Abstract
Humans and their ancestors have traversed the Ethiopian landscape for millions of years, and present-day Ethiopians show great cultural, linguistic, and historical diversity, which makes them essential for understanding African variability and human origins. We genotyped 235 individuals from ten Ethiopian and two neighboring (South Sudanese and Somali) populations on an Illumina Omni 1M chip. Genotypes were compared with published data from several African and non-African populations. Principal-component and STRUCTURE-like analyses confirmed substantial genetic diversity both within and between populations, and revealed a match between genetic data and linguistic affiliation. Using comparisons with African and non-African reference samples in 40-SNP genomic windows, we identified “African” and “non-African” haplotypic components for each Ethiopian individual. The non-African component, which includes the SLC24A5 allele associated with light skin pigmentation in Europeans, may represent gene flow into Africa, which we estimate to have occurred ∼3 thousand years ago (kya). The non-African component was found to be more similar to populations inhabiting the Levant rather than the Arabian Peninsula, but the principal route for the expansion out of Africa ∼60 kya remains unresolved. Linkage-disequilibrium decay with genomic distance was less rapid in both the whole genome and the African component than in southern African samples, suggesting a less ancient history for Ethiopian populations.
Introduction
Much of the key fossil evidence for human origins and evolution is found in modern-day Ethiopia. Early putative hominin fossils such as Ardipithicus kadabba (5.2–5.8 million years ago [mya])1 and Ardipithecus ramidus (4.4 mya; e.g., “Ardi”),2 as well as the earliest indisputable hominin species, Australopithecus anamensis (3.9–4.2 mya) and the better-known Australopithecus afarensis (3.0–3.9 mya; e.g., “Lucy”),3 have all been found there. It is also the homeland of the earliest known anatomically modern human remains: Omo 1 (195 thousand years ago [kya])4 and Homo sapiens idaltu (154–160 kya).5 Perhaps for these reasons and because of Ethiopia's geographical position between Africa and Eurasia, its capital, Addis Ababa, is often used in genetic studies as a proxy embarkation point for modern human range expansions.6,7 However, such studies have seldom included Ethiopians; they are absent from widely used collections, such as the Human Genome Diversity Project (HGDP),8 HapMap,9 and 1000 Genomes10 sets. In practice, our understanding of genome-wide patterns of diversity in Africa has been limited to populations from central and western Africa. Indeed, with a few exceptions,11,12 studies of African genetic diversity that have included Ethiopians have been restricted to mtDNA13–16 and the Y chromosome.14,17 This deficiency has led to an incomplete picture of African genetic diversity that has implications for the study of our origins as a species, including the route followed during the dispersal(s) out of Africa and more recent demographic events involving East Africa.
In linking present-day genetic diversity to the Middle and Late Stone Age populations of Africa, it is important to consider the possibility of long-term population discontinuity in the region and the sparseness of information relating to Ethiopia over the past 200 thousand years (ky). Although archaeological studies focusing on the past few millennia document indigenous Ethiopian developments, including the early cultivation of local species such as teff (Eragrostis tef, a cereal), enset (Musa ensete), and coffee (Coffea arabica),18 they also reveal some cultural influences from outside, such as the cultivation of wheat and barley, which originated in the Fertile Crescent and reached Ethiopia presumably through Egypt during the first documented trade links, around 5 kya.19,20 External contacts with the Ethiopian region are also evident in the historical record from the first millennium BCE onward, wherein Sudanese, Egyptian, South Arabic, and Mediterranean influences are documented.19,21 Another line of evidence for the variegated history of the Ethiopian people comes from linguistic studies. The spread of the two major language families spoken in Ethiopia today—Afro-Asiatic and Nilotic—is considered to be the outcome of cultural and demographic events over the past 10 ky.22 The presence of three diverse Afro-Asiatic branches (Omotic, Semitic, and Cushitic) makes the Horn of Africa one potential source of this family, although the Ethio-Semitic branch is likely to have originated at a later stage in the Middle East.23 The Nilotic languages, represented in Ethiopia by the East Sudanic, Kunama, and Koman branches, are more widespread in Sudan, and their presence in Ethiopia is probably the result of recent demographic processes.24 Similarly, genetic studies indicate that a major component of recent Ethiopian ancestry originates outside Africa: for example, half of the mtDNA haplotypes16 and more than one-fifth of Y haplotypes17 found in Ethiopia belong to lineages that, on the basis of phylogeographic criteria, have been attributed to a non-African rather than a sub-Saharan African origin. These historical admixture events are themselves of interest to historians, anthropologists, and linguists, as well as to geneticists.
Our current study is motivated by four questions. First, where do the Ethiopians stand in the African genetic landscape? Second, what is the extent of recent gene flow from outside Africa into Ethiopia, when did it occur, and is there evidence of selection effects? Third, do genomic data support a route for out-of-Africa migration of modern humans across the mouth of the Red Sea? Fourth, assuming temporal stability of current populations, what are the estimated ages of Ethiopian populations relative to other African groups? In order to address these questions, we generated genome-wide SNP genotypes from Ethiopian individuals.
Given that little genetic information on Ethiopian populations was available in advance, we sought to analyze a broad sample of 188 Ethiopians from ten diverse populations, chosen from a collection of > 5,000 samples assembled by N.B.25,26 The samples genotyped included representatives of a range of geographical regions and all four linguistic groups (Semitic, Cushitic, Omotic, and Nilotic). For comparative studies, we combined our Ethiopian data with published data from the HGDP27 and HapMap39 projects, as well as more focused studies.28,29 Furthermore, to compensate for the lack of published data of populations immediately surrounding Ethiopia, we additionally genotyped 24 South Sudanese and 23 Somali samples.
Material and Methods
Samples and Genotyping
The Ethiopian and Sudanese DNA samples used in this study were extracted from buccal swabs collected in various Ethiopian and Sudanese locations from apparently healthy, anonymous male donors who provided their informed consent. The collection was performed by members of The Centre for Genetic Anthropology at University College London (UCL) and of Addis Ababa University in Ethiopia, and samples were enrolled into the current study when self-reported ethnicity matched that reported for the donor's parents, paternal grandfather, and maternal grandmother. The populations sampled (numbers) were the Semitic-speaking Amhara (26) and Tigray (21); the Cushitic-speaking Oromo (21), Ethiopian Somali (17), and Afar (12); the Omotic-speaking Ari Cultivators (24), Ari Blacksmiths (17), and Wolayta (8); and the Nilotic-speaking Gumuz (19) and Anuak (23). In addition to these groups, we also generated South Sudanese data from mixed populations (24) and Somali data from Somali populations (23). Additional information, together with the sampling locations of these populations, is available in Table S1 available online. The use of the samples for the present study was approved by the UK research ethics committee (approval numbers 99/0196 and 0489/001). The Somali DNA samples (previously obtained from Somali expatriates in Islamabad, Pakistan) were extracted from lymphoblastoid cell lines in the collection created by S.Q.M.
All the samples were whole-genome amplified with the GE GenomiPhi HY DNA Amplification Kit (catalog no. 25-6600-25, General Electric) and genotyped on the Illumina Omni 1M SNP array at the Wellcome Trust Sanger Institute. SNP calls and quality checks were performed by the Sanger genotyping facility with the use of GenoSNP.30 Y-chromosomal haplogroups were also determined at both UCL and Sanger labs. The above 235 genotypes were pooled with data from published sources,9,27–29 providing ∼280,000 overlapping markers in 4,442 individuals.
