Abstract
Purpose
To evaluate the effectiveness of a fully automated postprocessing filter algorithm in pulsed arterial spin labeling (PASL) MRI perfusion images in a large clinical population.
Materials and Methods
A mean and standard deviation-based filter was implemented to remove outliers in the set of perfusion-weighted images (control – label) before being averaged and scaled to quantitative cerebral blood flow (CBF) maps. Filtered and unfiltered CBF maps from 200 randomly selected clinical cases were assessed by four blinded raters to evaluate the effectiveness of the filter.
Results
The filter salvaged many studies deemed uninterpretable as a result of motion artifacts, transient gradient, and/or radiofrequency instabilities, and unexpected disruption of data acquisition by the technologist to communicate with the patient. The filtered CBF maps contained significantly (P < 0.05) fewer artifacts and were more interpretable than unfiltered CBF maps as determined by one-tail paired t-test.
Conclusion
Variations in MR perfusion signal related to patient motion, system instability, or disruption of the steady state can introduce artifacts in the CBF maps that can be significantly reduced by postprocessing filtering. Diagnostic quality of the clinical perfusion images can be improved by performing selective averaging without a significant loss in perfusion signal-to-noise ratio.
Keywords: Clinical Perfusion MRI, Arterial Spin Labeling, PASL Filtering
Arterial Spin Labeling (ASL) is a noninvasive magnetic resonance imaging (MRI) method for measuring cerebral blood flow (CBF). Recent studies (1) have shown great potential for ASL in clinical imaging. ASL is completely noninvasive, repeatable, quantitative, and has shown close correlations with other blood flow imaging techniques including dynamic susceptibility contrast MRI, computed tomography perfusion, positron emission tomography, and single photon emission computed tomography (2).
While other imaging modalities use an exogenous contrast agent, ASL uses the water in the blood as an endogenous tracer to measure blood flow from a set of tagged (label) and untagged (control) images. The control and label image pairs are subtracted from each other to eliminate the static tissue signal while retaining the signal from the blood that has perfused into the imaging slices. Since the static tissue signal is much larger than the signal from the blood, instabilities between the control/label images resulting from patient motion, physiological conditions, hardware instabilities, and unexpected mid-scan interruptions by the technologist to communicate with the patient can lead to large subtraction errors. While these instabilities may occur in only a small percentage of the control/ label images, the effect can have a drastic and detrimental affect on the final perfusion-weighted image (PWI) obtained by averaging all of the individual PWIs together in the time series. These subtraction errors between the control and label images can manifest themselves in the individual PWIs as hyper- or hypoin-tense perfusion signal involving just a few slices or the entire brain volume. By selectively discarding these inconsistent images in the perfusion time series, the final averaged PWI can be improved with only a minor penalty in the signal-to-noise ratio (SNR).
A number of techniques have been developed to improve CBF maps acquired with ASL: physiological noise reduction for functional perfusion experiments (3,4), applying a low pass filter to the subtracted control/ label images (5), static tissue background suppression methods (6), and selective averaging of control/label images based on motion criteria (7). While these methods have been shown to be of importance when examining the spectrum of the perfusion time series for functional MRI ASL studies, they may not be able to correct for dramatic signal variations caused by hardware instability, large and abrupt patient motion, and unexpected disruption of the steady state.
One previous study (7) has shown that the quality of CBF maps can be improved by removing control/label pairs that were acquired during periods of motion exceeding 2 mm of translations and 1.5 degrees of rotations. The remaining control/label images were subtracted from each other and averaged to create a final PWI. The final PWIs for all cases were visually inspected. Those with severe residual artifacts were removed from the study for further analysis. For research studies with a dedicated staff team, visual inspection is certainly a viable method for postprocessing and improving perfusion image quality. However, as perfusion imaging becomes readily available for clinical applications, it is neither feasible nor practical to manually process each ASL case. An automated method is necessary for improving the perfusion image quality in a clinical environment.
