had no effect (Fig. 2D) suggesting a potential steric-dependent but enzyme-independent mechanism.
Having shown concentration and time-dependent uptake of soluble NE by MDA-MB-231 cells, we next investigated whether these cells could take up cell-associated NE. MDA-MB-231 cells were co-cultured with irradiated neutrophils as a source of NE or irradiated lymphocytes, which lack NE. Radiation induced cell death in approximately 50% of neutrophils and the concentration of NE in the culture at four hours was 16μg/ml. Uptake of NE was again determined using flow cytometry which showed greater uptake of the cell-associated NE then soluble NE (P < .01) (Fig. 3).
Figure 3. MDA-MB-231 breast cancer cells are able to take up cell-associated neutrophil elastase.
MDA-MB-231 cells were cultured with soluble NE (10μg/ml), irradiated neutrophils (PMN) or irradiated mononuclear cells (MNC) at a 1:3 ratio for 4 hours. Cells were permeabilized and stained with FITC-conjugated NE antibodies then analyzed using flow cytometry. MDA-MB-231 cells co-cultured with irradiated PMN showed significantly greater NE uptake compared with cells cultured in soluble NE-supplemented media NE (ANOVA with Tukey’s multiple comparison test). Experiments were performed in triplicate and mean MFI ± SD is shown. MFI = median fluorescence intensity.
Neutrophil elastase localization following uptake
Confocal images had shown uptake of soluble NE into distinct cellular compartments (Supplementary Fig. 3). We therefore sought to determine the subcellular compartment to which NE localized after uptake. MDA-MB-231 cells were cultured in NE-supplemented (10μg/ml) media. At increasing time points, cells were harvested and co-stained for NE and either early endosomal antigen-1 (EEA-1) or lysosome-associated membrane protein (LAMP)-2 as markers for early endosomes or lysosomes, respectively. These experiments confirmed early uptake of NE as it was detected intracellularly within 10 minutes, and showed that soluble NE localizes to an early endosomal compartment (Fig. 4A). There was no evidence of NE uptake into lysosomes (Fig. 4B). Experiments were repeated following uptake of cell-associated NE which showed early uptake of NE with perinuclear localization (Fig 4C and 4D).
Figure 4. Intracellular localization of neutrophil elastase (NE) following uptake by breast cancer cells.
MDA-MB-231 cells were cultured in NE-supplemented (10μg/ml) media after which they were permeabilized and stained with 647-conjugated NE antibody and FITC-conjugated antibodies to EEA-1, an endosomal marker (A), or LAMP, a lysosomal marker (B). Merged images demonstrate localization of NE in an endosomal compartment within 10 minutes. There was no evidence of NE localization in lysosomes. MDA-MB-231 cells were also co-cultured with irradiated neutrophils to evaluate localization following uptake of cell-associated NE. Cells were stained for NE and EEA-1 (C) or LAMP (D). Images demonstrate perinuclear accumulation of cell-associated NE. Red = NE, green = EEA-1 (A) and LAMP (B), blue = DAPI, yellow = merge of NE with EEA-1.