For the fixation index (FST), mtDNA, and genomic minimum pairwise distance, we chose to reference non-African populations along the two putative routes: Bedouin, Druze, Palestinian, Syrian, Lebanese, Jordanian, Iranian, Greek, French, Pathan, Han, and Surui populations representing the northern route; Yemeni, Saudi Arabian, Dravidian, and Papuan populations representing the southern route.
Summary Statistics
SNP frequencies, heterozygosity, and linkage disequilibrium (LD, r and r2) were calculated for each group with PLINK,31 and pairwise FST values were calculated with an in-house script implementing the Weir and Cockerham formula.32 The FST and heterozygosity values were interpolated and plotted on a geographic map with Surfer (Golden Software). The merged data set was pruned to remove SNPs in high LD (r2 > 0.1), and ADMIXTURE analyses were run as described33 after removal of samples showing high relatedness (PLINK identity-by-descent score ≥ 0.125) with any other sample in the same population (1 Amhara, 2 Ari Cultivators, 6 Ari Blacksmiths, 3 South Sudanese, and 1 Gumuz).34 Cross validation was used to estimate the optimum number of clusters (K). Principal-component analysis (PCA) was implemented with EIGENSTRAT35 on the same pruned data set.
We phased the one million Ethiopian SNPs with BEAGLE,36 incorporating information from the HapMap3 YRI (Yoruba in Ibadan, Nigeria from the CEPH collection) trios.9 Candidate population-specific signals of positive selection were identified with the integrated haplotype score (iHS) statistic.37
Genome Partitioning
We implemented the following approach, modified from published chromosome-painting methodology,28 to partition each individual genome into windows that were more similar to the African and non-African populations, respectively. To obtain a list of SNPs that were independent in each of the reference populations, we LD pruned34 the data in three steps, using 20 French, 20 Han Chinese, and 20 Yoruba samples, sequentially. The pruned markers were then divided into 40-SNP, nonoverlapping windows covering the whole genome. Every window was then phased independently within each population with the PHASE program,38 and the phased haplotypes were used in the following steps.
Each test haplotype was compared with haplotypes from the corresponding genomic window taken from 20 individuals from each of the three reference populations (Han Chinese, French, and Yoruba). The comparison was performed by running a PCA with the use of the “princomp” function of the R package. Three reference clouds (Han Chinese, French, and Yoruba) were defined by the median and 50% confidence radius, calculated from the relevant haplotypes. The Euclidean distance between the principal component (PC) coordinates of the test haplotype and the confidence perimeter of each cloud were then calculated. Due to the similarity between the European and Asian haplotypes relative to the African haplotypes and the consequent difficulty in drawing a clear separation between the two non-African clouds, we then labeled each test 40-SNP haplotype as either “African” or “non-African” according to its position in the PCA plot, or “NA” if there was no separation between the reference clouds. The “NA” haplotypes (less than 1% of the total) were removed from the downstream analyses.
Analyses of Partitioned African and Non-African Genomic Components
The resulting genome partitions were used in a series of analyses whereby either the African or the non-African component of a set of populations was taken into consideration. In order to compare various populations with different levels of African and non African components, we pooled together either the African or non-African haplotypes to create ten mosaic haploid genomes per population. Each mosaic haploid genome would then include either African or non-African haplotypes from different individuals of the same population.
To analyze the LD of the African component of each genome, we included all available SNPs and calculated LD decay over a range of distances as described.28
The minimum pairwise distance between African and non-African populations was calculated using ten mosaic non-African haploid genomes (made of either African or non-African haplotypes only) from each Ethiopian, Somali, and Sudanese population (together, “Ethiopian+”). For each Ethiopian+ 40-SNP window, we calculated the shortest distance to the same window in the non-African population, and averaged the distance over all windows in each population.
A Z-score based on the number of chromosomes in the non-African state was assigned to each 40-SNP window in each of the five Semitic-Cushitic populations. The Z-score was calculated for each 40-SNP window in each population on the basis of the average and SD of the full set of regions for that population. We then binned the Z-scores and counted the number of regions occurring for a given bin in a given number of the examined populations. Any region showing a Z-score > 2 or < −2 in more than two of the five populations examined was flagged as an outlier, and its gene content was examined for functional interest.
Assuming that the African and non-African components of the Ethiopian genomes result from a single admixture event, we used ROLLOFF39 to estimate the midpoint of the period of admixture. However, if there were multiple or continuous admixture events, as with the North African populations, this method detected39 the most recent event or the admixture midpoint, respectively. ROLLOFF computes the correlation between (1) a (signed) statistic for LD between a pair of markers and (2) a weight that reflects their allele-frequency differentiation in the ancestral populations. We used as putative ancestral populations either CEU (Utah residents with ancestry from northern and western Europe) and YRI (as previously described39) or CEU and Ari, chosen because of their extremal positions in a PC plot (Figure S4). Because of the lack of publicly available code at the time of the analyses, the ROLLOFF algorithm was recoded in-house (details available upon request) from the description provided,39 following advice kindly provided by its authors, and was shown to give similar age estimates (r2 > 0.9, data not shown) for a set of test populations previously analyzed with the use of this approach39 (African Americans, Palestinians, Sardinians, Bedouins, and Druze; all treated as a mixture of CEU and YRI). Before running the analyses, we performed a PCA on the Ethiopian, North African, and Middle Eastern individuals, together with YRI and CEU, to identify and remove outlier individuals (1 Amhara, 1 South Sudanese, 1 Bedouin, 2 Egyptian, 3 Moroccan, 4 Mozabite, 1 Saudi, and 1 Yemeni) and to split those populations forming more than one cluster (e.g., Oromo was divided into Oromo1 and Oromo2), as recommended by the authors.
Results
In the following sections, we consider sequentially the four questions identified in the Introduction, and thus move from more recent to more ancient events.
Modern Ethiopians in the African Genetic Landscape
The first PC of the African samples separates sub-Saharan Africans from North Africans, with Ethiopians positioned between them (Figure 1A), whereas the second and third components separate the hunter-gatherers (click speakers and Pygmies) and the East Africans, respectively (Figures 1A and 1B). Both plots separate the Ethiopian samples according to their linguistic origin. This linguistic clustering appears to be more important than geographical structure, especially for the Semitic and Cushitic populations (Figure 1D), and is also supported by the neighbor-joining tree of Figure S2. Remarkably, the Ethiopian clusters, taken together, span half of the space delimited by all the African populations and surround the Maasai from Kenya. To investigate this high diversity further, we performed an African-only PCA (Figure S1A) using five randomly chosen samples from each Ethiopian population, in order to eliminate bias that might arise from including a large number of Ethiopian samples, and a worldwide PCA using the full data set (Figure S1B). Both plots confirmed the high diversity in Ethiopia; Ethiopians spanned most of the African branch in the worldwide PCA (Figure S1A) and showed similar internal structure in both PCA plots (Figures 1B and S1B).
Figure 1.
