Summary
Background
Mycobacterium leprae was the only known cause of leprosy until 2008, when a new species, named Mycobacterium lepromatosis, was found to cause diffuse lepromatous leprosy (DLL), a unique form of leprosy endemic in Mexico.
Methods
We sought to differentiate the leprosy agents among 120 Mexican patients with various clinical forms of leprosy and to compare their relative prevalence and disease features. Archived skin biopsy specimens from these patients were tested for both M. leprae and M. lepromatosis using polymerase chain reaction-based species-specific assays.
Results
Eighty-seven (72.5%) patients were confirmed for etiologic species, including 55 with M. lepromatosis, 18 with M. leprae, and 14 with both organisms. The endemic regions of each agent differed but overlapped. Patients with M. lepromatosis were younger and from more states, and their clinical diagnoses included 13 DLL, 34 lepromatous leprosy (LL), and eight other forms of leprosy. By contrast, the diagnoses of patients with M. leprae included none DLL, 15 LL and three other forms. Thus, M. lepromatosis caused DLL specifically (p=0.023). Patients with M. lepromatosis also showed more variable skin lesions and the extremities were the commonest biopsy sites. Finally, patients with dual infections manifested all clinical forms and accounted for 16.1% of all species-confirmed cases.
Conclusions
M. lepromatosis is another cause of leprosy and is probably more prevalent than M. leprae in Mexico. It mainly causes LL and also specifically DLL. Dual infections caused by both species may occur in endemic area.
Keywords: leprosy, diffuse lepromatous leprosy, Lucio’s phenomenon, Mycobacterium lepromatosis, Mycobacterium leprae
Introduction
Leprosy is a chronic dermatologic infection that has plagued humans for at least 100,000 years.1 Worldwide the disease still affects millions of people in the forms of new diagnosis and neurologic disability.2,3 The disease manifests a spectrum of clinicopathologic forms, ranging from tuberculoid leprosy (TL), to borderline forms, to lepromatous leprosy (LL), and can be paucibacillary or multibacillary in skin lesions. One form of the disease may markedly dominate others within a geographic region.2 It has been long thought that these variations are due to the individual host immune effects.4
Mycobacterium leprae was the only organism known to cause leprosy until 2008, when a new species, namely Mycobacterium lepromatosis, was found to be the cause of diffuse lepromatous leprosy (DLL) in two patients of Mexican origin who died of the disease.5 Further analysis of 22,814 nucleotides revealed a 9.1% difference between the two organisms to substantiate a species-level divergence that occurred approximately 10 million years ago.6 This divergence was much earlier (~88 times) than the well-known divergence between M. tuberculosis and M. bovis (113,000 years ago).
Neither M. leprae nor M. lepromatosis has been cultivated, which has hindered the care and research for leprosy. In recent decades, however, the use of animal passage and molecular techniques has enabled many in-depth studies on M. leprae. For instance, genomes of four M. leprae strains from India, Thailand, Brazil, and the United States have been sequenced (3.3 × 106 nucleotides), which uncovered a genome-wide difference of only ~200 nucleotides (0.005%).7,8 Similarly, worldwide M. leprae strains (400 strains or samples from 30 countries of all continents) have also been shown to be clonal, differing only in single nucleotide polymorphism and tandem repeats.1,8 Therefore, clonal M. leprae strains are not responsible for the clinical and geographic variations of leprosy.