Uptake of soluble elastase increases LMW CCNE expression and susceptibility to CCNE-CTL mediated cytotoxicity
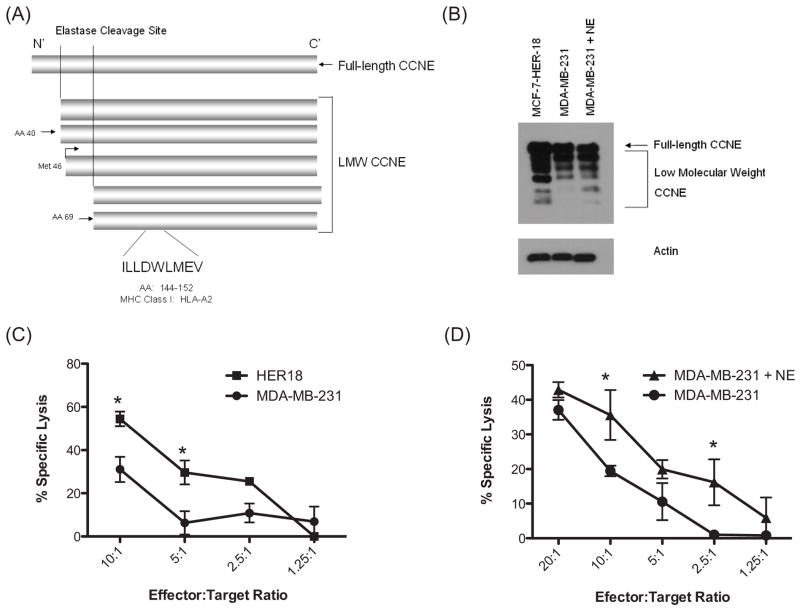
NE has been shown to cleave full-length CCNE at two sites giving rise to LMW isoforms which subsequently undergo phosphorylation to generate two sets of doublets (Fig. 5A) (18, 29). LMW isoforms of CCNE lack a nuclear localization sequence and therefore accumulate in the cytoplasm (23, 24) which may facilitate ubiquitination and proteasomal processing for presentation on HLA-I molecules (30–33). We therefore hypothesized that cells with increased LMW CCNE would be more susceptible to lysis by CCNE-CTL, by virtue of increased HLA/CCNE144-152 surface expression. To test this hypothesis, we expanded CCNE144-152-CTL from PBMC from HLA-A2+ healthy donors. Lysis was tested using cytotoxicity assays. Initially, HER18 and MDA-MB-231 (both HLA-A2+), were used as targets because of differences in baseline LMW CCNE expression (Fig. 5B) (34). These assays demonstrated that CCNE-CTL more effectively lysed HER18, which express more LMW CCNE than MDA-MB-231 (Fig. 5C). CCNE-CTL specific cytolysis was confirmed using unpulsed T2 cells and T2 cells pulsed with CCNE (Supplementary Fig. 5). We next investigated the effect of NE uptake on LMW CCNE expression by MDA-MB-231 cells and whether this impacted susceptibility to lysis by CCNE-specific CTL. Western blot analysis confirmed that uptake of soluble NE resulted in increased expression of LMW CCNE (Fig. 5B). Processing of CCNE by NE to LMW CCNE was confirmed using recombinant CCNE incubated with NE over a range of concentrations (5μg/ml – 100μg/ml) (Supplementary Fig. 6). Importantly, CCNE-CTL specific lysis of NE-pulsed MDA-MB-231 cells was greater than that versus unpulsed cells (Fig. 5D). The cytotoxicity assays were performed multiple times using CTL generated from different healthy donors with variable precursor frequencies of CCNE-CTLs thereby explaining differences in the absolute levels of CCNE-specific killing. Taken together, these data show that exogenous NE such as may be present in the tumor microenvironment, can be taken up by breast cancer cells exposing the CCNE-derived epitope and rendering the cells susceptible to CCNE-CTL-mediated cytolysis.
Figure 5. Extent of low molecular weight (LMW) cyclin E (CCNE) expression impacts susceptibility to CCNE-CTL mediated cytotoxicity.
