Abstract
During tumor initiation, oncogene-induced senescence (OIS) is proposed to limit the progression of preneoplasms to invasive carcinoma unless circumvented by the acquisition of certain tumor suppressor mutations. Using a variety of biomarkers, OIS has been previously reported in a wide range of human and murine precursor lesions, including the pancreas, lung, colon and skin. Here, we have characterized a panel of potential OIS biomarkers in human and murine pancreatic intraepithelial neoplasia (PanIN), and found that only senescence-associated β-galactosidase (SAβgal) activity is specifically enriched in these precursors, compared with pancreatic ductal adenocarcinoma (PDA). Indeed, many of the other proposed OIS biomarkers are detected in actively proliferating PanIN epithelium and in cells within the microenvironment. Surprisingly, acinar to ductal metaplasia (ADM), a distinct preneoplasm that is potentially a precursor for PanIN, also exhibits SAβgal activity and contains a higher content of p21 and p53 than PanIN. Therefore, SAβgal activity is the only biomarker that accurately identifies a small and heterogeneous population of non-proliferating premalignant cells in the pancreas, and the concomitant expression of p53 and p21 in ADM supports the possibility that PanIN and ADM each exhibit discrete senescence blocks.
Keywords: senescence, Kras, PanIN, ADM, PDA, SAβgal
Introduction
Amongst the proffered hypotheses regarding the long latency of tumor development, oncogene-induced senescence (OIS) has emerged as a barrier to tumorigenesis (Campisi and d'Adda di Fagagna, 2007; Collado et al., 2007; Courtois-Cox et al., 2008; Halazonetis et al., 2008; Prieur and Peeper, 2008). OIS arrested cells have been identified in the non-invasive precursor stages of cancer development (Braig et al., 2005; Collado et al., 2005; Michaloglou et al., 2005; Bartkova et al., 2006; Courtois-Cox et al., 2006; Di Micco et al., 2006; Dankort et al., 2007; Sun et al., 2007; Halazonetis et al., 2008; Majumder et al., 2008; Dhomen et al., 2009), following an initiating mutation, but prior to the development of frank carcinoma. It is at this critical juncture that OIS is thought to functionally constrain tumorigenesis. In the case of pancreatic ductal adenocarcinoma (PDA), several molecular alterations have been identified and an activating point mutation in the proto-oncogene Kras is a frequent initiating event. As pancreatic intraepithelial neoplasias (PanINs) progress in a stepwise manner from grade 1 to grade 3, additional mutations occur in key tumor suppressor genes, including p16, p53 and SMAD4 (Hruban et al., 2000). Applying the OIS framework to the PanIN progression model suggests that any senescence that occurs would be detectable at the earliest stage PanIN 1, when Kras mutations are already present, but prior to the acquisition of mutations capable of circumventing senescence such as p16 or p53.
Elucidation of a number of potential biomarkers for the senescent state has facilitated the detection of OIS arrested cells in vivo (Collado and Serrano, 2006). Indeed, prior to the discovery of the putative OIS biomarkers p15INK4b, Dec1 and DcR2 (Collado et al., 2005), the most commonly used biomarker was senescence-associated β-galactosidase (SAβgal; Dimri et al., 1995). Additional biomarkers frequently associated with OIS include p16INK4b, p19ARF, p53 and p21 due to their functional role in the induction of senescence (Collado et al., 2005; Dankort et al., 2007; Morton et al., 2009). Other potential biomarkers include proteins related to the senescence-associated secretory phenotype such as IL6, IL8 and CXCR2 (Acosta et al., 2008; Chien and Lowe, 2008; Kuilman et al., 2008; Wajapeyee et al., 2008; Kuilman and Peeper, 2009); proteins related to autophagy (Young et al., 2009), and the characteristic heterochromatin condensation observed under senescent conditions in vitro (Narita et al., 2003). Lastly, the DNA damage response due to the activation of replicative stress has been implicated in senescence induction; therefore, the expression and/or activation of proteins related to the DNA damage response including γH2AX, CHK1, CHK2, ATM or ATR have also been shown to be associated with OIS (Bartkova et al., 2006; Mallette et al., 2007; Halazonetis et al., 2008; d'Adda di Fagagna, 2008). The elevated presence of these potential biomarkers in a precursor lesion has been used to support the conclusion that senescence is occurring in vivo. However, it is still unclear which of these markers truly define a senescent phenotype.
