Abstract
A γ-AApeptide-based tracer for positron emission tomography imaging of integrin αvβ3 is reported. Despite its shorter sequence and linear nature, this tracer had comparable integrin αvβ3 binding affinity as the cyclic arginine-glycine-aspartic acid peptide but significantly higher resistance to enzymatic degradation and better stability.
Positron emission tomography (PET) has been widely used in clinical oncology for tumor diagnosis, staging, and treatment monitoring.1 Development and clinical translation of novel molecularly targeted PET tracers will facilitate future personalized medicine of cancer patients, such as patient stratification and monitoring the therapeutic response for anti-cancer drugs.2 Non-invasive PET imaging of tumor angiogenesis (i.e. new blood vessel formation) has gained tremendous interest over the last decade,3 since the development and metastasis of all solid tumors depend on tumor angiogenesis.4 Among the many proteins that are involved in tumor angiogenesis, integrin αvβ3 is one of the most intensively studied and several PET tracers targeting this cell adhesion molecule have entered clinical investigation.5 Frequently overexpressed on the tumor neovasculature, as well as cancer cells of many tumor types (e.g. lung/prostate/breast cancer and glioblastoma), integrin αvβ3 is an attractive target for both cancer diagnosis and therapy.6
Since integrin αvβ3 binds tightly to extracellular matrix proteins that contain the Arg-Gly-Asp (RGD) tripeptide epitopes, a wide variety of peptides/peptidomimetics based on the RGD motif have been investigated for anti-cancer drug development and/or cancer imaging.5a, 6 The Achilles' heel of peptides is that they are susceptible to degradation by a variety of enzymes hence are not very stable in vivo. Many approaches have been employed to improve the in vivo stability of peptide-based imaging/therapeutic agents, such as cyclization and the use of unnatural amino acids.5a Recently, a PET tracer based on peptoids (i.e. poly-N-substituted glycines) was reported for PET imaging,7 which opened the door to a fertile area of future research and development.
We recently designed a new class of peptidomimetics that are termed “γ-AApeptides”,8 which have shown promising potential for various biological applications such as effective disruption of protein-protein interactions,8 recognition of specific nucleic acids,9 and as novel antimicrobial agents.10 In addition, these γ-AApeptides are resistant to proteolytic degradation and are amenable for limitless diversification. Since γ-AApeptides project the same number of side chains as that of peptides of the same length, they are expected to be ideal candidates for short peptide mimicry. To further explore their potential applications in biomedical research, and to develop novel molecular imaging agents, herein we report a short and linear γ-AApeptide-based RGD mimetics (Fig. 1), which can be employed for in vivo PET imaging of integrin αvβ3 expression. Our results demonstrate that this new class of RGD mimetics are comparable to the commonly used c(RGDyK) (where y denotes D-tyrosine) peptide in terms of integrin αvβ3 binding affinity and specificity. Furthermore, they are much more protease-resistant, hence are more suitable for PET imaging applications.
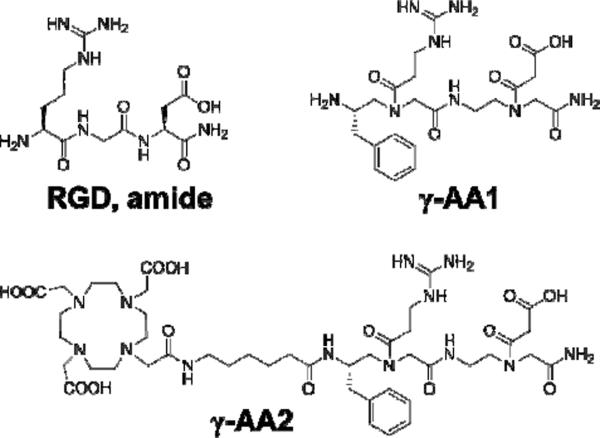
Fig. 1.
The RGD tripeptide and the γ-AApeptides. γ-AA1 is a γ-AApeptide that mimics RGD. γ-AA2 is a DOTA conjugated γ-AA1 for 64Cu-labeling and PET imaging.