At our institution, a pulsed ASL (PASL) CBF map is acquired as part of the standard clinical MRI protocol for patients with an indication of vascular abnormalities (stroke, transient ischemic attack, or carotid stenosis), neoplasm, or seizure. Over 10,000 clinical CBF maps (8–10) with a rate of 15–20 cases per day have been acquired in a time period of 2 years using an automated image processing pipeline (11). This represents the largest experience with PASL in clinical practice to date. In this article we describe an automated postprocessing filtering procedure that effectively improves the overall quality and stability of the clinical perfusion measurement.
Materials and Methods
PASL Acquisition and Reconstruction
After obtaining Institutional Review Board (IRB) approval from our institution, this study used data from our clinical patient population undergoing an MRI examination. The purpose was to evaluate the effectiveness of a postprocessing filter to improve perfusion images acquired for diagnostic purposes. Image acquisitions were done using five clinical MRI scanners (three 1.5T GE Signa Software Platform 12M5, one 1.5T GE Twinspeed Software Platform 14M5, and one 3T GE Twinspeed Software Platform 12M5, GE Healthcare, Milwaukee, WI). Body coil transmit with receive-only head coils (HD 8 Channel HiRes Brain Array Coil and 4 Channel Neurovascular Coil manufactured by Invivo Devices, Gainesville, FL) were used for data collection.
CBF was measured with QUantitative Imaging of Perfusion using a Single Subtraction with Thin Slice TI1 Periodic Saturation (QUIPSS II TIPS a.k.a. Q2TIPS) (12,13) with Flow-sensitive Alternating Inversion Recovery (FAIR) (14) acquiring 60 control/label pairs in 6.3 minutes using a single shot, ramp sampled gradient echo EPI. The pulse sequence was developed and validated internally. The acquisition parameters are listed in Table 1. A proton density weighted (M0) image was acquired at the beginning of the Q2TIPS-FAIR sequence for scaling the final PWI to a quantitative CBF map.
Table 1. Q2TIPS-FAIR Implementation Parameters.
| Parameter | Value |
|---|---|
| FOV / slice thickness | 240mm / 8mm |
| TI1 / TI1s / TI2 / TR | 800ms / 1200ms / 2000ms / 3000ms |
| Acquisition matrix | 64 × 64 |
| TE | 24.1 ms |
| Number of slices | Varying from 7 - 17 |
| b (diffusion gradient) | 5.25 mm2/sec |
The imaging data were automatically transferred from the MRI scanners to the offline workstations for fully automated reconstruction using the Sun Grid Engine (Sun Microsystems, Santa Clara, CA) for distributed processing. The individual control and label imaging volumes were realigned with the software package SPM2 (Statistical Parametric Mapping, Wellcome Department of Imaging Neuroscience, London, UK). The PWI was then scaled to a quantitative CBF map using the General Kinetic Model as described by Buxton et al (15). Magnetic field strength differences were accounted for during quantification by assuming different relaxation times for the blood (T1 of the blood was assumed to be 1200 msec at 1.5T and 1490 msec at 3T (16)). As part of the automated processing pipeline, the final CBF maps were transferred to the clinical Picture Archiving and Communications System (PACS, AGFA, Mortsel, Belgium) for system-wide review.
ASL Filtering
In PASL, the overall MR signal by the delivered blood is about 1% of the total signal caused by the tissue, resulting in very low SNR for a single subtraction image (17). The control/label pairs are thus acquired multiple times such that the subtraction images are averaged together to achieve proper SNR for diagnostic purposes. Since the control/label images are acquired under identical conditions, the resulting PWIs should be identical, with the exception of noise. However, we have observed that a few PWIs can have significant signal variation leading to a variety of imaging artifacts, and thereby rendering CBF results of poor diagnostic quality. The quality of the CBF map can be significantly improved by selectively removing those corrupted PWIs. Our ASL filtering technique was to improve the quality of the final perfusion image by automatically identifying and discarding the inconsistent PWIs from the control/label subtractions.
Before filtering, a brain tissue mask was created by averaging all the control and label images together. Pixels of values greater than 5% of the maximum of the averaged image were classified as brain tissue, whereas the remaining pixels were classified as background. The tissue mask was dilated with a 4-pixel wide kernel to ensure entire brain coverage.