(A) Schematic of full-length and LMW isoforms of CCNE (Adapted from Akli S and Keyomarsi K. 2003 Cancer Biol Ther 2:S38-47). The CCNE-derived HLA-A2-restricted peptide CCNE144-152 (ILLDWLMEV) is present in the FL protein and LMW isoforms. (B) Whole cell lysates were obtained from HER18, MDA-MB-231 and MDA-MB-231 cells cultured in NE-supplemented (10μg/ml) media for 24 hours. Western blot showed greater LMW CCNE expression in HER18 cells than MDA-MB-231 cells. MDA-MB-231 cells maintained in NE-supplemented media had increased LMW CCNE expression. (C) CCNE-CTL were used in cytotoxicity assays at various effector to target (E:T) ratios versus HER18 and MDA-MB-231 breast cancer cells. CCNE-CTL more effectively lysed HER18 cells. CCNE-specific lysis was confirmed using T2 cells unpulsed or pulsed with CCNE peptide. Cytotoxicity assays were done in triplicate and results are representative of three separate experiments. (D) Cytotoxicity experiments were performed with CCNE-CTL versus MDA-MB-231 cells that were cultured in 0.5% serum media or media supplemented with NE. Uptake of NE by MDA-MB-231 breast cancer cells resulted in increased CCNE-specific lysis at all E:T ratios. Assays were done in triplicate; results are representative of five separate experiments. (* denotes P<.05)
CCNE144-152 tetramer positive CD8+ T cells are present in peripheral blood of breast cancer patients
Having shown that breast cancer cells expressing CCNE are susceptible to lysis by CCNE-CTL, we next sought to confirm whether immunity to CCNE144-152 is detected in breast cancer patients. PBMC were obtained from 11 HLA-A2+ breast cancer patients and seven HLA-A2+ healthy donors and stained with CCNE144-152 tetramer to assess the frequency of CCNE144-152-specific CTL. Figure 6A demonstrates our gating strategy. All breast cancer patients had CCNE144-152-specific CTL present at a low precursor frequency with the median number of CCNE144-152-specific CTL=.074±.02 (Fig. 6B). The frequency of CCNE144-152-specific CTL in breast cancer patients was greater than in healthy donors (P=.001). These data suggest that the CCNE144-152 peptide is naturally processed in breast cancer patients resulting in immunity to CCNE and that vaccination with a CCNE144-152 peptide could potentially augment the immunologic response against CCNE-expressing breast cancer targets.
Figure 6. CCNE-specific cytotoxic T lymphocytes are detected in breast cancer patients.
PBMC from breast cancer patients and healthy donors were stained with lineage markers (anti-CD3, CD14, CD16, CD19, CD20 and CD56), anti-CD4, anti-CD8 and CCNE tetramer. CD8+ T cells were analyzed for CCNE tetramer expression. (A) A representative histogram from one patient is shown to demonstrate gating strategy. Abbreviations: FSC-A, forward scatter-area; SSC-A, side scatter-area; FSC-W, forward scatter-width. (B) The extent of CCNE-specific CD8+ T cells in breast cancer patients (n=11) was greater than in healthy donors (n=7). A Mann-Whitney test was performed to compare the two groups.
Discussion
In this report, we identified a novel function of NE, a serine protease in the tumor microenvironment. We showed NE is present in TANs and that breast cancer cells do not produce NE, suggesting TANs as the primary source of NE in breast cancer. We also showed that NE is specifically taken up by breast cancer cells. Importantly, after NE uptake, LMW isoforms of CCNE increase and breast cancer cells become susceptible to cytolysis by CTL specific for an HLA-A2-restricted peptide (CCNE144-152) that is contained within each LMW isoform. This data confirms that CCNE-derived peptides are naturally processed and presented on breast cancer cells. Therefore, we have established a novel mechanism linking NE, a protease secreted by cells of the innate immune system, to an adaptive immune response against a novel tumor antigen in breast cancer.
Our results, combined with previous studies that show NE cleaves CCNE into LMW isoforms, strongly suggest that after uptake, NE increases substrate availability of CCNE fragments in breast cancer cells, which could augment antigen processing of CCNE peptides. In support of this hypothesis, we showed that HER18 cells are more susceptible to cytolysis by CCNE-CTL compared to MDA-MB-231, which express similar amounts of full-length CCNE but less of the LMW isoforms compared to HER18. Furthermore, after uptake of soluble NE from culture media, LMW isoform expression increased in MDA-MB-231 cells and the cells were more susceptible to lysis by CCNE-CTL. Additional studies must be done to confirm whether full-length CCNE or the LMW isoforms are the predominant source of CCNE144-152 peptide. However, our data support the discovery of CCNE as a breast cancer antigen with potential implications for immunotherapy strategies targeting CCNE. Because the LMW isoforms are tumor-specific and their overexpression drives breast cancer cell proliferation (12, 16–19), they have characteristics of an ideal tumor associated antigen. Importantly, using tetramer staining of PBMC from breast cancer patients, we provide evidence of a low precursor frequency of CCNE144-152 CTL, suggesting that adaptive immunity against the peptide is increased in patients. A peptide vaccine incorporating the CCNE144-152 peptide with an immunoadjuvant may therefore be effective in augmenting a CCNE-specific CTL response. High grade, triple negative tumors have been found to have an intense immune cell infiltrate (35) suggesting that patients with such tumors might be candidates for a CCNE-targeting vaccine.