Senescence has been detected in the pancreas, but under contrasting circumstances. Initial reports of OIS in PanIN suggest widespread senescence induction in response to endogenous expression of KrasG12V, as measured by the immunohistochemical detection of the biomarkers p16INK4a, p15INK4b, Dec1 and DcR2 (Collado et al., 2005). Functional studies then demonstrated that the OIS block, which occurred in PanIN in response to KrasG12D activation, was dependant on an intact p21-p53 signaling axis (Morton et al., 2009) or intact LKB1 expression (Morton et al., 2010). In these studies, OIS was detected with p21, p53 and SAβgal as biomarkers. In contrast, a recent study demonstrated that the endogenous expression of KrasG12D protected the wild-type pancreatic ductal compartment from inflammation-induced senescence (Lee and Bar-Sagi, 2010). However, implicit to the results of this study is the notion that endogenous KrasG12D expression in the pancreas was insufficient to induce senescence in the ductal lineage. This latter finding is consistent with the initial characterization of the murine PanINs (mPanINs) from KrasG12D;Cre mice (Hingorani et al., 2003), although a more recent study has demonstrated that cerulean-induced inflammation is essential for senescence bypass in the pancreas (Guerra et al., 2011). Indeed, the differences in senescence phenotype between mouse models have often been attributed to technical and methodological issues (Collado et al., 2007; Kuilman et al., 2010; Lee and Bar-Sagi, 2010). Therefore, it remains uncertain what role senescence has in shaping the evolution of PanIN to PDA.
We have evaluated the senescence phenotype in parallel in both human and mPanIN and PDA to establish its potential relevance, using the Pdx1-cre;KrasG12D model, which is amongst the most widely used murine models of cancer currently available. We demonstrate the absence of specificity of most potential OIS biomarkers in human and mPanIN, due to the lack of enrichment of biomarker expression in precursor lesions and the presence of the same biomarkers in dividing cells. In contrast, only SAβgal activity specifically identified non-proliferating cells and labeled 10% of all mPanIN cells. Together, these two findings indicate an analysis, deeper than putative OIS biomarkers alone, is required to accurately identify a senescence block in preneoplastic lesions in vivo. In addition, SAβgal was as readily detectable in acinar-ductal metaplasia (ADM) as in PanIN, with ADM exhibiting a significant increase in the number of p21 and p53 positive cells relative to PanIN. These unique OIS characteristics suggest that ADM may represent a distinct lineage from PanIN, and underscores prior work reporting the morphological and genetic distinctions between ADM and PanIN (Hruban et al., 2006; Shi et al., 2009). Therefore, senescence does occur during the development of murine PDA (mPDA), but in multiple preneoplastic lineages, and no biomarkers currently available readily enable its detection on fixed human tissues.
Results and discussion
OIS biomarkers are expressed in human pancreatic preneoplasms
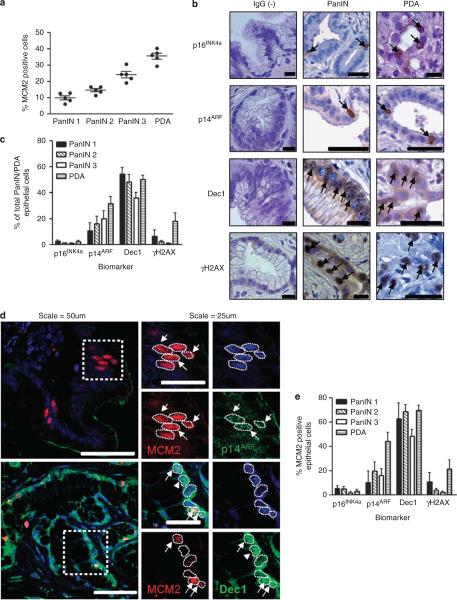
Senescence barriers in preneoplasms have been characterized by decreased proliferation and increased OIS biomarker expression relative to invasive neoplasms. To determine if this feature was present in human PanIN (hPanIN) relative to human PDA (hPDA), immunohistochemical detection was undertaken for proliferation and putative OIS biomarker antigens (Table 1). We found that 10% of hPanIN 1A cells are proliferating, with an increase in proliferation seen with advancing hPanIN grade and frank hPDA (Figure 1a). In contrast, when hPanIN and hPDA were evaluated for the presence of putative OIS biomarkers, p15INK4b and DcR2 were undetectable (data not shown) and enrichment of p16INK4a, p14ARF, Dec1 and γH2AX in hPanIN relative to hPDA was not observed (Figures 1b and c). Immunofluorescent detection of MCM2 and of each potential OIS biomarker was performed to distinguish the proliferating and senescent subpopulations, respectively. Surprisingly, each of these biomarkers colocalized to varying extents with MCM2, most prominently p14ARF and Dec1 (Figures 1d and e), suggesting that this cohort of biomarkers does not specifically identify senescent cells in vivo.