Two criteria, the mean and the standard deviation, were implemented in the filter. The mean criterion detected PWIs with artificial hyper- or hypointensity. The mean intensity of voxels classified as tissue from the PWI, denoted ΔMi, was calculated. The mean (μΔM) and the standard deviation (σΔM) of ΔMi for all the PWIs were calculated. Individual PWIs with mean tissue value ΔMi that satisfied:
| [1] |
were classified as outliers and were removed from the set of PWIs to be averaged together. The parameter n represents a scaler. A threshold of n = 2.5 was chosen based on empirical performances of the filter from over 100 initial clinical cases prior to the statistical evaluation (discussed below).
In the ideal situation, the signal variation among tissue voxels in a single PWI is expected to be similar to the background noise due to the low SNR. The standard deviation criterion was designed to identify and remove PWIs with large signal variation in the region classified as tissue when compared with the average PWIs. The standard deviation of the tissue voxels in the PWI, denoted Si, was calculated. The mean (μS) and the standard deviation (σS) of Si across the set of PWIs were calculated. Individual PWIs with standard deviation value Si that satisfied:
| [2] |
were classified as outliers and removed from the set of PWIs to be averaged together. The parameter m represents a scaler and m = 1.5 was determined based on empirical performances of the filter from same initial dataset as the mean criterion that was used to determine the scaler variable n.
To minimize overfiltering, a PWI was considered “stable” when the natural log of the difference value between max(Si) and min(Si) was less than 1, that is:
| [3] |
When this criterion was satisfied, all of the individual PWIs were included in the final averaging step. The software for the ASL filter was written in MatLab (Math-Works, Natick, MA) and inserted into our automated PASL postprocessing pipeline.
The overall design of the ASL filtering process is illustrated in Fig. 1. Two criteria, the mean and the standard deviation across the set of PWIs, were executed in parallel to remove inconsistent PWIs as previously described. It was also observed that instability could affect all or only a few slices within the brain volume. Hence, the filtering procedure was performed on both a volume-by-volume basis and a slice-by-slice basis. This allowed removal of an entire volume, or just a few slices within a single volume of the corrupted dataset. In the volume-based approach, all the slices were treated together as a single volume for which ΔMi and Si were calculated along the temporal domain. In the slice-based approach, ΔMi and Si were calculated for each individual slice along the temporal domain. The remaining images that were not identified as outliers were used for the calculation of the final averaged PWI. The SNR of the final averaged PWI undergoing filtering is slightly reduced according to:
Figure 1.
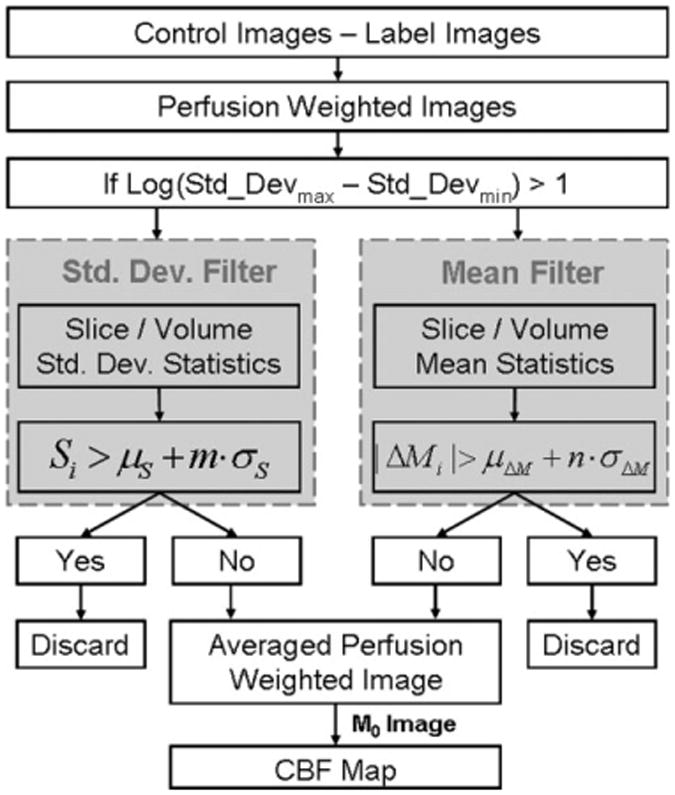
PASL filter processing steps. The tissue data are extracted by masking out the background noise. The filtering based on the mean and standard deviation is performed in both volume-based and slice-based fashions, for which the union of the results determines the subtraction images that are discarded from the final calculation of the perfusion-weighted image.
| [4] |
where Ntotal is the total number subtraction images before filtering and Nfiltered is the actual number of volumes used for averaging after filtering. On average, the ASL filter removed five images per slice, corresponding to an SNR loss of 4%.