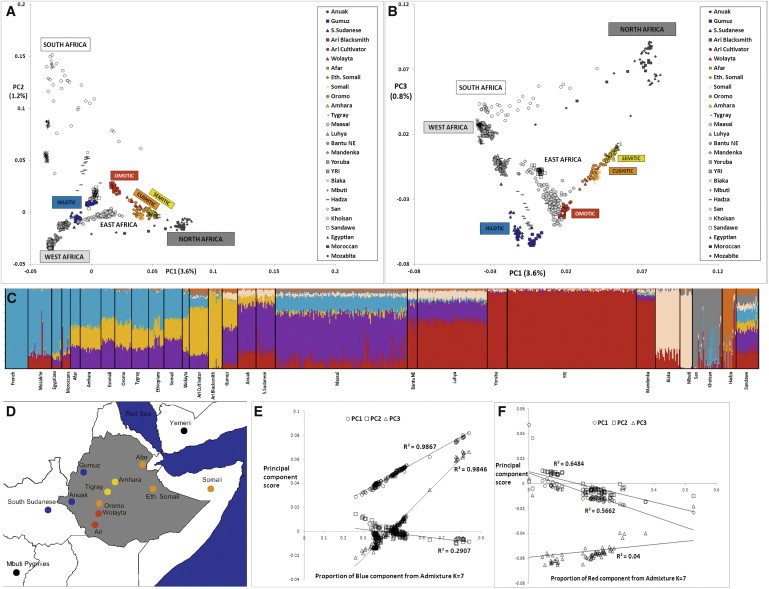
Principal Components and STRUCTURE-like Analyses of the Full African Data Set
The first three PCs are represented in bidimensional plots (first versus second in A and first versus third in B). The samples genotyped in this study are represented in yellow (Semitic), orange (Cushitic), red (Omotic), or blue (Nilotic); the rest of the African samples are shown with the use of a gray scale. The proportion of explained variance is reported next to each axis.
(C) displays the best fit (K = 7) ADMIXTURE result, including all the African samples and with the addition of French as a non-African population. The colors in (C) do not match those in (A) and (B).
(D) shows the sampling locations in Ethiopia. Each population is colored according to the linguistic family to which it belongs.
(E) Correlation between the proportion of “non-African” admixture (x axis, blue component from C) and the first three PCs for the Semitic, Cushitic, Omotic, and Egyptian samples.
(F) Correlation between the proportion of Nigerian-Congolese admixture (x axis, red component from C) and the first three PCs for the Anuak, Gumuz, and South Sudanese samples.
ADMIXTURE34 was applied to the same African data set, with the addition of the HGDP French27 as a reference group for the non-African component (Figure S1C). The best-supported34 clustering (K = 7, Figure 1C) divided the Ethiopians into two main groups: the Semitic-Cushitic Ethiopians stand out as a relatively uniform set of individuals characterized by a strong (40%–50%) non-African component (light blue in Figure 1C) and an African component split between a broad East African (purple in Figure 1C) and an apparently Ethiopia-specific component (yellow); the Nilotic and Omotic Ethiopians show little or no non-African component and are instead characterized by eastern (purple and yellow) or western (dark red) African components, with some traces of additional components. The yellow and purple components represent the major proportion of the African component in the Egyptian Afro-Asiatic population, but are less predominant than the red West African component among northwestern African populations who also speak Afro-Asiatic languages. However, it is striking that North Africans share substantially more variation with non-African populations (80%) than do Ethiopians (40%–50%).
To investigate the role played by the non-African component in the PCA clustering of the Semitic and Cushitic samples, we looked for correlations between the former (obtained from ADMIXTURE, K = 7) and the first three PCs. As shown in Figure 1E, both PC1 and PC3 strongly correlate (both r2 values are above 0.98) with the blue component of Figure 1C, whereas PC2 shows a weaker correlation (r2 = 0.29). The strong PC1 and PC3 correlations therefore seem to indicate that the proportion of non-African admixture is the main driver of the Ari-Egyptian cline formed by the Semitic-Cushitic samples in the PCA plot, regardless of their population of origin. However, when looking for correlations between the Nigerian-Congolese component (blue in Figure 1C) and the first three PCs in the Nilotic populations, we found a much weaker correlation (Figure 1F) than observed for the Semitic-Cushitic component. The Ari-Yoruba cline observed for the Nilotic samples cannot therefore be explained as a simple admixture event between Ethiopians and Nigerian-Congolese populations.
To compare the level of genetic variation in the populations investigated, we estimated average SNP heterozygosity in the pruned genomes of ten individuals from each population and the pairwise FST between African and worldwide populations (Figure 2 and Table S2). The Semitic-Cushitic and North African populations showed the highest values of heterozygosity worldwide, which may reflect a combination of SNP ascertainment bias and the mixture of African and non-African components in these populations. The observed pattern of uniform decline of FST values away from North, West, or East Africa is consistent with previous interpretations of a single exit, followed by “isolation by distance.”6,40,41
Figure 2.
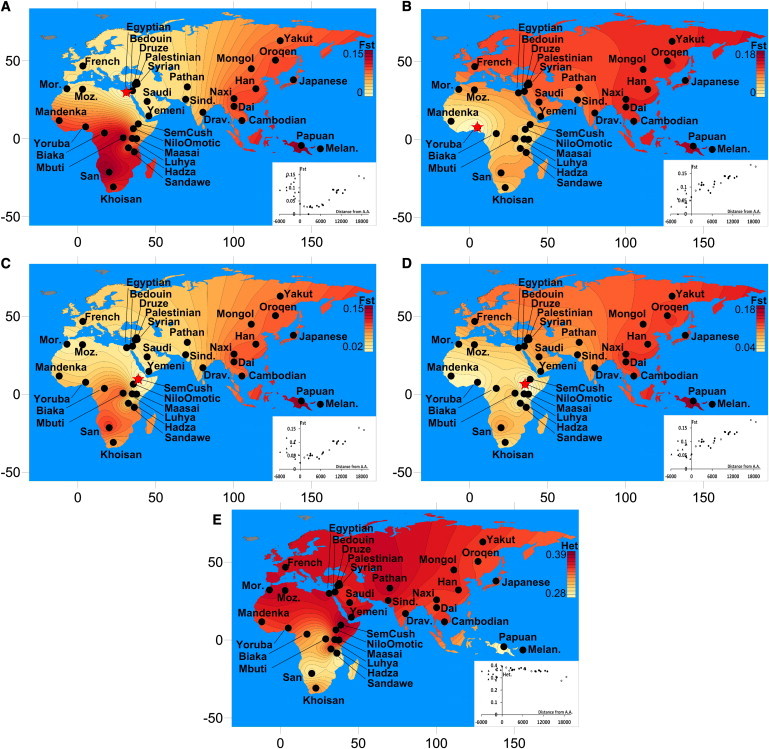
Pairwise FST and SNP Heterozygosity in a Set of Worldwide Populations
FST was calculated with the use of ten individuals from each worldwide population and Egyptians (A), Yoruba (B), and Semitic-Cushitic (C) and Nilotic-Omotic Ethiopians (D), and is displayed as a heat surface, produced with the Surfer software. Values in (C) and (D) are the averages for all the Semitic-Cushitic or Nilotic-Omotic populations. (E) shows the average genomic heterozygosity calculated for the same samples with the use of the available SNPs. The bottom-right section of each panel includes a scatter plot displaying the actual values of either Fst or heterozigosity over the geographic distance (in km) from Addis Ababa (negative for sub-Saharan populations). Filled and empty circles represent non-African populations along the putative northern or southern routes, respectively. Triangles represent sub-Saharan populations.
Back to Africa
Before considering questions related to ancient demographic events, we needed to separate the probable ancient African components from that which might have originated from more recent (<60 kya) gene flow back to Africa (light blue in Figure 1C).
In order to perform this partitioning, we modified a PCA-based method,28 dividing the genome into haploid windows of 40 SNPs and labeling each as either African or non-African (see Material and Methods). The effectiveness of this method was assessed through comparison of the proportion of each individual genome assigned to an African or non-African origin by PCA with the ADMIXTURE K = 2 clustering. The patterns are very similar (Figures S3A and S3B), and the correlation between the proportions is high (r2 > 0.99; Figure S3C). The added value of the PCA approach is that it locates the African and non-African haplotype windows within each genome, and thus allows their subsequent analysis.