Although NE uptake could promote anti-tumor immunity by enhancing susceptibility to a CCNE-specific immune response, it is also possible that NE uptake in the setting of chronic inflammation in the tumor microenvironment could promote tolerance resulting from anergy if antigen presentation occurred in the absence of adequate co-stimulation. Although breast cancer cells express MHC class I molecules and can present CCNE144-152 and other peptides, they lack costimulatory molecules and may therefore be unable to stimulate naïve T cells. Nevertheless, strategies such as adoptive T-cell therapy or antibodies that target the CCNE144-152 /HLA-A2 conformational epitope (36, 37) might overcome such tolerance.
Within tumors, the major effects of TAN are thought to promote tumor growth. For instance, neutrophil-derived cytokines enhance tumor growth by producing factors such as interleukin-8 (IL-8) and oncostatin M that promote angiogenesis, invasion and metastasis (38–40). Neutrophil-derived proteases, including NE, degrade cytokines, chemokines, and their receptors and are important for remodeling the extracellular matrix (7,8). By these mechanisms, neutrophil-derived products impact tumor proliferation, vessel density and metastatic potential (41). A study by Houghton et al, showed NE uptake by lung adenocarcinoma cells, where it localized to endosomes and induced tumor cell proliferation by cleaving insulin receptor substrate-1 inducing hyperactivity of the phosphatidylinositol-3-kinase pathway and uncontrolled proliferation (22). Our study confirms that cancer cells take up exogenous NE. The study by Houghton et al showed NE is taken up into clatharin-coated vesicles and we show that NE uptake is antigen-specific, and time and dose dependent, suggesting a receptor-mediated mechanism. Our study provides an additional potential mechanism of NE-induced tumor cell growth by increasing CCNE LMW isoforms. Conversely, NE can also potentiate anti-tumor immunity by increasing the susceptibility of breast cancer cells to lysis by CCNE-CTL. Thus, the net effect of NE uptake in tumor cells likely depends upon additional factors. Because the timing and subcellular localization of NE after uptake is similar in distinct cancer cells, it is possible that there is a common uptake mechanism. If so, such a mechanism could be critical for controlling cell growth and modulating responsiveness to adaptive immunity. Experiments are ongoing in our laboratory to explore this possibility.
Inflammatory cells comprise a significant component of the tumor microenvironment (42–46). Included in these inflammatory cells are neutrophils that are derived from myeloid hematopoietic progenitor cells and produce proteases, including NE. Although neutrophils have a short life span in circulation, they survive longer within the inflammatory environment, possibly as a result of the effects of cytokines on their survival (47). In addition, upregulation of neutrophil-chemotactic substances such as IL-8, results in continuous recruitment of neutrophils to the tumor site (41). Therefore, there is a growing interest in studying TANs. Jensen et al demonstrated that the presence of CD66b+ intra-tumoral neutrophils was an independent prognostic factor for shorter DFS and OS in clear cell renal cell carcinoma (48). Increased neutrophil infiltration is also associated with poor outcomes in bronchioalveolar carcinoma (49). To date, there have been no studies investigating intratumoral neutrophils in breast cancer. Two groups have shown that the presence of immune-reactive NE in whole tumor extracts in breast cancer patients correlates with poor clinical outcomes (4–6). However, NE is synthesized by bone marrow precursor cells that give rise to neutrophils, with NE mRNA being demonstrated in these early myeloid progenitors (1, 50). To our knowledge, no study has definitely demonstrated NE mRNA in epithelial cells providing further support for our conclusion that NE in breast tumors is derived from TANs.