DLL is a unique, severe form of leprosy initially recognized by Lucio and Alvarado in 1852 and further described by Latapi and Chevez-Zamora in 1948, both in Mexico.9,10 It is thus also known as diffuse leprosy of Lucio and Latapi,11,12 leprosy with Lucio’s phenomenon,13,14 or merely Lucio’s leprosy. DLL is characterized clinically by diffuse, non-nodular cutaneous infiltration, manifesting recurrent crops of large sharply demarcated skin lesions called Lucio’s phenomenon, which is considered as an unusual reaction to the infection. In the advanced stage, the lesions may become ulcerated, particularly on the lower extremities, or even generalized, leading to fatal secondary bacterial infection and sepsis.2 The diagnosis of DLL may be delayed, especially in non-endemic areas, resulting in death.5,15–17 The usual hallmark of leprosy – mycobacterial invasion into the skin and nerves, is readily visible by microscopy. In addition, the mycobacterium uniquely invades into the endothelium, causing endothelial proliferation, vascular occlusion, and/or vasculitis in the dermis and subcutis.5,10,12,14 DLL is predominantly seen in patients from western and central Mexico and the Caribbean countries,2,18 but rare cases have been reported elsewhere, including Asia (India, Iran, Malaysia, and Singapore),15–17,19,20 Europe (France),21 North America (United States),22 South America (Brazil),23,24 and northern Africa (Tunisia).25
The discovery of M. lepromatosis has raised such questions as whether it is the sole cause of DLL, whether it also causes or contributes to other clinical forms of leprosy, and its overall prevalence and significance in Mexico and beyond. In this study, we attempted to answer some of these questions by determining the etiologic agent(s) of 120 cases of leprosy from Mexico. We tested the DNA extracted from archived skin biopsy specimens for both M. lepromatosis and M. leprae by using polymerase chain reactions (PCR) that targeted at the unique 16S rRNA genes of the organisms.
Methods
This study used archived formalin-fixed and paraffin-embedded skin biopsy specimens from Mexico, with one specimen per patient. One hundred biopsy specimens were obtained from the National Leprosy Control Program and 20 from a hospital in Sinaloa. This number was chosen for statistical consideration, reasonable workload for the testing, and as much representation of the endemic states as possible. They were mostly consecutive recent accessions from both institutions without selection of particular leprosy forms. These tissue blocks, which measured 2 to 120 mm2 (median 16 mm2) in cut surface area and had been archived for 1–20 years (median 3.5 years), were retrieved in August 2008, wrapped individually for containment, and sent to Houston for testing.
The specimens were sectioned at a research-only histology laboratory. The cutting blade and forceps were changed for each specimen to prevent cross contamination. Two to four sections of seven micron thickness of tissue were used depending on the size, and those tested negative tissue samples were cut and tested once or twice more to minimize sampling bias. DNA extraction was done using a tissue kit (QIAamp DNA Mini Kit, Qiagen, Valencia, CA). Tissue sections were de-paraffinized by xylene treatment, digested with proteinase K, spiked with non-specific carrier DNA, and loaded onto the silica-based mini-column. After washing, DNA was eluted in a buffer for testing. The exceedingly low DNA quantity only allowed one to three PCR analyses for each sample.
The 16S rRNA gene, known for all described bacteria (~9000 species), was selected as PCR target, and we aimed at the most variable first 500 bp of this gene.5,26–28 Two rounds of semi-nested PCRs were designed to maximize detection sensitivity and specificity. The first-round PCR used primers AFBFO (5′gcgtgcttaacacatgcaagtc) and MLER4 (5′ccacaagacatgcgccttgaag) that are common to all known mycobacteria (>100 species). The resulting amplicons (171 bp in size) were diluted and further amplified by two separate second-round PCRs using MLER4 and LPMF2 (5′gtctcttaatacttaaacctattaa) for M. lepromatosis (142-bp) and MLER4 and LERF2 (5′ctaaaaaatcttttttagagatac) for M. leprae (135-bp). The amplicons were examined by electrophoresis. As defined or noted previously,5,27,28 the inner primers LPMF2 and LERF2 were unique for M. lepromatosis and M. leprae respectively, and the lack of these inserted sequences in all other known mycobacteria ensured test specificity. A previous PCR study also showed that a primer similar to LERF2 detected M. leprae specifically, not other mycobacteria, in experimental DNA as well as tissue DNA from leprosy patients.28 The semi-nested PCRs amplified each target through 70 doubling cycles to enable detection of several copies, a sensitivity shown previously in an M. leprae PCR study.29 The design of small amplicons aimed to avoid the problem of DNA fragmentation (to 200 bp to 500 bp) that occurs during formalin fixation and paraffin embedment of tissue. The method also used each extracted DNA once (in the first round) for both organisms. To prevent amplicon contamination, benches for PCR setup and examination were kept separate along with separate sets of equipments and reagents. We also followed good molecular testing practice as required by regulation.