Table 1.
Table of primary antibodies used
| Antibody | Species | Supplier | Dilution | Tissue origin |
|---|---|---|---|---|
| p16 | Rabbit | Santa Cruz M156, sc-1207 (Santa Cruz, CA, USA) | 1:500 | M |
| p19 | Rabbit | Abcam ab80 (Cambridge, UK) | 1:2000 | M |
| DcR2 | Rabbit | Stressgen AAP-371 # 03160608 (Exeter, UK) | 1:500 | M, H |
| Dec1 | Rabbit | Adrian Harris (Oxford, UK) | 1:100 | M, H |
| p15 | Rabbit | Marcos Malumbres (Madrid, Spain) | 1:1000 | M, H |
| γH2AX (Ser139) | Mouse | Upstate Cell Signalling Solutions | 1:500 | M, H |
| 05-636 Lot# 32526 Clone JBW301 (Billerica, MA, USA) | ||||
| p53 | Mouse | BioVision 3036-100 (Mountainview, CA, USA) | 1:100 | M |
| p21 | Mouse | Santa Cruz Biotechnology (F-5) sc-6246 | 1:500 | M |
| GFP | Chicken | Abcam ab 13970 | 1:2000 | M |
| Ki67 | Rat | DakoCytomation Clone TEC-3 M7249 (Ely, UK) | 1:25 | M |
| Cleaved Caspase 3 (Asp175) | Rabbit | Cell Signalling 9664 (Danvers, MA, USA) | 1:100 | M |
| Smooth muscle actin | Mouse | DakoCytomation Clone 1A4 M0851 | 1:200 | M, H |
| F4/80 | Rat | Serotec MCA1957 (Kidlington, UK) | 1:100 | M |
| MCM2 | Goat | Santa Cruz Biotechnology (N-19) sc-9839 | 1:100 | H |
| p16 | Mouse | Santa Cruz Biotechnology (F-12) sc-1661 | 1:200 | H |
| p15 | Rabbit | Cell Signalling 4822 | 1:100 | H |
| p14 | Rabbit | Abcam ab3642 | 1:100 | H |
| CD68 | Mouse | DakoCytomation Clone PG-M1 M0876 | 1:100 | H |
Abbreviations: H, human; M, mouse.
Figure 1.
Absence of enrichment of putative OIS biomarkers in hPanIN and presence of markers in dividing cells. Formalin-fixed paraffin-embedded human pancreatic PanIN and PDA samples were acquired from Addenbrookes' Teaching Hospitals NHS Trust, Cambridge, UK, or from the Department of Pathology, Johns Hopkins University School of Medicine in Baltimore, MD, USA. Ethical approval was granted by the local research and ethics committee (LREC Number: 08/H0306/32). (a) Proliferation content of hPanIN and PDA by using marker MCM2; each data point represents the average of at least four fields from the human sample (N = 5). (b) p16INK4a, p19ARF, Dec1 and γH2AX are present and measurable in hPanIN and PDA. Species-specific IgG was used as a negative control (left column). Arrows denote biomarker positive cells. After heat-induced antigen retrieval for 12.5 min in 0.01 m sodium citrate, immunohistochemistry was performed using antibodies at dilution denoted in Table 1. Primary antibodies were incubated overnight and species-specific biotinylated secondary antibodies followed by streptatvidin-peroxidase conjugate, developed with DAB and counterstained with hematoxylin. Scale = 200 μm. (c) Quantification of putative OIS biomarker abundance in hPanIN and PDA. Each bar represents at least four hPanIN or PDA samples. (d) Coimmunofluorescence experiments for potential OIS biomarker (green) and proliferating antigen MCM2 (red) demonstrate colocalization of these two biomarkers in PanIN. Nuclei outlined by white dotted line. OIS/MCM2 double-positive cells are denoted by white arrows. OIS biomarker positive only cells are denoted by white arrowheads. After heat-mediated antigen retrieval in 0.01 m sodium citrate for 12.5 min and blocking in donkey serum for 30 min, primary OIS biomarker and proliferation biomarker were applied together overnight. The next day, secondary antibodies conjugated to the appropriate fluorophore were applied for 30 min at 37 °C, followed by two washes in phosphate-buffered saline and mounting in Prolong Gold antifade reagent containing 4,6-diamidino-2-phenylindole (Invitrogen, Carlsbad, CA, USA). Scale = 200 μm. (e) Quantification of OIS biomarker expression in proliferating cells. Each bar represents at least four hPanIN or PDA samples. Error bars indicate s.e.m.