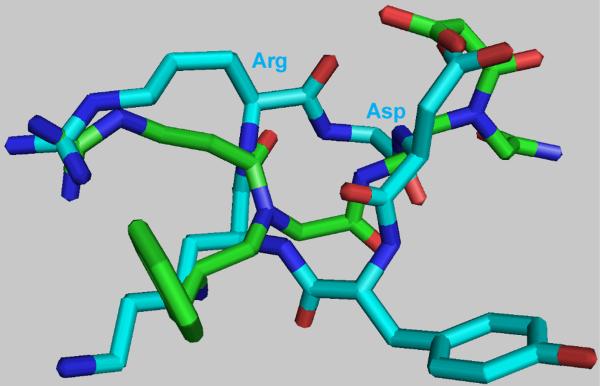
γ-AA1 is a γ-AApeptide designed to mimic the tripeptide RGD. An additional phenyl moiety was included in the molecule to provide a balance of hydrophilicity and hydrophobicity, as was found in many RGD-containing peptides for imaging and/or therapeutic applications,6 which is not expected to interfere with integrin αvβ3 binding. To rationalize such design, structural studies were carried out by superimposing the energy-minimized structure of γ-AA1 onto that of c(RGDyK). As shown in Figure 2, the guanidino and carboxyl groups within γ-AA1 superimpose very well with the functional groups of Arg and Asp residues from c(RGDyK), which are responsible for the recognition of integrin αvβ3. T hus, computer modeling supports the straightforward and effective design of γ-AApeptides for RGD motif mimicry. Future systematic studies will be carried out to investigate the effect of different functional groups on integrin αvβ3 binding of the γ-AApeptide-based RGD mimetics.
Fig. 2.
The superimposition of energy-minimized γ-AA1 (green) and c(RGDyK) (cyan). The energy minimization and superimposition was carried out using the ChemBioOffice program.
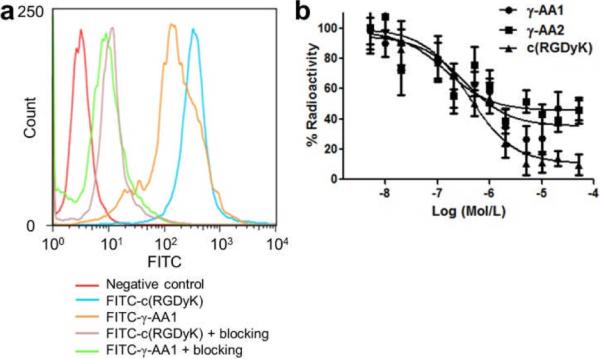
To further evaluate the capability of the γ-AApeptides for RGD mimicry, γ-AA1 and the c(RGDyK) peptide, an extensively studied and validated high affinity antagonist for integrin αvβ3,5a were each conjugated to FITC. After purification by high performance liquid chromatography (HPLC), FITC-γ-AA1 and FITC-c(RGDyK) were compared for integrin αvβ3 binding affinity and specificity in U87MG human glioblastoma cells that express high level of integrin αvβ3.11 At a 5 μg/mL concentration which is under a non-saturating condition (i.e. the fluorescence signal was in the 102–103 range instead of 104), FITC-γ-AA1 has similar uptake in the U87MG cells as FITC-c(RGDyK), as evidenced by flow cytometry (Fig. 3a). Blocking the receptor with 2 μM of unconjugated c(RGDyK) significantly reduced the uptake of both FITC-γ-AA1 and FITC-c(RGDyK) to a similar extent. U87MG cell binding assay using 64Cu-DOTA-c(RGDyK) as the radioligand revealed that the IC50 values were 831 and 897 nM for γ-AA1 and γ-AA2, respectively (Fig. 3b). These values are slightly lower but comparable to that of c(RGDyK), with an IC50 value of 639 nM in the same assay. Together, these findings indicated that γ-AA1/γ-AA2 and the c(RGDyK) peptide have similar binding affinity and specificity to integrin αvβ3 in vitro.