The clinicopathologic data that accompanied initial workup of the biopsy specimens years ago included the patient’s age and sex, the address, the biopsy date and site, the clinicopathologic diagnoses, the features of the skin lesions, and the load of mycobacterium in the tissue smear (if available).
Statistical analysis primarily used χ2 or Fisher’s exact method when indicated.
Results
From the 120 biopsy specimens from patients diagnosed with leprosy by clinicopathologic criteria, the PCR analyses detected mycobacteria in 87 specimens (72.5%): M. lepromatosis in 55 specimens, M. leprae in 18, and both organisms in 14. There were 33 PCR-negative specimens. Thus, of the 87 species-confirmed cases, M. lepromatosis caused 63.2%, M. leprae caused 20.7%, and both species were present in 16.1%.
Geographic Distributions
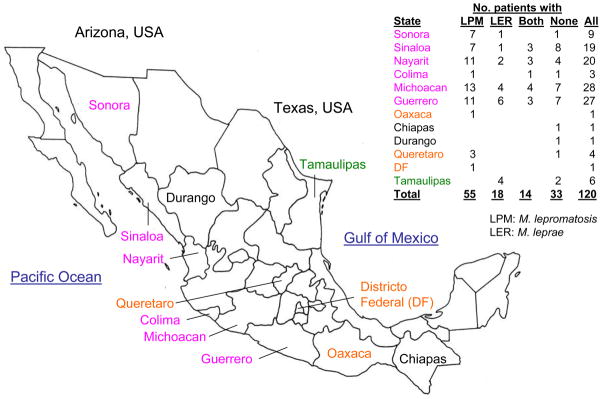
The 120 specimens were from 12 Mexican states, and the 87 species-confirmed specimens were from 10 states (Figure 1). M. lepromatosis was found in nine states whereas M. leprae in six states, and there were pockets of disease by different species. For instance, only four M. leprae cases were found in Tamaulipas, significantly different from the other nine states where M. lepromatosis cases outnumbered M. leprae cases by 55 to 14 (p = 0.0028, Fisher’s exact test). Tamaulipas borders the Gulf of Mexico and Texas (USA) and is distant from those Pacific and central states. In particular, M. lepromatosis was predominant (ratio 7:1) in Sonora, a western Pacific state bordering Arizona (USA) where this species was initially discovered in two Mexican immigrants.
Figure 1. Map of Mexico showing states where cases of leprosy in this study occurred.
Orange states had M. lepromatosis infections only; green states had M. leprae infections only; purple had infections by both species; black states had only cases without species confirmation.
The 14 dually infected cases were found in Sinaloa, Nayarit, Colima, Michoacan, and Guerrero, paralleling more leprosy cases and the dominance of M. lepromatosis in these states. Together, the detection of more cases of leprosy involving M. lepromatosis and their wider distribution suggest higher prevalence of this organism than M. leprae in Mexico.
Age of Patients
Table 1 lists the demographic and clinicopathologic features of the patients. There was a significant difference in the ages of the two groups of patients infected with a single organism: the ages of the M. lepromatosis group spread from 17 to 86 (median 51) with 19 of the 55 patients (34.5%) aged below 46 years (a dividing line), whereas the M. leprae group had a median age of 57 and only one of 18 (5.6%) aged below 46 (p= 0.017). Similarly, the median age of the dually infected group was in between, or 52 years old, and five of 14 (35.7%) were aged below 46 years (p= 0.042, Fisher’s). Thus, early infection by M. lepromatosis, singly or dually, also indicated a higher prevalence of this organism. The gender difference was not statistically significant (p=0.26).
Table 1.