OIS biomarker expression in mPanIN
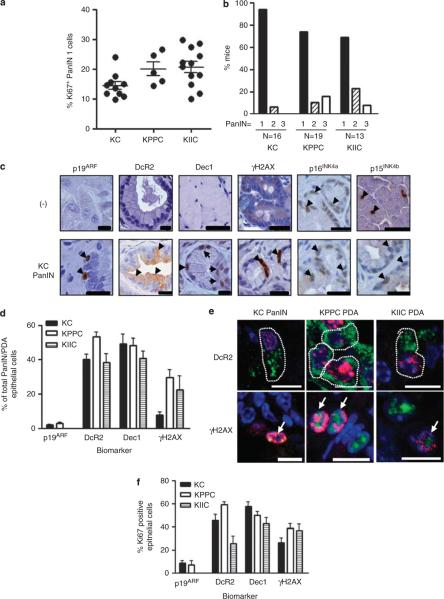
Loss of tumor suppressor gene expression has been functionally linked to senescence bypass; therefore, mPanIN and mPDA sections taken at endpoint from KC mice (8–14 months old) were compared with KPPC and KIIC mice (2 months old) for proliferation, mPanIN grade and OIS biomarker expression. mPanIN 1 from KPPC and KIIC pancreata were more proliferative than mPanIN from KC mice (Figure 2a), and KPPC and KIIC mPanIN lesions were more advanced than mPanIN from KC mice (Figure 2b), supporting a functional role for these tumor suppressor genes in mPanIN biology. Using genetically null tissue as a negative control, immunohistochemical analysis did not reveal specific p16INK4a and p15INK4b protein expression with the antibodies employed (Figure 2c). The remaining biomarkers, including p19ARF, Dec1, DcR2 and γH2AX were specific and detectable in the murine pancreas (Figure 2c), but were not enriched in KC mPanIN relative to KPPC and KIIC mPDA (Figure 2d). In addition, coimmunofluorescent analysis of markers for proliferation and OIS in KC mPanIN, and KPPC and KIIC mPDA (Figure 2e) revealed that more than a quarter of Dec1-, DcR2- and γH2AX-positive cells were also positive for Ki67 (Figure 2f). Therefore, the majority of biomarkers commonly associated with senescence in vivo can be detected in proliferating cells and therefore cannot be used as definitive biomarkers for the senescent state. Rather, these proposed biomarkers may identify cell stress states that are irrespective of the proliferative potential of pancreatic preneoplasms.
Figure 2.
Putative OIS biomarker expression is unchanged in mPanIN, conditionally mutant for p53 or Cdkn2a. All animals were housed in accordance with the UK Home Office, within the designated vivarium at the Cancer Research UK Cancer Research Institute. Pancreatic-specific expression of a conditional point-mutated allele of Kras (LSL-KrasG12D) elicits mPanIN and PDA that exhibit pathology, consistent with the human condition (Hingorani et al., 2003). Acceleration of tumor development was accomplished by interbreeding these KrasG12D;Cre (KC) mice with mice harboring a conditional floxed allele of Trp53 or Ink4A/ARF to generate the KrasG12D;p53flox/flox;Cre (KPPC) or KrasG12D;Ink4A/ARFflox/flox;Cre (KIIC) strains, respectively. Rosa26-RYFP allele (Srinivas et al., 2001) was bred to KC mice to label cre-mediated recombination in the pancreas. Archival formalin-fixed paraffin-embedded KC, KPPC and KIIC pancreata were sectioned for immunohistochemical analyses. (a) Proliferation of KC, KPPC and KIIC PanIN samples. Each data point represents the average of at least five fields from a single mouse. Statistical analyses were carried out using Graphpad Prism version 5.0 (GraphPad Software, San Diego, CA, USA). Mann–Whitney U-test was used to determine significance: KC versus KPPC, P = 0.0400; KC versus KIIC, P = 0.0029. (b) KPPC and KIIC mice harbor more high-grade PanIN, compared with KC mice. (c) Immunohistochemical analysis of potential OIS biomarker expression in mouse. Knockout mouse tissue was used as a negative control to evaluate the specificity of all markers, except DcR2. DcR2 negative control is species-specific IgG (top row). Arrowheads denote positive cells. Scale = 200 μm. (d) Quantification of potential OIS biomarker expression in murine KC PanIN, and KPPC and KIIC PDA. At least three representative images from each mouse were quantified, the average of which represents a single data point. (e) Immunofluorescent detection reveals that Dec1, DcR2, γH2AX and p19ARF (green) can be colocalized in Ki67 (red)-positive epithelial cells in mPanIN and PDA. Scale = 10 μm. White arrows denote double positive cells. (f) Quantification of coimmunofluorescent staining indicates that a significant fraction of Ki67-positive cells harbor putative OIS biomarkers. Error bars indicate s.e.m. (N = at least 4). At least three representative images from each mouse were quantified, the average of which represents a single data point.