Image Quality Assessment and Statistical Analysis
CBF maps of 200 clinical cases randomly selected from a total of 540 cases performed in February, 2008, were to be included in the study. The patient population of those 200 cases was representative of the clinical patients who had CBF maps acquired as part of their MRI examinations. Briefly, the study included 88 males and 112 females ranging from 0.3–91 years old with a mean age of 48. From these 200 cases the MRI examinations as determined by a board-certified radiologist revealed that 33% were considered normal (ie, no clinical findings); 14.5% had tumor; 11% had undergone brain surgical procedures; the remaining cases consist of pathologies including seizure, infarct, atrophy, metastases, bleed, arachnoid cyst, arteriovenous malformation, and anoxia.
The filtered and unfiltered CBF maps from these patients were independently analyzed in a random order by three board-certified neuroradiologists experienced in clinical PASL interpretation and one board-certified physician with significant radiology training and experience with research PASL analyses. The observers were blinded to the medical history of the patients and evaluated the CBF images on image quality criteria only. Intraobserver variability was measured by having 50 cases randomly selected out of the 200 to be rated twice. Prior to the individual analysis, all observers had a training session discussing the baseline rules for quality assessment followed by a trial dataset for quality assurance. The observers were asked to rate the individual images based on the overall image interpretability and three representative artifacts including edge, ring, and CSF shine-through (see Fig. 3; details of the artifacts were described in (8)). Adopted from McConnell et al (18), each potential artifact was rated on a scale of 0-3 where 0 indicated no gross artifacts and 3 indicated severe artifacts, except for CSF shine-through, which was either present or absent. To maintain consistency with artifact scores for the overall evaluation, we decided to use 0 for the most interpretable case and 3 for the least interpretable case. Rating details are described in Table 2.
Figure 3.
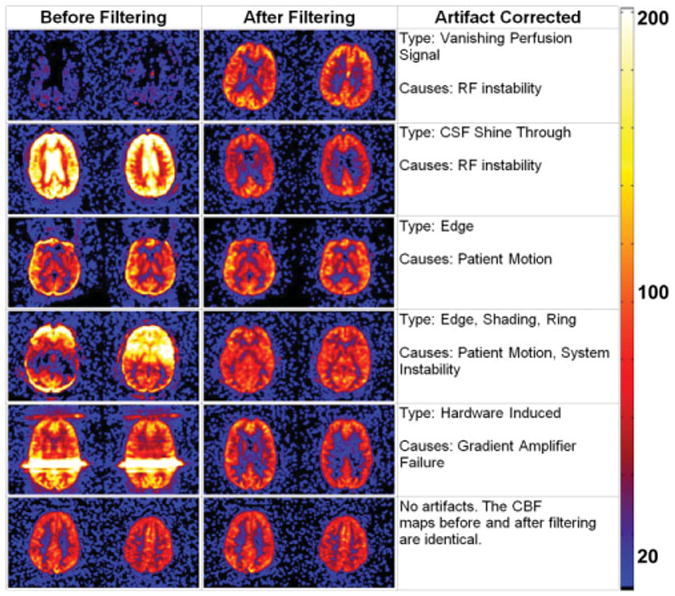
Examples of clinical CBF maps before and after ASL filtering for both unstable and stable cases. The scale for the color map is in milliliters of blood / 100 g of tissue / minute.