Nevertheless, a report by Nguyen et al showed NE protein in MDA-MB-231 cells by indirect immunofluorescence (20), suggesting NE in tumors could be partially derived from breast cancer cells. However, in MDA-MB-231 cells, we were unable to identify NE protein by confocal microscopy or in immunoblots of whole cell lysates, and we were unable to find NE mRNA transcripts. We also found no NE in the supernatants of cultured breast cancer cells by an anti-NE ELISA (data not shown). Most importantly, we were unable to amplify NE mRNA transcripts from RNA isolated from primary breast cancer cells with single cell LCM. Moreover, breast cancer biopsies stained with an anti-NE antibody and examined by immunohistochemistry confirmed that NE was present in neutrophils but not in breast cancer cells. We therefore believe our study is the first to show the cellular origin of NE in breast tumors is from TAN within the tumor microenvironment.
In conclusion, we show that breast cancer cells rapidly take up NE derived from neutrophils. After NE uptake, LMW CCNE isoforms are increased and the susceptibility of breast cancer to lysis by CTL specific for the novel HLA-A2-restricted CCNE144-152 peptide is increased. Thus, we propose a previously undescribed indirect mechanism linking NE derived from neutrophils, a component of the innate immune system, to an adaptive immune response against a novel breast cancer antigen that is cleaved after specific uptake of NE. Further investigation into the mechanisms regulating NE uptake and the subsequent effects on antigen processing are warranted to improve our understanding of the link between inflammation and breast cancer.
Supplementary Material
Acknowledgments
The authors would like to thank Jieqing Chen for her assistance with immunohistochemical staining of the primary breast tissue.
Grant Support
This work was supported by grant R00CA133244 (E.A.M.), Leukemia SPORE CA 100632 (J.J.M) and P01 CA 148600-01A1 (J.J.M) from the National Cancer Institute as well as Leukemia and Lymphoma Society Specialized Center of Research grant #7262-08 (J.J.M) and #6030-12 (J.J.M.). The Flow Cytometry and Cellular Imaging core is supported by Cancer Center Support Grant NCI #P30CA16672. STR DNA fingerprinting was done by the Cancer Center Support Grant funded Characterized Cell Line core, NCI #CA16672.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.Fouret P, du Bois RM, Bernaudin JF, Takahashi H, Ferrans VJ, Crystal RG. Expression of the neutrophil elastase gene during human bone marrow cell differentiation. J Exp Med. 1989;169:833–45. doi: 10.1084/jem.169.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molldrem JJ, Komanduri K, Wieder E. Overexpressed differentiation antigens as targets of graft-versus-leukemia reactions. Curr Opin Hematol. 2002;9(6):503–8. doi: 10.1097/00062752-200211000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Pulford KA, Erber WN, Crick JA, Olsson I, Micklem KJ, Gatter KC, et al. Use of monoclonal antibody against human neutrophil elastase in normal and leukaemic myeloid cells. J Clin Pathol. 1988;41:853–60. doi: 10.1136/jcp.41.8.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akizuki M, Fukutomi T, Takasugi M, Takahashi S, Sato T, Harao M, et al. Prognostic significance of immunoreactive neutrophil elastase in human breast cancer: long-term follow-up results in 313 patients. Neoplasia. 2007;9:260–4. doi: 10.1593/neo.06808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foekens JA, Ries C, Look MP, Gippner-Steppert C, Klijn JG, Jochum M. The prognostic value of polymorphonuclear leukocyte elastase in patients with primary breast cancer. Cancer Res. 2003;63:337–41. [PubMed] [Google Scholar]