OIS biomarker expression in tumor microenvironment cells
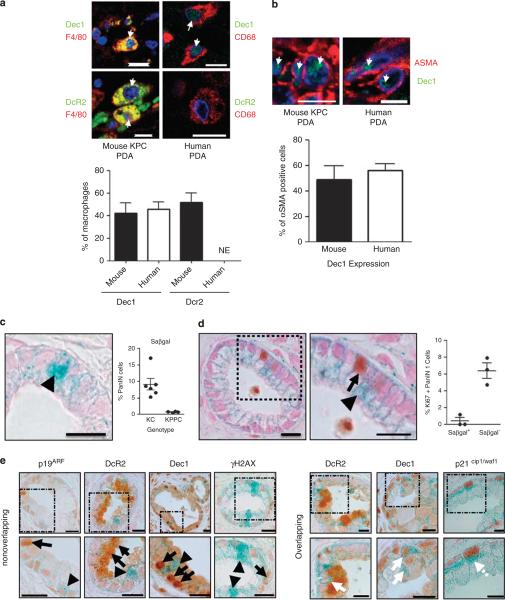
Many of the biomarkers frequently associated with senescence in both murine and human pancreatic samples were visible in non-epithelial cell types adjacent to PanIN and PDA. Cancer-associated fibroblasts and tumor-associated macrophages comprise nearly 50% of the cell types present in murine and hPDA (data not shown). Coimmunofluorescence revealed that Dec1 was expressed in half of the cancer-associated fibroblasts and tumor-associated macrophages in mouse and hPDA (Figures 3a and b). DcR2 was present in half of murine tumor-associated macrophages, but was undetectable in human samples (Figures 3a and b). Therefore, these two biomarkers are neither specific nor selective for the growth-arrested epithelial cells in vivo. This is consistent with the original characterization of Dec1 expression, where many normal tissues, including macrophages in the lung, were reported to express Dec1 (Ivanova et al., 2005).
Figure 3.
SAβgal labels a small population of growth-arrested PanIN cells. After heat-mediated antigen retrieval in 0.01 m sodium citrate for 12.5 min and blocking in donkey serum for 30 min, two primary antibodies were applied together overnight. The next day, secondary antibodies conjugated to the appropriate fluorophore were applied for 30 min at 37 °C, followed by two washes in phosphate-buffered saline (PBS) and mounting in Prolong Gold antifade reagent containing 4,6-diamidino-2-phenylindole. (a) Dec1 (green) is found in a significant fraction of murine and human tumor-associated macrophages. DcR2 (green) is found in murine, but not human tumor-associated macrophages. NE, not expressed (human N=5, mouse N=4). Arrows denote double positive cells. (b) Coimmunofluorescence for Dec1 (green) and αSMA (red) shows Dec1 is expressed in cancer-associated fibroblasts. Quantification is shown below. Scale=10 μm. Error bar indicates s.e.m. (c) Freshly dissected pancreas was cut into 1-3 mm pieces and fixed in 4% paraformaldehyde (PFA) (pH 6.0) for 2 h on ice. Samples were washed with PBS and tissue was incubated overnight in SAβgal staining solution at 37 °C. The following day, the samples were washed with PBS then fixed in 10% formalin overnight. The next day the tissues were washed with PBS and processed using reduced xylene exposure times. 4 μm sections were cut, counterstained with nuclear fast red (Sigma, St Louis, MO, USA) and coverslipped for immediate imaging or first subjected to immunohistochemistry. Representative image of SAβgal stain (left). Arrowhead denotes Saβgal-positive cell. SAβgal expression in KC PanIN and conditional biallelic deletion of Trp53 rescues the majority of PanIN SAβgal content (right). Each dot represents the average of at least five images for a single mouse of the indicated genotype. (d) Slides were dewaxed in xylenes (1 min shaking) and rehydrated in 100% ethanol (40 s shaking) and 70% ethanol (20 s shaking). Antigen unmasking was performed in 10 mm sodium citrate buffer, pH 6.0. Immunohistochemistry was performed as described above with the exception that slides were counterstained for 10 s with nuclear fast red (Sigma). Ki67 immunohistochemistry overlaid on SAβgal stained KC PanIN tissue reveals a mutually exclusive staining pattern (inset at right). Arrowhead denotes SAβgal positive cells. Arrow denotes Ki67 positive cell. Scale 200 μm. Quantification demonstrating SAβgal is largely excluded from Ki67-positive cells at the right. (e) Additional putative OIS biomarkers are not enriched in SAβgal expressing PanIN cells. Arrows denote OIS-associated biomarker-positive cells. Arrowheads denote SAβgal-positive cells. White arrows denote OIS/SAβgal double-positive cells. Scale=200 μm.