We calculated the genetic distance (FST) between Semitic and Cushitic Ethiopians and populations of the Levant, North Africa, and the Arabian Peninsula using two approaches: (1) the whole genome and (2) only the non-African component. In the whole-genome analysis, Ethiopian Semitic and Cushitic populations appear to be closest to the Yemeni (Figure 3A); when only the non-African component is used, they are closer to the Egyptians and populations inhabiting the Levant (Figure 3B). We explored this finding further by calculating the minimum pairwise difference (see Material and Methods) between Africans and non-Africans for their whole genome, and for the non-African component only. The results are concordant with the results of the FST analyses in showing that the Egyptians are closer than Yemeni to Ethiopians in their non-African component (Table S3). A possible explanation for this result is that there has been gene flow into Ethiopia from the Levant and Egypt, although we cannot say whether the gene flow was episodic or continuous. The Ethiopian similarity with the Yemeni detected throughout the genome could be explained as an Ethiopian contribution to the Yemeni gene pool, consistent with that observed with mtDNA.16
Figure 3.
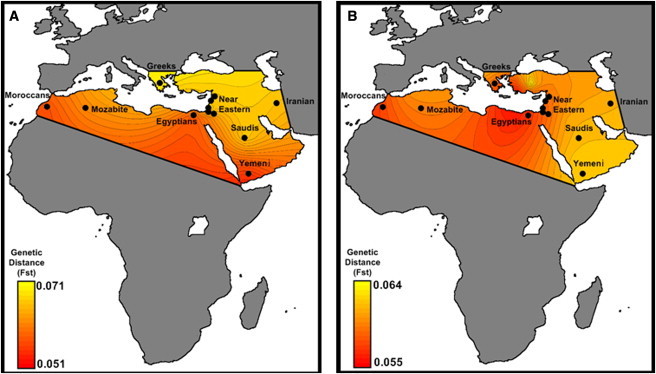
Pairwise FST between Semitic-Cushitic Ethiopians and Surrounding Populations
Contour plots derived from FST were calculated with (A) ten haploid genomes from the Semitic-Cushitic Ethiopians, showing that modern Yemeni, Egyptians, and Moroccans are closest to the Ethiopians, and (B) ten haploid non-African genomes from the same groups, showing instead a prevalence of Egyptian and Middle Eastern contributions to the non-African Ethiopian gene pool.
We considered two sources (western and eastern) for the African component of the Ethiopian genomes. The distinction between the East and West African components is supported by the PCA, wherein our samples formed a triangle (Figure S4) with the three corners represented by West Africans (YRI), non-Africans (CEU), and East Africans (Ari Cultivators and Blacksmiths). The other populations were distributed along the three sides of the triangle in a way that could imply different patterns of admixture. We applied ROLLOFF to estimate admixture dates for the Ethiopian populations, considered as a combination of West Africans with non-Africans or East Africans with non-Africans, depending on their position in the PC plot (Figure S4). The dates of admixture (assuming 30 years per generation)42 are reported in Table 1. Notably, in most of the Semitic, Cushitic, and Omotic populations, the admixture of African and non-African ancestry components dates to 2.5–3 kya, whereas in North Africa, the admixture dates are ∼2 ky more recent, clustering around 1 kya, consistent with previous reports.43 The consistency between the Ethiopian estimates and the appearance in the area of a linguistic family (Ethio-Semitic) with a West Asian origin23 support the hypothesis of a recent gene flow from the Levant. Although ROLLOFF estimated a date for an admixture event involving the Nilotic populations, examination of the relationship between the correlation coefficient and genetic distance (Figure 4) revealed no exponential decay for these populations, implying less support for an admixed origin of the Nilotic populations than of the Semitic, Cushitic, and Omotic populations.
Table 1.
Admixture Date Estimates in East and North African Populations
| Region | Population | YRI-CEU Admixture Date | Ari-CEU Admixture Date |
|---|---|---|---|
| East Africa | Ari Blacksmith | −1228 | NA |
| East Africa | Ethiopian Somali | −1094 | −1201 |
| East Africa | Ari Cultivator | −1017 | NA |
| East Africa | Somali | −953 | −1996 |
| East Africa | Amhara | −637 | −1502 |
| East Africa | Tygray | −425 | −1319 |
| East Africa | Wolayta | −209 | −1418 |
| East Africa | Afar | −170 | −1039 |
| East Africa | Oromo1 | −168 | −1062 |
| East Africa | Anuak | 71 | NA |
| East Africa | Oromo2 | 96 | −906 |
| West Asia | Druze | 767 | 958 |
| East Africa | Maasai | 883 | NA |
| West Asia | Saudi2 | 1109 | 1232 |
| North Africa | Egyptian | 1117 | 1283 |
| West Asia | Bedouin2 | 1130 | 1122 |
| West Asia | Palestinian | 1159 | 1137 |
| West Asia | Saudi | 1164 | 1466 |
| North Africa | Moroccan | 1176 | 1407 |
| West Asia | Bedouin1 | 1256 | 1365 |
| North Africa | Mozabite | 1267 | 1388 |
| West Asia | Yemeni | 1548 | 1548 |
| East Africa | Gumuz | 1588 | NA |
| East Africa | South Sudanese | 1839 | NA |
| North America | African American | 1855 | NA |
The date of admixture for each populations reported in the table was calculated with an in-house version of the ROLLOFF algorithm.39 To facilitate the interpretation of results, we converted the number of generations into years using 30 years per generation, and then into a CE or BCE date by subtracting 2011. Column 3 reports this date, and models the populations as a mixture of CEU and YRI (Utah residents with ancestry from northern and western Europe and Yoruba in Ibadan, Nigeria, respectively, from the CEPH collection).39 Column 4 reports corresponding estimates, modeled assuming admixture between CEU and the Ari Ethiopians. The rationale for these two analyses is provided in Figure S4. NA, not available.
Figure 4.
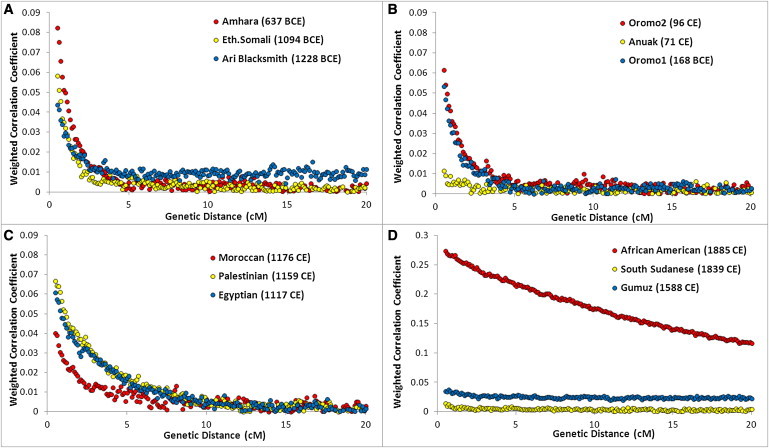
ROLLOFF Plots
Three populations from each of the four historical periods of admixture (A: <500 BCE, B: ∼0 CE, C: ∼1000 CE, and D: >1500 CE) are plotted to show their LD decay (represented by a weighted correlation coefficient as previously described39) with genetic distance. The legend reports the name of each population, with the estimated date of admixture in brackets. Notably, all three Nilotic populations (Gumuz, South Sudanese, and Anuak) have very flat decay curves compared to those of the other populations in the same plot.