Table 2. Image Quality and Artifact Assessments.
| Scale | Interpretability Grading |
Edge & ring | CSF shine-through | ||
|---|---|---|---|---|---|
|
|
|
||||
| Scale | Grading | Scale | Grading | ||
| 0 | Interpretable | 0 | None | 0 | Nonexist |
| 1 | Interpretable with minor artifacts | 1 | Minor | 1 | Exist |
| 2 | Interpretable with major artifacts | 2 | Moderate | ||
| 3 | Uninterpretable | 3 | Severe | ||
The image quality scores in each category for the filtered and unfiltered CBF maps were calculated by averaging rating scores from all raters. A lower score indicated a more interpretable image with fewer artifacts. Since the primary function for the ASL filter was to remove outliers in the perfusion time series based on certain characteristics, the resulting time series would have less variation and, hence, be more stable. We wanted to focus on detecting improvements that the ASL filter has provided. To do so, a one-tail, paired t-test was performed to compare the ratings on the filtered and unfiltered CBF maps. P < 0.05 was considered significant for filtered CBF maps being easier to interpret with fewer artifacts. A two-sample t-test assuming equal variance was used to examine the intraobserver variability on the repeated 50 cases. If P > 0.05, then there was no significant difference between the rating scores from the first and second repeated trial for the same observer. The intraobserver statistical tests were two-tailed. The rating process was done via internally developed software.
Results
Filtering Results
As previously discussed, signal across PWIs should contain little variation for a stable case under ideal situations. Any unexpected fluctuation in the system could cause drastic signal changes in the perfusion time series producing various artifacts. Those fluctuations were seen by computing the standard deviation of the perfusion signal across all timepoints. The coefficients of variation of gray matter voxels were calculated along the temporal domain and averaged for all the cases to demonstrate how system fluctuations could affect the CBF maps (Fig. 2). The causes for those system fluctuations were speculated to be primarily associated with patient motion, hardware instability, or a disruption of the steady state by the technologist to communicate with the patient. The ASL filter has removed those artifacts in a significant number of cases. Examples of unstable and stable clinical perfusion cases before and after filtering are illustrated in Fig. 3. Many of the unstable cases before filtering presented little usable information. After removing a few corrupted volumes (>95% of the original data were kept), the perfusion information was recovered without significant loss of SNR.
Figure 2.
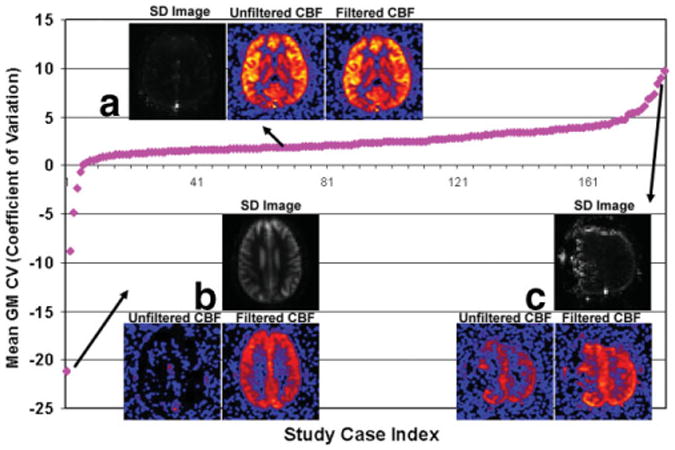
Mean coefficient of variation (CV) values were calculated for gray matter voxels for each perfusion case. Standard deviations (SD images) were calculated across perfusion time series to visually represent cases with large CV (magnitude) values. All SD images were normalized for comparison purposes. Three sample cases from which the ASL filter had no influence in the final CBF, completely recovered the perfusion signal, and recovered partial perfusion signal are shown in subfigure A, B, and C, respectively. It should be noted that CV can be used as one, but not an exclusive, factor to quantify the stability of a perfusion case. Although there was a trend that cases with high (magnitude) CV were less stable, exceptions may be made for patient with hypoperfusion. CV values were calculated for 185 cases out of 200. The remaining cases were excluded because the gray matter mask was either unavailable or visually inaccurate.
Unstable perfusion cases for which the ASL filter was ineffective were usually associated with continuous large movements from the patient throughout the data acquisition. Artificial hyper- and hypoperfusions could be observed among most PWIs in the perfusion time series, leaving little usable information and those cases were usually “unrecoverable.” Such scenarios usually required a rescan. It should be noted that not all hypoperfusion cases were artificial. Some may be associated with true physiological conditions that would be inherently difficult to measure with ASL methods. In many of these cases, the low ASL signal in the unfiltered and filtered maps was the reason why the filtered CBF maps were rated lower than the unfiltered CBF maps.