SAβgal labels senescent PanIN subpopulation
We next assessed whether SAβgal could denote senescent cells in mPanIN, and found that approximately 10% of KC PanIN 1 cells were SAβgal positive, with KPPC PanIN-1 exhibiting a significant reduction in SAβgal positivity (Figure 3c), consistent with the proposed staining pattern for OIS biomarkers. To confirm that SAβgal indeed denoted senescent cells, a modified SAβgal staining protocol was devised to enable the coincident immunohistochemical detection of biomarkers in tissue sections. Importantly, SAβgal was rarely expressed in Ki67 positive mPanIN cells (Figure 3d), although the pancreatic tissue demonstrated decreased sensitivity to antigen detection as the Ki67 index was decreased from 14% (Figure 2a) to 7% (Figure 3d). Lastly, although some overlapping occurred, there was no clear enrichment for the putative OIS biomarkers with SAβgal (Figure 3e and data not shown), further suggesting that these proteins cannot be used to definitively specify senescent cells in PanIN.
Our results should prompt an evaluation of the experimental methodologies used to detect OIS in vivo. Indeed, 10% of PanIN 1 cells are senescent in this study, based on SAβgal positivity, and presumably, this reflects the same proportion of cells detected by immunohistochemical methods in other reports, although quantification in these studies was not provided (Collado et al., 2005; Morton et al., 2009). Senescence in PanIN appears to be balanced by proliferation, as each comprises approximately 10% of all PanIN 1 cells. Analyses of future OIS biomarkers should ensure that they are not expressed in normal or premalignant proliferating cells.
Subpopulations of senescent cells are wild type or mutant for Kras
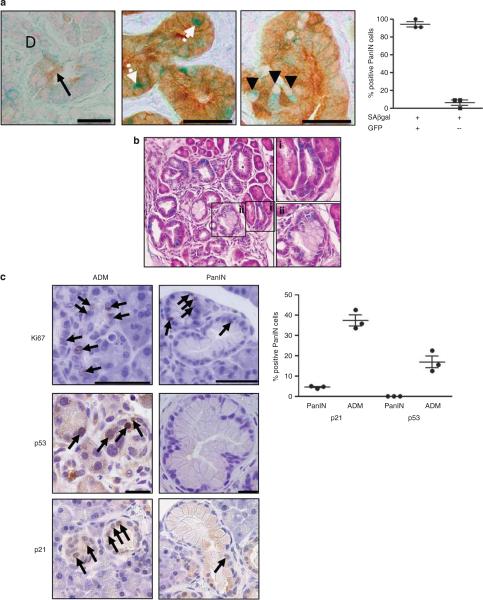
Senescence has also been shown to occur under physiological stress, in which cells are otherwise genotypically wild type. Indeed, senescence of liver stellate cells impairs carbon tetrachloride-induced hepatic fibrosis (Krizhanovsky et al., 2008). Therefore, to determine if wild-type pancreatic epithelial cells contributed to the population of senescent PanIN cells, a Cre green fluorescent protein reporter allele was used to denote recombination and acted as a surrogate for KrasG12D expression in mPanIN. On the basis of this approach, the SAβgal positive mPanIN cells were subdivided into a larger population of green fluorescent protein-positive PanIN cells (91.5%) and a smaller green fluorescent protein-negative population (8.5%; Figure 4a). This result suggests that mPanIN are heterogenous for Cre activation, and may therefore contain a proportion of co-opted wild-type pancreatic epithelial cells that have undergone senescence, potentially as a result of stress-associated secreted factors (Kuilman and Peeper, 2009).