Selection Following Admixture
An intriguing consequence of admixture between populations is the opportunity for packages of genes to be “tested” in different environments. As a result, the genomic regions containing functionally divergent genes might experience either positive or negative selection, depending on whether their adaptive contribution was beneficial or damaging in the new environment, or whether it affected social factors such as sexual selection. To look for such outlier regions of admixture in Ethiopian populations (Semitic and Cushitic) where the estimated proportions of African and non-African ancestries were roughly equal, we listed those regions showing an excess or a deficit (see Material and Methods) of non-African haplotypes (Table S4). Of the fourteen 40-SNP windows observed with a Z-score > 2, we noted one that contained SLC24A5 (MIM 113750). This gene is a major contributor to the pigmentation differences between Africans and Europeans and a strong candidate for positive selection in Europe.44,45 Given that SLC24A5 is one of the most highly differentiated genes between African and European populations,10,46 we then looked for other highly differentiated genes10 among the outlier windows, but found none. We also checked whether the 24 large Z-score windows reported in Table S4 showed enrichment for regions with extreme distances between the African and non-African clouds. After ranking all the 40-SNP windows by the distance between the African and European cloud centers divided by the SD of the European cloud around its center, none of the large Z-score windows were present within the top 1%. We therefore speculate that the excess of non-African SLC24A5 haplotypes must be linked to the biological function of that gene.
The iHS scan performed on the Semitic-Cushitic populations (considered as a whole) confirmed that SLC24A5 was within the top 5% of selection signals, whereas the gene was not detected as an outlier in the other groups of Ethiopians. The unusual history of this gene was further supported by the presence of the derived A allele of the SNP rs1834640, associated with the light skin pigmentation of Europeans and western Asians,47 at higher frequencies in Semitic-Cushitic groups compared with Omotic, Nilotic, or Nigerian-Congolese groups (0.55 versus 0.23, 0.07, and 0.04, respectively). To further investigate the effect of admixture on the genetic landscape of skin pigmentation in Ethiopia, we also looked at other genes associated with pigmentation in Europe;46 however, none were found in our outlier regions.
Source of the Major Out-of-Africa Migration
Consistent with previous studies' reports of a steady decline in genetic similarity among non-African populations as a function of geographical traveling distance from East Africa, we found that the FST values estimated between either Ethiopian or North African populations and non-African populations followed the same pattern (Figure 2, Table S2). This steady decline has been argued27 to be compatible with a single exit followed by isolation-by-distance, rather than with two distinct African sources contributing to the non-African diversity. Neither including nor excluding the Ethiopian data altered the pattern. To follow the thread left by this dispersal in more detail, we used the genome partitioning performed earlier to calculate the minimum pairwise difference between the African component of the Egyptian and Ethiopian populations and the equivalent genomic segment in non-Africans. The partitioning would remove noise, caused by recent backflows into Africa, which might otherwise mask the original out-of-Africa signal. If the mouth of the Red Sea had been a major migration route out of Africa, we might observe a closer affinity of Ethiopians, rather than Egyptians, with non-Africans.
As a proof of principle, we first applied the approach to a genetic system with a well-understood phylogeographic structure: mtDNA. Virtually all indigenous sub-Saharan African mtDNA lineages belong to L haplogroups, whereas the presence of haplogroups M and N in North and East Africa has been interpreted as a signal of gene flow back to Africa.48,49 With the full set of 18 mtDNA SNPs used in our genome-wide data set, Egyptians and Moroccans proved to be the closest African population to any non-African population examined (Table 2A). However, when we first partitioned the mtDNA lineages into African and non-African (i.e., L and non-L) and considered only the L component, a different pattern emerged: Ethiopians were the closest population to the non-Africans (Table 2B), consistent with inferences drawn from more detailed mtDNA analyses.50
Table 2.
Minimum Pairwise Difference between Africans and Non-Africans Calculated for the Whole Genome or mtDNA and for their African Component Only
| Population | Cushitic-Semitic | Omotic | Nilotic | Egyptian | Moroccan | Mozabite |
|---|---|---|---|---|---|---|
| Whole Genome | ||||||
| Han | 0.0407 | 0.0418 | 0.0422 | 0.0402 | 0.0407 | 0.0406 |
| Bedouin | 0.0385 | 0.0402 | 0.0409 | 0.0365 | 0.0375 | 0.0379 |
| Druze | 0.0386 | 0.0403 | 0.0412 | 0.0365 | 0.0376 | 0.0379 |
| French | 0.0391 | 0.0409 | 0.0419 | 0.0372 | 0.0381 | 0.0378 |
| Greek | 0.0389 | 0.0408 | 0.0416 | 0.0369 | 0.0378 | 0.0378 |
| Iranian | 0.0389 | 0.0406 | 0.0413 | 0.0372 | 0.0382 | 0.0382 |
| Jordanian | 0.0386 | 0.0402 | 0.0410 | 0.0365 | 0.0379 | 0.0379 |
| Lebanese | 0.0385 | 0.0403 | 0.0411 | 0.0368 | 0.0376 | 0.0377 |
| Moroccan Jews | 0.0386 | 0.0403 | 0.0412 | 0.0364 | 0.0375 | 0.0376 |
| Palestinian | 0.0387 | 0.0403 | 0.0411 | 0.0370 | 0.0377 | 0.0379 |
| Saudi | 0.0386 | 0.0404 | 0.0412 | 0.0367 | 0.0377 | 0.0378 |
| Syrians | 0.0387 | 0.0404 | 0.0414 | 0.0364 | 0.0377 | 0.0379 |
| Yemeni | 0.0384 | 0.0396 | 0.0399 | 0.0372 | 0.0373 | 0.0384 |
| Yemeni Jews | 0.0385 | 0.0404 | 0.0412 | 0.0367 | 0.0375 | 0.0380 |
| AVERAGE | 0.0388 | 0.0404 | 0.0412 | 0.0370 | 0.0379 | 0.0381 |
| Whole mtDNA Pool | ||||||
| Bedouin | 0.0024 | 0.0033 | 0.0041 | 0.0024 | 0.0012 | 0.0024 |
| Palestinian | 0.0020 | 0.0023 | 0.0028 | 0.0017 | 0.0006 | 0.0011 |
| Saudi | 0.0008 | 0.0015 | 0.0012 | 0.0025 | 0.0019 | 0.0025 |
| Yemeni | 0.0031 | 0.0046 | 0.0062 | 0.0044 | 0.0040 | 0.0040 |
| Yemeni Jews | 0.0018 | 0.0022 | 0.0022 | 0.0017 | 0.0022 | 0.0022 |
| French | 0.0019 | 0.0014 | 0.0023 | 0.0000 | 0.0000 | 0.0006 |
| Pathan | 0.0020 | 0.0017 | 0.0028 | 0.0011 | 0.0006 | 0.0017 |
| Dravidian | 0.0008 | 0.0008 | 0.0011 | 0.0000 | 0.0000 | 0.0006 |
| Papuan | 0.0006 | 0.0006 | 0.0006 | 0.0006 | 0.0006 | 0.0006 |
| AVERAGE | 0.0017 | 0.0020 | 0.0026 | 0.0016 | 0.0012 | 0.0017 |
| African Component | ||||||
| Han | 0.0420 | 0.0414 | 0.0412 | 0.0415 | 0.0418 | 0.0434 |
| Bedouin | 0.0397 | 0.0395 | 0.0394 | 0.0364 | 0.0382 | 0.0406 |