- 6.Yamashita J, Ogawa M, Shirakusa T. Free-form neutrophil elastase is an independent marker predicting recurrence in primary breast cancer. J Leukoc Biol. 1995;57:375–8. doi: 10.1002/jlb.57.3.375. [DOI] [PubMed] [Google Scholar]
- 7.Mainardi CL, Dixit SN, Kang AH. Degradation of type IV (basement membrane) collagen by a proteinase isolated from human polymorphonuclear leukocyte granules. J Biol Chem. 1980;255:5435–41. [PubMed] [Google Scholar]
- 8.Sun Z, Yang P. Role of imbalance between neutrophil elastase and alpha 1-antitrypsin in cancer development and progression. Lancet Oncol. 2004;5:182–90. doi: 10.1016/S1470-2045(04)01414-7. [DOI] [PubMed] [Google Scholar]
- 9.Desmedt C, Ouriaghli FE, Durbecq V, Soree A, Colozza MA, Azambuja E, et al. Impact of cyclins E, neutrophil elastase and proteinase 3 expression levels on clinical outcome in primary breast cancer patients. Int J Cancer. 2006;119:2539–45. doi: 10.1002/ijc.22149. [DOI] [PubMed] [Google Scholar]
- 10.Yamashita JI, Ogawa M, Ikei S, Omachi H, Yamashita SI, Saishoji T, et al. Production of immunoreactive polymorphonuclear leucocyte elastase in human breast cancer cells: possible role of polymorphonuclear leucocyte elastase in the progression of human breast cancer. Br J Cancer. 1994;69:72–6. doi: 10.1038/bjc.1994.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akli S, Keyomarsi K. Cyclin E and its low molecular weight forms in human cancer and as targets for cancer therapy. Cancer Biol Ther. 2003;2(4 Suppl 1):S38–47. [PubMed] [Google Scholar]
- 12.Akli S, Zheng PJ, Multani AS, Wingate HF, Pathak S, Zhang N, et al. Tumor-specific low molecular weight forms of cyclin E induce genomic instability and resistance to p21, p27, and antiestrogens in breast cancer. Cancer Res. 2004;64:3198–208. doi: 10.1158/0008-5472.can-03-3672. [DOI] [PubMed] [Google Scholar]
- 13.Dulic V, Drullinger LF, Lees E, Reed SI, Stein GH. Altered regulation of G1 cyclins in senescent human diploid fibroblasts: accumulation of inactive cyclin E-Cdk2 and cyclin D1-Cdk2 complexes. Proc Natl Acad Sci U S A. 1993;90:11034–8. doi: 10.1073/pnas.90.23.11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 15.Keyomarsi K, Tucker SL, Buchholz TA, Callister M, Ding Y, Hortobagyi GN, et al. Cyclin E and survival in patients with breast cancer. N Engl J Med. 2002;347:1566–75. doi: 10.1056/NEJMoa021153. [DOI] [PubMed] [Google Scholar]
- 16.Keyomarsi K, Herliczek TW. The role of cyclin E in cell proliferation, development and cancer. Prog Cell Cycle Res. 1997;3:171–91. doi: 10.1007/978-1-4615-5371-7_14. [DOI] [PubMed] [Google Scholar]
- 17.Keyomarsi K, O’Leary N, Molnar G, Lees E, Fingert HJ, Pardee AB. Cyclin E, a potential prognostic marker for breast cancer. Cancer Res. 1994;54:380–5. [PubMed] [Google Scholar]
- 18.Porter DC, Zhang N, Danes C, McGahren MJ, Harwell RM, Faruki S, et al. Tumor-specific proteolytic processing of cyclin E generates hyperactive lower-molecular-weight forms. Mol Cell Biol. 2001;21:6254–69. doi: 10.1128/MCB.21.18.6254-6269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wingate H, Zhang N, McGarhen MJ, Bedrosian I, Harper JW, Keyomarsi K. The tumor-specific hyperactive forms of cyclin E are resistant to inhibition by p21 and p27. J Biol Chem. 2005;280:15148–57. doi: 10.1074/jbc.M409789200. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen HH, Aronchik I, Brar GA, Nguyen DH, Bjeldanes LF, Firestone GL. The dietary phytochemical indole-3-carbinol is a natural elastase enzymatic inhibitor that disrupts cyclin E protein processing. Proc Natl Acad Sci U S A. 2008;105:19750–5. doi: 10.1073/pnas.0806581105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He H, Ishiyama K, Alatrash G, Kondo Y, Lu S, Mollrem JJ. T-cell immunity to two HLA-A2-restricted self determinants of cyclin E may contribute to remission after stem cell transplantation. Blood (ASH Annual Meeting Abstracts) 2009;114:686. [Google Scholar]