ASL Filter Evaluation Results
Excellent agreement was found within the repeated cases for all four observers (P > 0.5). The image quality scores for filtered CBF maps were better than unfiltered CBF maps across all categories. The individual scores for each observer were plotted in Fig. 4, and it illustrates that the filtered CBF maps had consistently better scores than the unfiltered CBF maps, although there existed some variations in the baseline scores among different observers. Overall, four raters (22%, 52%, 49%, and 26%) determined that on average 37% of all CBF maps (filtered and unfiltered) contained image artifacts. That is to say that a rater gave a score greater than 0 (no artifacts) for at least one of the three artifacts (edge, ring, and CSF shine-through) being assessed. On average among those images affected by artifacts, 39% were improved by the filtering process (40%, 37%, 41%, and 38%), 45% were unaffected by the filtering process (46%, 47%, 42%, and 46%). The remaining 16% of the cases where artifacts appeared to be worse with filtering were usually associated with hypoperfusion and severe artifacts (the interpretability scores for those cases were rated equal or greater than 2) and belonged to the “unrecoverable” category.
Figure 4.
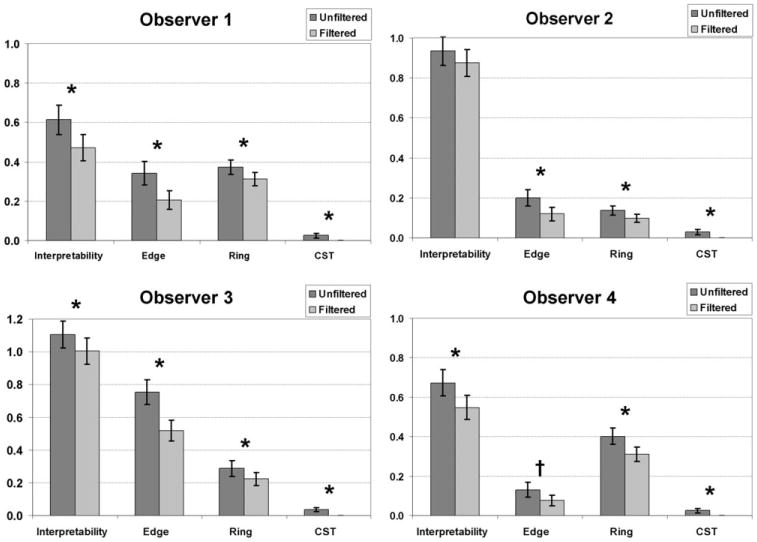
Bar charts showing image quality scores based on each of the four categories, interpretability, edge artifact, ring artifact, and CSF shine-through artifact, for all observers. *The rating score is significant that the filtered CBF map is better than the unfiltered CBF map. †The rating score indicates there is a trend that the filtered CBF map is better than the unfiltered CBF map.
According to the one-tail paired t-test, the ASL filter significantly reduced the ring artifact (P < 0.05) and completely eliminated CSF shine-through (P = 0). Three observers found that the ASL filter significantly reduced edge artifacts (P < 0.03), while one observer found there was a tendency for the ASL filter to reduce edge artifact (P ≈0.06). Three observers found that the ASL filter significantly improved the interpretability of the CBF maps (P < 0.04), while one neuroradiologist disagreed on the same issue (P > 0.1). The P-values for each rater are shown in Table 3. We observed that when the SNR of the CBF maps were low, some observers preferred unfiltered images with slightly higher SNR compared to the filtered image when artifacts were minor. This may explain the fluctuation in the results for image interpretability.
Table 3. Significance Scores (P-values) of Each Rater for All Categories.
| Observer | Interpretability | Edge | Ring | CSF shine-through |
|---|---|---|---|---|
| One | 0.019 | 0.009 | 0.034 | 0.012 |
| Two | 0.039 | 0.001 | 0.023 | 0.004 |
| Three | 0.141 | 0.023 | 0.031 | 0.007 |
| Four | 0.017 | 0.063 | 0.003 | 0.012 |
Discussion
The purpose of this work was to evaluate a method to improve the quality and stability of the absolute quantitative perfusion maps in a clinical environment where PASL is used routinely. The conventional PASL sequence was used in conjunction with an additional postprocessing filtering. The key design for the filter was to automatically detect an inconsistency within the PASL time series and to improve the quality and robustness of the CBF map by removing corrupted data.