| Druze | 0.0399 | 0.0398 | 0.0396 | 0.0365 | 0.0388 | 0.0410 |
| French | 0.0406 | 0.0404 | 0.0402 | 0.0379 | 0.0392 | 0.0416 |
| Greek | 0.0403 | 0.0401 | 0.0400 | 0.0375 | 0.0389 | 0.0412 |
| Iranian | 0.0402 | 0.0400 | 0.0397 | 0.0375 | 0.0389 | 0.0412 |
| Jordanian | 0.0399 | 0.0397 | 0.0395 | 0.0371 | 0.0385 | 0.0408 |
| Lebanese | 0.0399 | 0.0396 | 0.0394 | 0.0371 | 0.0381 | 0.0407 |
| Moroccan Jews | 0.0400 | 0.0398 | 0.0395 | 0.0367 | 0.0385 | 0.0409 |
| Palestinian | 0.0400 | 0.0399 | 0.0395 | 0.0366 | 0.0382 | 0.0408 |
| Saudi | 0.0399 | 0.0398 | 0.0394 | 0.0366 | 0.0385 | 0.0408 |
| Syria | 0.0401 | 0.0398 | 0.0397 | 0.0366 | 0.0387 | 0.0411 |
| Yemeni | 0.0394 | 0.0391 | 0.0387 | 0.0367 | 0.0378 | 0.0403 |
| Yemeni Jews | 0.0399 | 0.0397 | 0.0395 | 0.0364 | 0.0384 | 0.0407 |
| AVERAGE | 0.0401 | 0.0399 | 0.0397 | 0.0372 | 0.0388 | 0.0411 |
| L-mtDNA Only | ||||||
| Bedouin | 0.0036 | 0.0034 | 0.0051 | 0.0077 | 0.0050 | 0.0058 |
| Palestinian | 0.0026 | 0.0024 | 0.0035 | 0.0056 | 0.0032 | 0.0032 |
| Saudi | 0.0023 | 0.0016 | 0.0015 | 0.0041 | 0.0027 | 0.0035 |
| Yemeni | 0.0054 | 0.0047 | 0.0077 | 0.0102 | 0.0072 | 0.0072 |
| Yemeni Jews | 0.0029 | 0.0023 | 0.0028 | 0.0037 | 0.0032 | 0.0032 |
| French | 0.0025 | 0.0015 | 0.0029 | 0.0038 | 0.0033 | 0.0033 |
| Dravidian | 0.0013 | 0.0009 | 0.0014 | 0.0019 | 0.0016 | 0.0016 |
| Pathan | 0.0032 | 0.0017 | 0.0035 | 0.0056 | 0.0040 | 0.0048 |
| Papuan | 0.0008 | 0.0006 | 0.0007 | 0.0010 | 0.0008 | 0.0008 |
| AVERAGE | 0.0027 | 0.0021 | 0.0032 | 0.0048 | 0.0034 | 0.0037 |
Applying the same principle, we then calculated the shortest distance between the African and non-African populations on the basis of either full genome data or the African component of this data set. In contrast to the mtDNA results, the Egyptians proved to be the closest to the non-Africans in both cases (Tables 2A and 2B).
Relative Ages of the Ethiopian and Other African Populations
The decay of LD with time provides a robust proxy for the “age” of a population of a constant size: that is, the length of time that the ancestors of the sampled individuals have been evolving as part of the same breeding unit. To assess how relatively “old” the patterns of LD are in Ethiopian populations, we compared the LD at different distances between the Ethiopian populations and a range of other African populations (Figure 5).28 We also performed the same analyses on the African components of each population to reduce the bias introduced by the recent genetic back-flow (Figure 5B). In both cases, the Ethiopians displayed less LD decay than did the click speakers, Pygmies, or Nigerian-Congolese groups, suggesting a younger age, a smaller long-term effective population size, or a combination of these.
Figure 5.
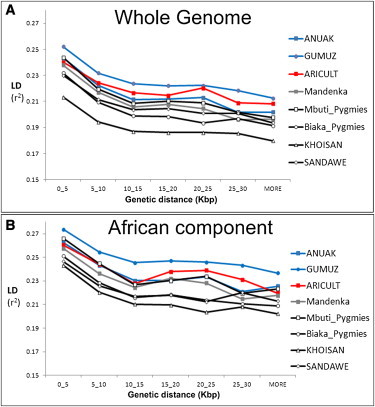
LD Decay over Distance
Analyses were performed with the use of 12 individuals from a set of African populations (A), including Ethiopians (red-yellow scale), west-central Africans (gray scale), and click speakers (blue scale). A modified version of the same analyses (B) was performed with the use of only ten haploid African-genome equivalents. In both cases, the Ethiopian samples show less-rapid LD decay than the other African populations in the figure.
Discussion
We present an extensive genome-wide data set representing Ethiopian geographical, linguistic, and ethnic diversity. Its study has allowed us to cast light on a number of questions, some long-standing, about both ancient and recent demographic events in human evolution. In the Discussion, we again follow a roughly chronological path from the more recent to the older events.
The Ethiopian populations show high genetic diversity, with stratification matching the linguistic families (Figure 1B), except for the overlap in both PCA and FST analyses of populations belonging to two mutually unintelligible linguistic groups (Semitic and Cushitic). This overlap reflects both the similar amount of non-African genome present in these individuals and the similar African component (Figures 1C and 1E). It may also reflect factors such as the recent expansion of some Cushitic and Semitic groups and landscape such as highland and lowland environments. Of particular interest is the distinctiveness of the Omotic groups, whose position in Figures 1A and S3 is intriguingly compatible with being a putative ancestral Ethiopian population. One insight provided by the ADMIXTURE plot (Figure 1C) concerns the origin of the Ari Blacksmiths. This population is one of the occupational caste-like groups present in many Ethiopian societies that have traditionally been explained as either remnants of hunter-gatherer groups assimilated by the expansion of farmers in the Neolithic period or as groups marginalized in agriculturalist communities due to their craft skills.51 The prevalence of an Ethiopian-specific cluster (yellow in Figure 1C) in the Ari Blacksmith sample could favor the former scenario; the ancestors of this occupational group could have been part of a population that inhabited the area before the spread of agriculturalists. Further study of multiple groups comparing agriculturists and caste-like groups would reveal whether there is a pattern of a greater Ethiopia-specific genomic profile associated with caste-like occupations, an observation which would support the absorption rather than the exclusion hypothesis.
ADMIXTURE analyses revealed a major (40%–50%) contribution to the Ethiopian Semitic-Cushitic genomes that is similar to that of non-African populations. Our estimates of genetic similarity between this component and extant non-African populations suggest that the source was more likely the Levant than the Arabian Peninsula. We estimate that this admixture event took place approximately 3 kya. The more recent admixture dates for the Oromo and Afar can be explained by the effect of a subsequent Islamic expansion that particularly impacted these groups, as well as the North Africans.52 Levant people may have arrived in Ethiopia via land or sea subsequently, leaving a similar signature also in modern Egyptians, or the similarity between Ethiopians and Egyptians may be a consequence of independent genetic relationships. This putative migration from the Levant to Ethiopia, which is also supported by linguistic evidence, may have carried the derived western Eurasian allele of SLC24A5, which is associated with light skin pigmentation. Although potentially disadvantageous due to the high intensity of UV radiation in the area, the SLC24A5 allele has maintained a substantial frequency in the Semitic-Cushitic populations, perhaps driven by social factors including sexual selection. The “African” component of the Ethiopian genomes may also result in part from recent migrations into Ethiopia from other parts of Africa, a possibility that we have not examined here.