Fig. 3.
(a) Flow cytometry analysis of FITC-conjugated γ-AA1 and c(RGDyK) peptide in U87MG cells at a 5 μg/mL concentration. Blocking experiments with 2 μM of c(RGDyK) peptide were also performed to confirm the specificity for integrin αvβ3. (b) U87MG cell binding assay demonstrated that both γ-AA1 and γ-AA2 bind to integrin αvβ3, similar to the c(RGDyK) peptide.
To enable 64Cu-labeling and PET imaging, DOTA (1, 4, 7, 10-tetraazacyclododecane-1, 4, 7, 10-tetraacetic acid) was linked to γ-AA1 via a 6-aminohexanoic acid linker, which was termed γ-AA2 (i.e. DOTA-γ-AA1, Fig. 1). 64Cu-labeling of γ-AA2, including final purification with HPLC, took 90 ± 15 min (n = 8). The decay-corrected radiochemical yield was 50 ± 15 %, based on 2 μg of γ-AA2 per 37 MBq of 64Cu, with a radiochemical purity of > 95%. The specific activity of 64Cu-γ-AA2 was measured to be ~9 GBq/mg of γ-AA2. The c(RGDyK) peptide was conjugated with DOTA, purified by HPLC, and labeled with 64Cu in a similar manner.
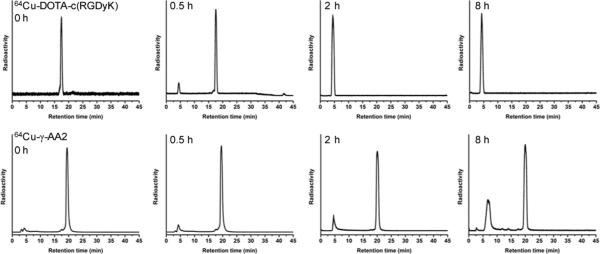
Before PET imaging was carried out to evaluate the in vivo behaviour of 64Cu-labeled γ-AA2, enzymatic stability of the tracer was investigated. Pronase, a mixture of proteinases isolated from the extracellular fluid of Streptomyces griseus, was used to compare the enzymatic stability of the two PET tracers.8, 12 After incubation with 0.1 mg/mL of pronase at 37 °C in 100 mM ammonium bicarbonate buffer (pH 7.8) for various time periods, stability of the two tracers was compared using radio-HPLC.
64Cu-γ-AA2 exhibited markedly better stability than 64Cu-DOTA-c(RGDyK) (Fig. 4). The stability of c(RGDyK) is expected to be significantly higher than the natural RGD peptide, attributed to the inclusion of a D-Tyr residue and head-to-tail cyclization, both of which are proven strategies for improving the stability of natural peptides. However, ~8% of 64Cu-DOTA-c(RGDyK) was already degraded at 0.5 h post-treatment, with 100% enzyme degradation at 2 h and 8 h post-treatment. In comparison, 64Cu-γ-AA2 only had 4%, 8%, and 15% degradation at 0.5 h, 2 h, and 8 h post-treatment, respectively. The UV traces were similar to the radio-HPLC results (see Supplementary Information). The fact that 64Cu-γ-AA2 is much more enzymatically stable than 64Cu-DOTA-c(RGDyK) makes γ-AApeptides a promising class of targeting ligands for PET imaging applications, which possess exceptional stability.
Fig. 4.
Serial radio-HPLC profiles of 64Cu-DOTA-c(RGDyK) and 64Cu-γ-AA2 before and after incubation in pronase at 37 °C.