Figure 4.
ADM exhibits additional senescence block by SAβgal, p21 and p53 expression. (a) Green fluorescent protein (GFP) immunohistochemistry overlaid on SAβgal stained PanIN tissue. Left panel demonstrates background from GFP antibody in acinar (black arrow), but not ductal tissues (labeled D) from a mouse lacking the GFP reporter. White arrows denote GFP/SAβgal-positive cells; arrowheads denote GFP-negative/SAβgal-positive cells. Quantification of cells expressing SAβgal and GFP reveals a small population of GFP-negative PanIN cells shown at right. (b) Hematoxylin and eosin counterstain over SAβgal stained KC pancreata. SAβgal is comparably detected in both (i) ADM and (ii) PanIN. (c) Ki67, p53 and p21 immunohistochemical detection in ADM and PanIN from KC mice. Arrows denote single-positive cells. Quantification of p21 and p53 positivity in PanIN and ADM regions, respectively, shown at right (N=3).
The absence of green fluorescent protein reporter expression can be explained by one of two scenarios: either these are Kras wild type cells, or instead signify unequal recombination by Cre at the Kras locus and the Rosa26-RYFP locus. It is not unprecedented for Kras wild type cells to contribute to PanIN structures, as only 45% of the lowest grade hPanIN harbor Kras muta tions (Hruban et al., 1993, 2000; Moskaluk et al., 1997). Indeed, the SAβgal negative PanIN cells in this report may mirror the inflammation-induced senescent population of wild-type ductal cells observed by Lee and Bar-Sagi (2010). Whether wild-type ductal or acinar cells contribute to this subpopulation of senescent cells remains to be determined; however, acinar cells have been described within PanIN structures (Zhu et al., 2007). Nonetheless, as this subpopulation of potentially Kras wild-type senescent PanIN cells represents less than 1% of all PanIN cells, the relevance of these cells is uncertain.
OIS biomarker expression in ADM versus PanIN
ADM is an additional pancreatic preneoplastic lesion that arises during inflammatory conditions and is considered morphologically and genetically distinct from PanIN in human samples (Hruban et al., 2006; Shi et al., 2009). In several mouse models, ADM has been reported to directly precede mPanIN formation (De La et al., 2008; Habbe et al., 2008). ADM are proliferative lesions consisting of swollen acinar cells, with a PanIN-like appearance, and surrounded by a reactive stroma (Hruban et al., 2006). In the KC mice, mPanIN arise in the middle of otherwise normal looking pancreatic epithelium, but ADM can also be observed nearby or separately. Using a modified histochemical staining method on pancreatic tissues previously processed for SAβgal activity, SAβgal expression was as readily detectable in ADM as in mPanIN (Figures 3c and 4b, and data not shown), suggestive of an additional independent senescence block.
In addition, although mPanIN have been previously reported to contain abundant p21 and p53 protein, we found that both proteins were either minimally present or undetectable in KC mPanIN (Figure 4c). Instead, ADM was highly positive for both of these biomarkers (Figure 4c). This discrepancy to reported results likely stems from differences in pathological evaluation. The high p21 and p53 protein content reported by Morton et al. (2009) appears to localize to ADM, rather than to actual mPanIN, which would reconcile their data with that presented in this study. Together, these results are consistent with the human data and suggest that the majority of PanIN cells do not harbor high levels of p53-activating signals. Nonetheless, we agree with Morton et al. (2009) that p53 does have a role in mPanIN OIS, as SAβgal is the only biomarker sufficient to delineate senescent cells in mPanIN, and conditionally, p53-deficient mPanIN are rarely SAβgal positive.
Our data confirms the utility of SAβgal as a biomarker of the senescent state in pancreatic preneoplasia. As this approach is cumbersome and requires fresh tissue for processing, our findings should promote the development of additional methods to detect senescence in pancreatic tissues and to further characterize the biology of oncogene-induced senescence. In addition, the presence of biomarkers frequently associated with OIS in both ADM and PanIN emphasizes the need to clarify the cell of origin for PanIN and PDA.