The estimated time (3 kya) and the geographic origin (the Levant) of the gene flow into Ethiopia are consistent with both the model of Early Bronze Age origins of Semitic languages and the reported age estimate (2.8 kya) of the Ethio-Semitic language group.23 They are also consistent with the legend of Makeda, the Queen of Sheba. According to the version recorded in the Ethiopian Kebra Nagast (a traditional Ethiopian book on the origins of the kings), this influential Ethiopian queen (who, according to Hansberry,53 reigned between 1005 and 955 BCE) visited King Solomon—ruler, in biblical tradition, of the United Kingdom of Israel and Judah—bringing back, in addition to important trading links, a son. The ancient kingdom of Axum adopted Christianity as early as the fourth century. Historical contacts established between Ethiopia and the Middle East were maintained across the centuries, with the Ethiopian church in regular contact with Alexandria, Egypt. These long-lasting links between the two regions are reflected in influences still apparent in the modern Ethiopian cultural and, as we show here, genetic landscapes.
An abundance of evidence suggests that all modern non-Africans descend predominantly from a single African source via a dispersal event some 50 to 70 kya.6,7,27,49 However, debate continues about whether the principal migratory route out of Africa was north of the Red Sea to the Levant, or across its mouth to the Arabian Peninsula. The actual source of the migrations within Africa is a different question, but we assume that the migrators would have left genetic signatures in Egypt if they took the northern route or in Ethiopia if they took the southern route. We chose reference non-African populations along the two putative routes. However, both the northern and eastern Africans have genetic distances (FST) that gradually increase with geographic distance along both routes. This also holds true when Ethiopian populations that show little evidence of recent non-African gene flow (Omotic and Nilotic) are used as a source. A minimum-pairwise-distance measure based on the African component of the genome found that the Ethiopian mtDNA component was closer to non-African populations than was the Egyptian mtDNA component, as previously reported,50 but that the autosomal genome of non-Africans was closer to the African component of the Egyptian rather than Ethiopian populations. This could be interpreted as supporting a northern exit route. However, the 80% non-African proportion of the Egyptian genome (Figure 1C) reduces the power of our comparisons and, taken together with the requirement for the African state in at least ten chromosomes, means that this conclusion is based on just ∼1,800 SNPs (compared to 18,960 for the Ethiopians, 30,798 for the Mozabite, and 5,920 for the Moroccans). Therefore, the question requires further investigation beyond the scope of the present study.
On a broader time scale, the LD analyses pointed to click speakers, Pygmies, and a Nigerian-Congolese group as all having a deeper population history than both the whole genome and the African component of the East Africans sampled. Although this result might seem inconsistent with the outstanding fossil record available from Ethiopia, it may illustrate that genetic diversity assessed from modern populations does not necessarily represent their long-term demographic histories at the site. Alternatively, the rich record of human fossil ancestors in Ethiopia, and indeed along the Rift Valley, may reflect biases of preservation and discovery, with more fossils being exposed in regions of geological activity. Fluctuations in effective population size in the past and dispersals within Africa may have further confounded our analyses and their correlation with the fossil record. The fact that the observed genetic diversity in Ethiopia is lower than in some other African populations does not negate the possibility that Ethiopia was the cradle of anatomically modern humans. However, interpretations of the LD-based analyses may be challenged by future work in two key respects. First, whole-genome sequences can provide an independent measure of the demographic history of the groups studied,54 but they have not yet been applied to Ethiopian samples. Second, there is a need for a better understanding of the implication for the genomic recombination landscape of the observed allelic differences in PRDM9 (MIM 609760).55 The higher frequencies of the active allele reported for the West African Yoruba compared with the Eastern African Maasai might therefore imply the need for rethinking the direct correlation between LD patterns and population age.
In conclusion, Ethiopian SNP genotypes give insights into evolutionary questions on several timescales. Whether or not modern Ethiopians can be identified as the best living representatives of an ancestral human population, or even of the out-of-Africa movement, the data presented here reveal imprints of historical events that accompanied the formation of the rich cultural and genetic diversity observed in the area. Furthermore, we observe strong genetic structuring in East Africa, including a strong match between the linguistic and genetic structures. This is exemplified by the three distinct PC clusters (Omotic, Nilotic, and Semitic-Cushitic), confirming Ethiopia as one of the most diverse African regions.
Acknowledgments
The authors would like to acknowledge all the Ethiopian donors and collaborators, as well as Sarah Edkins, Emma Gray, Sarah Hunt, Avazeh Tashakkori Ghanbarian, and the staff at the Sanger Institute who performed the genotyping. Great help has also been provided by Priya Moorjani, David Reich, and Nick Patterson for a better understanding of the mathematics underlying the ROLLOFF approach. This work was supported by grant number 098051 from the Wellcome Trust. L.P. would like to thank the providers of a Domestic Research Scholarship, the Cambridge European Trust, and Emmanuel College, Cambridge, UK for sponsoring his research. N.B. is the settlor and senior trustee of Melford Charitable Trust and owner of Cordell Homes Ltd., which have in part funded this research. Neither N.B., the charitable trust, nor the company have any intellectual property or other rights with respect to the results of the study.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
BEAGLE, http://faculty.washington.edu/browning/beagle/beagle.html
EIGENSTRAT, http://genepath.med.harvard.edu/∼reich/Software.htm
Illumina 1M Newly Generated Genotypes, http://mega.bioanth.cam.ac.uk/data/Ethiopia
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.omim.org
R: Principal Component Analysis, http://stat.ethz.ch/R-manual/R-patched/library/stats/html/princomp.html
References
- 1.Haile-Selassie Y. Late Miocene hominids from the Middle Awash, Ethiopia. Nature. 2001;412:178–181. doi: 10.1038/35084063. [DOI] [PubMed] [Google Scholar]
- 2.White T.D., Asfaw B., Beyene Y., Haile-Selassie Y., Lovejoy C.O., Suwa G., WoldeGabriel G. Ardipithecus ramidus and the paleobiology of early hominids. Science. 2009;326:75–86. [PubMed] [Google Scholar]
- 3.Johanson D.C., White T.D. A systematic assessment of early African hominids. Science. 1979;203:321–330. doi: 10.1126/science.104384. [DOI] [PubMed] [Google Scholar]
- 4.McDougall I., Brown F.H., Fleagle J.G. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature. 2005;433:733–736. doi: 10.1038/nature03258. [DOI] [PubMed] [Google Scholar]
- 5.White T.D., Asfaw B., DeGusta D., Gilbert H., Richards G.D., Suwa G., Howell F.C. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423:742–747. doi: 10.1038/nature01669. [DOI] [PubMed] [Google Scholar]
- 6.Prugnolle F., Manica A., Balloux F. Geography predicts neutral genetic diversity of human populations. Curr. Biol. 2005;15:R159–R160. doi: 10.1016/j.cub.2005.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramachandran S., Deshpande O., Roseman C.C., Rosenberg N.A., Feldman M.W., Cavalli-Sforza L.L. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl. Acad. Sci. USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cann H.M., de Toma C., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 9.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell M.C., Tishkoff S.A. The evolution of human genetic and phenotypic variation in Africa. Curr. Biol. 2010;20:R166–R173. doi: 10.1016/j.cub.2009.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovell A., Moreau C., Yotova V., Xiao F., Bourgeois S., Gehl D., Bertranpetit J., Schurr E., Labuda D. Ethiopia: between Sub-Saharan Africa and western Eurasia. Ann. Hum. Genet. 2005;69:275–287. doi: 10.1046/j.1529-8817.2005.00152.x. [DOI] [PubMed] [Google Scholar]