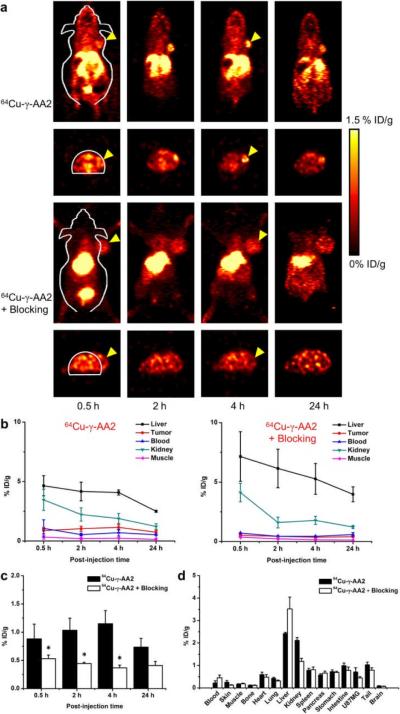
After demonstrating the excellent stability of 64Cu-γ-AA2 in vitro, serial in vivo PET imaging was carried out in the U87MG tumor model (which expresses high level of integrin αvβ3 on tumor vasculature and tumor cells11, 13) after intravenous injection of the tracer. PET scans at various time points post-injection (p.i.), as well as quantitative region-of-interest (ROI) analysis of the PET data, were performed as described previously.14 Coronal and transaxial PET slices that contain the U87MG tumors are shown in Fig. 5a and the quantitative data are shown in Fig. 5b.
Fig. 5.
Serial PET imaging and biodistribution studies of 64Cu-γ-AA2 in U87MG tumor-bearing mice. (a) Serial coronal and transaxial PET images of U87MG tumor-bearing mice at 0.5, 2, 4, and 24 h after injection of 64Cu-γ-AA2, or co-injection of c(RGDyK) and 64Cu-γ-AA2 (i.e., blocking). Arrowheads indicate tumors. (b) Time-activity curves of the liver, tumor, blood, kidney, and muscle in U87MG tumor-bearing mice after injection of 64Cu-γ-AA2 (left), or co-injection of 64Cu-γ-AA2 and a blocking dose of c(RGDyK) (right). (c) Comparison of U87MG tumor uptake of 64Cu-γ-AA2 between the two groups. (d) Biodistribution data at 24 h post-injection of the tracer. *: p < 0.05 (n = 3).
Tumor uptake of 64Cu-γ-AA2 was clearly visible as early as 0.5 h p.i., which remained persistent over time (0.9 ± 0.3, 1.0 ± 0.2, 1.1 ± 0.2, and 0.7 ± 0.2 %ID/g at 0.5, 2, 4, and 24 h p.i. respectively; n = 3; Fig. 5b). Excellent tumor contrast was observed, with tumor/muscle ratio of 3.8 ± 0.9, 4.8 ± 1.8, 5.8 ± 1.4, and 8.3 ± 3.9 at 0.5 h, 2 h, 4 h, and 24 h p.i. respectively (n = 3). Since 64Cu-γ-AA2 undergoes both hepatobiliary and renal clearance, tracer uptake was also observed in the liver/kidneys.
Administering a blocking dose of the c(RGDyK) peptide reduced the U87MG tumor uptake significantly to 0.5 ± 0.1, 0.4 ± 0.1, 0.4 ± 0.1, and 0.4 ± 0.1 %ID/g at 0.5, 2, 4, and 24 h p.i., respectively (n = 3; P < 0.05 at 0.5, 2, and 4 h p.i. when compared with mice injected with 64Cu-γ-AA2 only; Fig. 5a–c), which demonstrated integrin αvβ3 specificity of the tracer in vivo. Liver uptake of 64Cu-γ-AA2 was higher in the blocking group, being 7.2 ± 2.0, 6.1 ± 1.6, 5.3 ± 1.3, and 4.0 ± 0.6 %ID/g at 0.5, 2, 4, and 24 h p.i. respectively (n = 3). Radioactivity in the blood (0.5 ± 0.1, 0.4 ± 0.1, 0.4 ± 0.1, and 0.4 ± 0.1 %ID/g at 0.5, 2, 4, and 24 h p.i., respectively; n = 3) was lower for the blocking group at early time points, indicating faster blood clearance of the tracer (Fig. 5b). After the last PET scans, all mice were euthanized for biodistribution studies to validate the PET data. The %ID/g values of 64Cu-γ-AA2 in the tumor and major organs obtained from biodistribution studies (Fig. 5d) matched well with the results from ROI analysis of the PET scans, confirming that PET can enable accurate quantification of tracer distribution in vivo.