Acknowledgements
We thank P Krimpenfort, A Berns, R Taneja, F Alt, A Harris, M Malumbres C, Trejo, M McMahon, M Ferreira and F Watt for tissues and reagents. We thank Frances Connor and other members of the Tuveson lab for assistance and advice, the CRI animal care staff; S Vowler for statistical advice; Cancer Research UK Histopathology and In situ Hybridization Core, and the Microscopy and Imaging Core. This research was supported by the University of Cambridge and Cancer Research UK, The Li Ka Shing Foundation and Hutchison Whampoa Limited, the NIHR Cambridge Biomedical Research Centre and the NIH (grants CA101973, CA111294, CA084291 and CA105490) to DAT.
Footnotes
Conflict of interest The authors declare no conflict of interest.
References
- Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Chien Y, Lowe SW. Secreting tumor suppression. Cell. 2008;132:339–341. doi: 10.1016/j.cell.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- Collado M, Serrano M. The power and the promise of oncogene-induced senescence markers. Nat Rev Cancer. 2006;6:472–476. doi: 10.1038/nrc1884. [DOI] [PubMed] [Google Scholar]
- Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, et al. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois-Cox S, Jones SL, Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27:2801–2809. doi: 10.1038/sj.onc.1210950. [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21:379–384. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La OJ, Emerson LL, Goodman JL, Froebe SC, Illum BE, Curtis AB, et al. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci U S A. 2008;105:18907–18912. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, et al. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Collado M, Navas C, Schuhmacher AJ, Hernandez-Porras I, Canamero M, et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell. 2011;19:728–739. doi: 10.1016/j.ccr.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habbe N, Shi G, Meguid RA, Fendrich V, Esni F, Chen H, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A. 2008;105:18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Hruban RH, Adsay NV, Albores-Saavedra J, Anver MR, Biankin AV, Boivin GP, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 2006;66:95–106. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin Cancer Res. 2000;6:2969–2972. [PubMed] [Google Scholar]
- Hruban RH, van Mansfeld AD, Offerhaus GJ, van Weering DH, Allison DC, Goodman SN, et al. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol. 1993;143:545–554. [PMC free article] [PubMed] [Google Scholar]
- Ivanova A, Liao SY, Lerman MI, Ivanov S, Stanbridge EJ. STRA13 expression and subcellular localisation in normal and tumour tissues: implications for use as a diagnostic and differentiation marker. J Med Genet. 2005;42:565–576. doi: 10.1136/jmg.2004.029835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- Lee KE, Bar-Sagi D. Oncogenic KRas suppresses inflammation-associated senescence of pancreatic ductal cells. Cancer Cell. 2010;18:448–458. doi: 10.1016/j.ccr.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder PK, Grisanzio C, O'Connell F, Barry M, Brito JM, Xu Q, et al. A prostatic intraepithelial neoplasia-dependent p27 Kip1 checkpoint induces senescence and inhibits cell proliferation and cancer progression. Cancer Cell. 2008;14:146–155. doi: 10.1016/j.ccr.2008.06.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Morton JP, Jamieson NB, Karim SA, Athineos D, Ridgway RA, Nixon C, et al. LKB1 haploinsufficiency cooperates with Kras to promote pancreatic cancer through suppression of p21-dependent growth arrest. Gastroenterology. 2010;139:586–597. 597, e1–6. doi: 10.1053/j.gastro.2010.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci U S A. 2009;107:246–251. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57:2140–2143. [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- Prieur A, Peeper DS. Cellular senescence in vivo: a barrier to tumorigenesis. Curr Opin Cell Biol. 2008;20:150–155. doi: 10.1016/j.ceb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Shi C, Hong SM, Lim P, Kamiyama H, Khan M, Anders RA, et al. KRAS2 mutations in human pancreatic acinar-ductal metaplastic lesions are limited to those with PanIN: implications for the human pancreatic cancer cell of origin. Mol Cancer Res. 2009;7:230–236. doi: 10.1158/1541-7786.MCR-08-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Yoshizuka N, New L, Moser BA, Li Y, Liao R, et al. PRAK is essential for ras-induced senescence and tumor suppression. Cell. 2007;128:295–308. doi: 10.1016/j.cell.2006.11.050. [DOI] [PubMed] [Google Scholar]
- Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Shi G, Schmidt CM, Hruban RH, Konieczny SF. Acinar cells contribute to the molecular heterogeneity of pancreatic intraepithelial neoplasia. Am J Pathol. 2007;171:263–273. doi: 10.2353/ajpath.2007.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]