- 13.Quintana-Murci L., Semino O., Bandelt H.J., Passarino G., McElreavey K., Santachiara-Benerecetti A.S. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat. Genet. 1999;23:437–441. doi: 10.1038/70550. [DOI] [PubMed] [Google Scholar]
- 14.Passarino G., Semino O., Quintana-Murci L., Excoffier L., Hammer M., Santachiara-Benerecetti A.S. Different genetic components in the Ethiopian population, identified by mtDNA and Y-chromosome polymorphisms. Am. J. Hum. Genet. 1998;62:420–434. doi: 10.1086/301702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poloni E.S., Naciri Y., Bucho R., Niba R., Kervaire B., Excoffier L., Langaney A., Sanchez-Mazas A. Genetic evidence for complexity in ethnic differentiation and history in East Africa. Ann. Hum. Genet. 2009;73:582–600. doi: 10.1111/j.1469-1809.2009.00541.x. [DOI] [PubMed] [Google Scholar]
- 16.Kivisild T., Reidla M., Metspalu E., Rosa A., Brehm A., Pennarun E., Parik J., Geberhiwot T., Usanga E., Villems R. Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears. Am. J. Hum. Genet. 2004;75:752–770. doi: 10.1086/425161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semino O., Santachiara-Benerecetti A.S., Falaschi F., Cavalli-Sforza L.L., Underhill P.A. Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am. J. Hum. Genet. 2002;70:265–268. doi: 10.1086/338306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillipson D.W. British Museum Press; London: 1998. Ancient Ethiopia. Aksum: its antecedents and successors. [Google Scholar]
- 19.Pankhurst R. Blackwell Publishers Ltd; Oxford: 1998. The Ethiopians. [Google Scholar]
- 20.Phillipson D.W. The antiquity of cultivation and herding in Ethiopia. In: Shaw T., Sinclair P., Andah B., Okpoko A., editors. The Archaeology of Africa: Food, Metals and Towns. Routledge; London: 1993. pp. 344–357. [Google Scholar]
- 21.Levine D.N. The University of Chicago; Chicago: 1974. Greater Ethiopia. [Google Scholar]
- 22.Ehret C. University of California Press; Berkeley, CA: 1995. Reconstructing Proto-Afroasiatic (Proto-Afrasian): Vowels, Tone, Consonants, and Vocabulary. [Google Scholar]
- 23.Kitchen A., Ehret C., Assefa S., Mulligan C.J. Bayesian phylogenetic analysis of Semitic languages identifies an Early Bronze Age origin of Semitic in the Near East. Proc. Biol. Sci. 2009;276:2703–2710. doi: 10.1098/rspb.2009.0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blench R. AltaMira Press; Lanham, MD: 2006. Archaeology, Language, and the African Past. [Google Scholar]
- 25.Horsfall L.J., Zeitlyn D., Tarekegn A., Bekele E., Thomas M.G., Bradman N., Swallow D.M. Prevalence of clinically relevant UGT1A alleles and haplotypes in African populations. Ann. Hum. Genet. 2011;75:236–246. doi: 10.1111/j.1469-1809.2010.00638.x. [DOI] [PubMed] [Google Scholar]
- 26.Plaster, C.A. (2011). Variation in Y chromosome, mitochondrial DNA and labels of identity in Ethiopia. PhD thesis, University College London, London.
- 27.Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Myers R.M. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 28.Henn B.M., Gignoux C.R., Jobin M., Granka J.M., Macpherson J.M., Kidd J.M., Rodríguez-Botigué L., Ramachandran S., Hon L., Brisbin A. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc. Natl. Acad. Sci. USA. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behar D.M., Yunusbayev B., Metspalu M., Metspalu E., Rosset S., Parik J., Rootsi S., Chaubey G., Kutuev I., Yudkovsky G. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 30.Giannoulatou E., Yau C., Colella S., Ragoussis J., Holmes C.C. GenoSNP: a variational Bayes within-sample SNP genotyping algorithm that does not require a reference population. Bioinformatics. 2008;24:2209–2214. doi: 10.1093/bioinformatics/btn386. [DOI] [PubMed] [Google Scholar]
- 31.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cockerham C.C., Weir B.S. Estimation of inbreeding parameters in stratified populations. Ann. Hum. Genet. 1986;50:271–281. doi: 10.1111/j.1469-1809.1986.tb01048.x. [DOI] [PubMed] [Google Scholar]
- 33.Alexander D.H., Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics. 2011;12:246. doi: 10.1186/1471-2105-12-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 36.Browning B.L., Browning S.R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voight B.F., Kudaravalli S., Wen X., Pritchard J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephens M., Smith N.J., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moorjani P., Patterson N., Hirschhorn J.N., Keinan A., Hao L., Atzmon G., Burns E., Ostrer H., Price A.L., Reich D. The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet. 2011;7:e1001373. doi: 10.1371/journal.pgen.1001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu H., Prugnolle F., Manica A., Balloux F. A geographically explicit genetic model of worldwide human-settlement history. Am. J. Hum. Genet. 2006;79:230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barbujani G., Colonna V. Human genome diversity: frequently asked questions. Trends Genet. 2010;26:285–295. doi: 10.1016/j.tig.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 42.Fenner J.N. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am. J. Phys. Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- 43.Henn B.M., Botigué L.R., Gravel S., Wang W., Brisbin A., Byrnes J.K., Fadhlaoui-Zid K., Zalloua P.A., Moreno-Estrada A., Bertranpetit J. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 2012;8:e1002397. doi: 10.1371/journal.pgen.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamason R.L., Mohideen M.A., Mest J.R., Wong A.C., Norton H.L., Aros M.C., Jurynec M.J., Mao X., Humphreville V.R., Humbert J.E. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 45.Sabeti P.C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., Xie X., Byrne E.H., McCarroll S.A., Gaudet R., International HapMap Consortium Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pickrell J.K., Coop G., Novembre J., Kudaravalli S., Li J.Z., Absher D., Srinivasan B.S., Barsh G.S., Myers R.M., Feldman M.W., Pritchard J.K. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stokowski R.P., Pant P.V., Dadd T., Fereday A., Hinds D.A., Jarman C., Filsell W., Ginger R.S., Green M.R., van der Ouderaa F.J., Cox D.R. A genomewide association study of skin pigmentation in a South Asian population. Am. J. Hum. Genet. 2007;81:1119–1132. doi: 10.1086/522235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olivieri A., Achilli A., Pala M., Battaglia V., Fornarino S., Al-Zahery N., Scozzari R., Cruciani F., Behar D.M., Dugoujon J.M. The mtDNA legacy of the Levantine early Upper Palaeolithic in Africa. Science. 2006;314:1767–1770. doi: 10.1126/science.1135566. [DOI] [PubMed] [Google Scholar]
- 49.Underhill P.A., Kivisild T. Use of y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu. Rev. Genet. 2007;41:539–564. doi: 10.1146/annurev.genet.41.110306.130407. [DOI] [PubMed] [Google Scholar]
- 50.Soares P., Alshamali F., Pereira J.B., Fernandes V., Silva N.M., Afonso C., Costa M.D., Musilová E., Macaulay V., Richards M.B. The Expansion of mtDNA Haplogroup L3 within and out of Africa. Mol. Biol. Evol. 2012;29:915–927. doi: 10.1093/molbev/msr245. [DOI] [PubMed] [Google Scholar]
- 51.Freeman D., Pankhurst A. Hurst and Company; London: 2003. Peripheral People: The Excluded Minorities of Ethiopia. [Google Scholar]
- 52.Kaplan I. US Government Printing Office; Washington, D.C.: 1971. Area Handbook for Ethiopia. [Google Scholar]
- 53.Hansberry W.L. Howard University Press; Washington, D.C.: 1974. Pillars in Ethiopian History. [Google Scholar]
- 54.Li H., Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hinch A.G., Tandon A., Patterson N., Song Y., Rohland N., Palmer C.D., Chen G.K., Wang K., Buxbaum S.G., Akylbekova E.L. The landscape of recombination in African Americans. Nature. 2011;476:170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