- 22.Houghton AM, Rzymkiewicz DM, Ji H, Gregory AD, Egea EE, Metz HE, et al. Neutrophil elastase-mediated degradation of IRS-1 accelerates lung tumor growth. Nat Med. 2010;16:219–23. doi: 10.1038/nm.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delk NA, Hunt KK, Keyomarsi K. Altered subcellular localization of tumor-specific cyclin E isoforms affects cyclin-dependent kinase 2 complex formation and proteasomal regulation. Cancer Res. 2009;69:2817–25. doi: 10.1158/0008-5472.CAN-08-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore JD, Kornbluth S, Hunt T. Identification of the nuclear localization signal in Xenopus cyclin E and analysis of its role in replication and mitosis. Mol Biol Cell. 2002;13:4388–400. doi: 10.1091/mbc.E02-07-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molldrem J, Dermime S, Parker K, Jiang YZ, Mavroudis D, Hensel N, et al. Targeted T-cell therapy for human leukemia: cytotoxic T lymphocytes specific for a peptide derived from proteinase 3 preferentially lyse human myeloid leukemia cells. Blood. 1996;88:2450–7. [PubMed] [Google Scholar]
- 26.Molldrem JJ, Lee PP, Wang C, Felio K, Kantarjian HM, Champlin RE, et al. Evidence that specific T lymphocytes may participate in the elimination of chronic myelogenous leukemia. Nat Med. 2000;6:1018–23. doi: 10.1038/79526. [DOI] [PubMed] [Google Scholar]
- 27.Rezvani K, Yong AS, Mielke S, Savani BN, Musse L, Superata J, et al. Leukemia-associated antigen-specific T-cell responses following combined PR1 and WT1 peptide vaccination in patients with myeloid malignancies. Blood. 2008;111:236–42. doi: 10.1182/blood-2007-08-108241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yong AS, Rezvani K, Savani BN, Eniafe R, Mielke S, Goldman JM, et al. High PR3 or ELA2 expression by CD34+ cells in advanced-phase chronic myeloid leukemia is associated with improved outcome following allogeneic stem cell transplantation and may improve PR1 peptide-driven graft-versus-leukemia effects. Blood. 2007;110:770–5. doi: 10.1182/blood-2007-02-071738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mull BB, Cox J, Bui T, Keyomarsi K. Post-translational modification and stability of low molecular weight cyclin E. Oncogene. 2009;28:3167–76. doi: 10.1038/onc.2009.182. [DOI] [PubMed] [Google Scholar]
- 30.Kloetzel PM. Antigen processing by the proteasome. Nat Rev Mol Cell Biol. 2001;2:179–87. doi: 10.1038/35056572. [DOI] [PubMed] [Google Scholar]
- 31.Momburg F, Hammerling GJ. Generation and TAP-mediated transport of peptides for major histocompatibility complex class I molecules. Adv Immunol. 1998;68:191–256. doi: 10.1016/s0065-2776(08)60560-x. [DOI] [PubMed] [Google Scholar]
- 32.Reits EA, Vos JC, Gromme M, Neefjes J. The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature. 2000;404:774–8. doi: 10.1038/35008103. [DOI] [PubMed] [Google Scholar]
- 33.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–71. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 34.Mittendorf EA, Liu Y, Tucker SL, McKenzie T, Qiao N, Akli S, et al. A novel interaction between HER2/neu and cyclin E in breast cancer. Oncogene. 2010;29:3896–907. doi: 10.1038/onc.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2011;16 (Suppl 1):61–70. doi: 10.1634/theoncologist.2011-S1-61. [DOI] [PubMed] [Google Scholar]