A primary source of image artifacts in our CBF maps was related to patient motion that led to incomplete removal of the static tissue signal by imperfect subtraction of the control/label pairs. Background suppression methods such as ASSIST (6) have been introduced to help reduce these subtraction errors by applying additional inversion pulses to null out static tissue. Even with background suppression, a small amount of the static tissue signal (≈5%) is preserved in order to correct for patient motion. The residual static tissue signal after subtracting the control/label images, while significantly reduced, may still introduce some errors into the final perfusion image. Since the proposed filter in this study is completely done after image acquisition, we anticipate that this filter may further improve the diagnostic image quality of perfusion images acquired with background suppression techniques.
Our clinical experience with perfusion imaging has demonstrated that hardware system instability and disruption of the steady state occurs in a small percentage of our clinical cases. For example, in Fig. 3, image panel 5, a severe line artifact can be seen across the image. This was the result of the gradient fault of the phase encode gradient amplifier. In this particular case the scanner continued to acquire data. While background suppression would reduce the signal from the static tissue, it would not remove an unforeseen artifact caused by RF/gradient instabilities or technologists pausing and resuming the PASL acquisition due to uncooperative patients. While these instances do not occur often, they happen frequently enough to warrant the inclusion in the proposed filtering method to salvage these images.
We have developed and implemented a postprocessing filtering technique to optimize clinical perfusion imaging. The ASL filter was shown to significantly reduce motion and system-related image artifacts. Mechanisms were implemented to preserve good data and effectively prevent overfiltering. In stable perfusion cases the filter had little effect on the final CBF map. The filter processing time per case was on the order of seconds, adding no additional computational load to the processing pipeline. The implementation of the ASL filter was purely in the postprocessing stream, meaning the filter design could be incorporated into any ASL scheme without being restricted to a particular preparation and acquisition technique.
In conclusion, the ASL filtering technique has proven to be very clinically successful, salvaging perfusion images that would have been uninterpretable or difficult to interpret and making them clinically interpretable. In 2007–2008 we conducted over 10,000 clinical PASL scans. The ASL filter has become a necessity in improving the diagnostic quality and reducing image artifacts. The ASL filter salvaged ≈4% of the total cases in which CBF maps were completely uninterpretable to be diagnostically relevant. The ASL filter described here obviates the need to rescan patients in the event of an initially unusable scan, thus, implementation of such a filter can save time and resources for the patient and the hospital.
Acknowledgments
NIBIB R01EB004673, R01EB003880, Human Brain Project EB004673-02S2 NIAAA R01AA016748-02.
We thank Kathy Pearson and Ben Wagner for the help with computer programming and Dr. Hayasaka for helping with the statistical analysis.