In summary, the 64Cu-labeled γ-AApeptide-based RGD mimetic exhibited comparable integrin αvβ3 binding affinity as the c(RGDyK) peptide but significantly higher resistance to enzymatic degradation and better in vivo stability, despite its shorter sequence and linear nature. Integrin αvβ3 specificity, fast blood clearance, and good tumor contrast of 64Cu-γ-AA2 established γ-AApeptides as a novel class of enzymatically stable targeting ligands for molecular imaging applications.
Supported in part by W81XWH-11-1-0644, W81XWH-11-1-0648, the Elsa U. Pardee Foundation, and UW/USF startup funds.
Supplementary Material
Footnotes
† Electronic Supplementary Information (ESI) available: [synthesis, characterization, and experimental details]. See DOI: 10.1039/b000000x/
references
- 1.Gambhir SS, Czernin J, Schwimmer J, Silverman DH, Coleman RE, Phelps ME. J Nucl Med. 2001;42:1S–93S. [PubMed] [Google Scholar]
- 2.Weissleder R, Pittet MJ. Nature. 2008;452:580–589. doi: 10.1038/nature06917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai W, Chen X. J Nucl Med. 2008;49(Suppl 2):113S–128S. doi: 10.2967/jnumed.107.045922. [DOI] [PubMed] [Google Scholar]
- 4.Folkman J. N Engl J Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 5.(a) Cai W, Niu G, Chen X. Curr Pharm Des. 2008;14:2943–2973. doi: 10.2174/138161208786404308. [DOI] [PubMed] [Google Scholar]; (b) Haubner R, Weber WA, Beer AJ, Vabuliene E, Reim D, Sarbia M, Becker KF, Goebel M, Hein R, Wester HJ, Kessler H, Schwaiger M. PLoS Med. 2005;2:e70. doi: 10.1371/journal.pmed.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Mittra ES, Goris ML, Iagaru AH, Kardan A, Burton L, Berganos R, Chang E, Liu S, Shen B, Chin FT, Chen X, Gambhir SS. Radiology. 2011;260:182–191. doi: 10.1148/radiol.11101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai W, Chen X. Anti-Cancer Agents Med Chem. 2006;6:407–428. doi: 10.2174/187152006778226530. [DOI] [PubMed] [Google Scholar]
- 7.Hao G, Hajibeigi A, De León-Rodríguez LM, Öz OK, Sun X. Am J Nucl Med Mol Imaging. 2011;1:65–75. [PMC free article] [PubMed] [Google Scholar]
- 8.Niu Y, Hu Y, Li X, Chen J, Cai J. New J Chem. 2011;35:542–545. [Google Scholar]
- 9.Niu Y, Jones AJ, Wu H, Varani G, Cai J. Org Biomol Chem. 2011;9:6604–6609. doi: 10.1039/c1ob05738c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niu Y, Padhee S, Wu H, Bai G, Harrington L, Burda WN, Shaw LN, Cao C, Cai J. Chem Commun. 2011;47:12197–12199. doi: 10.1039/c1cc14476f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai W, Wu Y, Chen K, Cao Q, Tice DA, Chen X. Cancer Res. 2006;66:9673–9681. doi: 10.1158/0008-5472.CAN-06-1480. [DOI] [PubMed] [Google Scholar]
- 12.Hu Y, Li X, Sebti SM, Chen J, Cai J. Bioorg Med Chem Lett. 2011;21:1469–1471. doi: 10.1016/j.bmcl.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Cai W, Shin DW, Chen K, Gheysens O, Cao Q, Wang SX, Gambhir SS, Chen X. Nano Lett. 2006;6:669–676. doi: 10.1021/nl052405t. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Hong H, Engle JW, Yang Y, Barnhart TE, Cai W. Am J Nucl Med Mol Imaging. 2012;2:1–13. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.