- 36.Joseph RW, Peddareddigari VR, Liu P, Miller PW, Overwijk WW, Bekele NB, et al. Impact of clinical and pathologic features on tumor-infiltrating lymphocyte expansion from surgically excised melanoma metastases for adoptive T-cell therapy. Clin Cancer Res. 2011;17:4882–91. doi: 10.1158/1078-0432.CCR-10-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sergeeva A, Alatrash G, He H, Ruisaard K, Lu S, Wygant J, et al. An anti-PR1/HLA-A2 T-cell receptor-like antibody mediates complement-dependent cytotoxicity against acute myeloid leukemia progenitor cells. Blood. 2011;117:4262–72. doi: 10.1182/blood-2010-07-299248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imai Y, Kubota Y, Yamamoto S, Imai Y, Tahashi Y, Hurukawa H, et al. Neutrophils enhance invasion activity of human cholangiocellular carcinoma and hepatocellular carcinoma cells: an in vitro study. J Gastroenterol Hepatol. 2005;20:287–93. doi: 10.1111/j.1440-1746.2004.03575.x. [DOI] [PubMed] [Google Scholar]
- 39.Queen MM, Ryan RE, Holzer RG, Keller-Peck CR, Jorcyk CL. Breast cancer cells stimulate neutrophils to produce oncostatin M: potential implications for tumor progression. Cancer Res. 2005;65:8896–904. doi: 10.1158/0008-5472.CAN-05-1734. [DOI] [PubMed] [Google Scholar]
- 40.Shojaei F, Singh M, Thompson JD, Ferrara N. Role of Bv8 in neutrophil-dependent angiogenesis in a transgenic model of cancer progression. Proc Natl Acad Sci U S A. 2008;105:2640–5. doi: 10.1073/pnas.0712185105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregory AD, Houghton AM. Tumor-associated neutrophils: new targets for cancer therapy. Cancer Res. 2011;71:2411–6. doi: 10.1158/0008-5472.CAN-10-2583. [DOI] [PubMed] [Google Scholar]
- 42.Coffelt SB, Lewis CE, Naldini L, Brown JM, Ferrara N, De Palma M. Elusive identities and overlapping phenotypes of proangiogenic myeloid cells in tumors. Am J Pathol. 2010;176:1564–76. doi: 10.2353/ajpath.2010.090786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeNardo DG, Andreu P, Coussens LM. Interactions between lymphocytes and myeloid cells regulate pro- versus anti-tumor immunity. Cancer Metastasis Rev. 2010;29:309–16. doi: 10.1007/s10555-010-9223-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johansson M, Denardo DG, Coussens LM. Polarized immune responses differentially regulate cancer development. Immunol Rev. 2008;222:145–54. doi: 10.1111/j.1600-065X.2008.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 46.Murdoch C, Muthana M, Coffelt SB, Lewis CE. The role of myeloid cells in the promotion of tumour angiogenesis. Nat Rev Cancer. 2008;8:618–31. doi: 10.1038/nrc2444. [DOI] [PubMed] [Google Scholar]
- 47.Mantovani A. The yin-yang of tumor-associated neutrophils. Cancer Cell. 2009;16:173–4. doi: 10.1016/j.ccr.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 48.Jensen HK, Donskov F, Marcussen N, Nordsmark M, Lundbeck F, von der Maase H. Presence of intratumoral neutrophils is an independent prognostic factor in localized renal cell carcinoma. J Clin Oncol. 2009;27:4709–17. doi: 10.1200/JCO.2008.18.9498. [DOI] [PubMed] [Google Scholar]
- 49.Wislez M, Rabbe N, Marchal J, Milleron B, Crestani B, Mayaud C, et al. Hepatocyte growth factor production by neutrophils infiltrating bronchioloalveolar subtype pulmonary adenocarcinoma: role in tumor progression and death. Cancer Res. 2003;63:1405–12. [PubMed] [Google Scholar]
- 50.Niemann CU, Abrink M, Pejler G, Fischer RL, Christensen EI, Knight SD, et al. Neutrophil elastase depends on serglycin proteoglycan for localization in granules. Blood. 2007;109:4478–86. doi: 10.1182/blood-2006-02-001719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.