References
- 1.Brown GG, Clark C, Liu TT. Measurement of cerebral perfusion with arterial spin labeling. Part 2. Applications. J Int Neuropsychol Soc. 2007;13:526–538. doi: 10.1017/S1355617707070634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wintermark M, Sesay M, Barbier E, et al. Comparative overview of brain perfusion imaging techniques. J Neuroradiol. 2005;32:294–314. doi: 10.1016/s0150-9861(05)83159-1. [DOI] [PubMed] [Google Scholar]
- 3.Restom K, Behzadi Y, Liu TT. Physiological noise reduction for arterial spin labeling functional MRI. Neuroimage. 2006;31:1104–1115. doi: 10.1016/j.neuroimage.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 4.Pfeuffer J, Van de Moortele PF, Ugurbil K, Hu X, Glover GH. Correction of physiologically induced global off-resonance effects in dynamic echo-planar and spiral functional imaging. Magn Reson Med. 2002;47:344–353. doi: 10.1002/mrm.10065. [DOI] [PubMed] [Google Scholar]
- 5.Liu TT, Wong EC. A signal processing model for arterial spin labeling functional MRI. Neuroimage. 2005;24:207–215. doi: 10.1016/j.neuroimage.2004.09.047. [DOI] [PubMed] [Google Scholar]
- 6.Ye FQ, Frank JA, Weinberger DR, McLaughlin AC. Noise reduction in 3D perfusion imaging by attenuating the static signal in arterial spin tagging (ASSIST) Magn Reson Med. 2000;44:92–100. doi: 10.1002/1522-2594(200007)44:1<92::aid-mrm14>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 7.Miranda MJ, Olofsson K, Sidaros K. Noninvasive measurements of regional cerebral perfusion in preterm and term neonates by magnetic resonance arterial spin labeling. Pediatr Res. 2006;60:359–363. doi: 10.1203/01.pdr.0000232785.00965.b3. [DOI] [PubMed] [Google Scholar]
- 8.Deibler AR, Pollock JM, Kraft RA, Tan H, Burdette JH, Maldjian JA. Arterial spin-labeling in routine clinical practice. Part 1. Technique and artifacts. AJNR Am J Neuroradiol. 2008;29:1228–1234. doi: 10.3174/ajnr.A1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deibler AR, Pollock JM, Kraft RA, Tan H, Burdette JH, Maldjian JA. Arterial spin-labeling in routine clinical practice. Part 2. Hypoperfusion patterns. AJNR Am J Neuroradiol. 2008;29:1235–1241. doi: 10.3174/ajnr.A1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deibler AR, Pollock JM, Kraft RA, Tan H, Burdette JH, Maldjian JA. Arterial spin-labeling in routine clinical practice. Part 3. Hyperperfusion patterns. AJNR Am J Neuroradiol. 2008;29:1428–1435. doi: 10.3174/ajnr.A1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maldjian JA, Laurienti PJ, Burdette JH, Kraft RA. Clinical implementation of spin-tag perfusion magnetic resonance imaging. J Comput Assist Tomogr. 2008;32:403–406. doi: 10.1097/RCT.0b013e31816b650b. [DOI] [PubMed] [Google Scholar]
- 12.Wong EC, Buxton RB, Frank LR. Quantitative imaging of perfusion using a single subtraction (QUIPSS and QUIPSS II) Magn Reson Med. 1998;39:702–708. doi: 10.1002/mrm.1910390506. [DOI] [PubMed] [Google Scholar]
- 13.Luh WM, Wong EC, Bandettini PA, Hyde JS. QUIPSS II with thin-slice TI1 periodic saturation: a method for improving accuracy of quantitative perfusion imaging using pulsed arterial spin labeling. Magn Reson Med. 1999;41:1246–1254. doi: 10.1002/(sici)1522-2594(199906)41:6<1246::aid-mrm22>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 14.Kim SG. Quantification of relative cerebral blood flow change by flow-sensitive alternating inversion recovery (FAIR) technique: application to functional mapping. Magn Reson Med. 1995;34:293–301. doi: 10.1002/mrm.1910340303. [DOI] [PubMed] [Google Scholar]
- 15.Buxton RB, Frank LR, Wong EC, Siewert B, Warach S, Edelman RR. A general kinetic model for quantitative perfusion imaging with arterial spin labeling. Magn Reson Med. 1998;40:383–396. doi: 10.1002/mrm.1910400308. [DOI] [PubMed] [Google Scholar]
- 16.Stanisz GJ, Odrobina EE, Pun J, et al. T1, T2 relaxation and magnetization transfer in tissue at 3T. Magn Reson Med. 2005;54:507–512. doi: 10.1002/mrm.20605. [DOI] [PubMed] [Google Scholar]
- 17.Liu TT, Brown GG. Measurement of cerebral perfusion with arterial spin labeling. Part 1. Methods. J Int Neuropsychol Soc. 2007;13:517–525. doi: 10.1017/S1355617707070646. [DOI] [PubMed] [Google Scholar]
- 18.McConnell MV, Khasgiwala VC, Savord BJ, et al. Comparison of respiratory suppression methods and navigator locations for MR coronary angiography. AJR Am J Roentgenol. 1997;168:1369–1375. doi: 10.2214/ajr.168.5.9129447. [DOI] [PubMed] [Google Scholar]