Characteristics of 120 Mexican patients with leprosy and their etiologic agent(s)
| Features | Number of patients by organism(s) detected | Total | P value* | |||
|---|---|---|---|---|---|---|
| M. lepromatosis | M. leprae | Both | Neither | |||
| Total patients | 55 | 18 | 14 | 33 | 120 | |
| Men: women | 36:19 | 10:8 | 7:7 | 23:10 | 76:44 | |
| Age | ||||||
| Youngest-oldest (years) | 17–86 | 20–75 | 32–78 | 19–88 | 17–88 | |
| Mean/median (years) | 51.3/51 | 58.1/57 | 54.1/52 | 52.5/52 | 53.0/52 | |
| <46 years (No. of patients) | 19 | 1 | 5 | 8 | 33 | 0.017 |
| ≥46 years (No. of patients) | 36 | 17 | 9 | 25 | 87 | |
| Skin lesion(s) | ||||||
| Unique to M. lepromatosis | 17 | 0 | 2 | 2 | 21 | 0.007 |
| Diffuse nonnodular infiltration | 6 | 1 | 7 | |||
| Macules and nodules (mixed) | 3 | 1 | 4 | |||
| Skin anesthesia | 6 | 1 | 7 | |||
| Normal-appearing skin | 2 | 1 | 3 | |||
| Other lesion | 38 | 18 | 12 | 31 | 99 | |
| Macules only | 23 | 6 | 6 | 12 | 47 | |
| Nodules, plaques or unknown | 15 | 12 | 6 | 19 | 52 | |
| Site of biopsy | ||||||
| Face, ear, or chest | 10 | 10 | 6 | 8 | 34 | 0.004 |
| Abdomen, back or buttock | 10 | 0 | 1 | 7 | 18 | |
| Extremities (limbs) | 35 | 8 | 7 | 18 | 68 | |
| Arm or forearm | 17 | 4 | 6 | 10 | 37 | |
| Leg | 11 | 1 | 6 | 18 | ||
| Elbow, hand, ankle, knee or thigh | 7 | 3 | 1 | 2 | 13 | |
| Clinicopathologic diagnosis | ||||||
| Diffuse lepromatous leprosy | 13 | 0 | 3 | 2 | 18 | 0.023 |
| Other forms of leprosy | 42 | 18 | 11 | 31 | 102 | |
| Lepromatous | 34 | 15 | 7 | 20 | 76 | |
| Borderline (dimorphic) or tuberculoid | 8 | 3 | 4 | 11 | 26 | |
Two-tail p value based on significant χ2 tests between M. lepromatosis and M. leprae with bold numbers to show the 2 × 2 or 2 × 3 format.
Skin Lesions and Biopsy Sites
Of the eight types of skin lesions seen in this study, all were seen in the patients infected with M. lepromatosis, whereas those infected with M. leprae showed only macules, nodules, and plaques (Table 1). Diffuse infiltration, mixed macules and nodules, skin anesthesia (with or without macules), and normal-appearing skin were only associated with M. lepromatosis infections, being seen in 17 of the 55 patients (p=0.007). As observed clinically earlier, normal-appearing skin and mere skin anesthesia were early manifestations of DLL, although correct diagnosis might be difficult initially, and the four involved patients were diagnosed as LL and TL (three and one respectively). Overall, macules with or without concurrent skin anesthesia or nodules were most common, seen in 30 (54.5%) of the 55 patients with M. lepromatosis. Patients with dual infections showed features of both groups.
There were 13 diverse sites of skin biopsy among the patients: those with M. lepromatosis had lesions biopsied from all sites, whereas those with M. leprae had specimens taken from only eight of the 13 sites. In the M. lepromatosis group, 35 (63.6%) of the 55 biopsy specimens were taken from the extremities, whereas in the M. leprae group, the chest was the most common biopsy site (n=6), followed by the face/ear; these differences were significant (p=0.004, 2 × 3 contingency χ2).
The above results suggest that M. lepromatosis infection is associated with more variation of skin lesions and biopsy sites than is M. leprae infection.
Clinicopathologic Diagnoses
In the M. lepromatosis group, anesthetic non-nodular diffuse skin infiltration led to a clinical diagnosis of primary DLL for six patients, and seven patients were diagnosed with secondary DLL, ie, the progression of skin lesions from macules to diffuse infiltration. These 13 DLL diagnoses (23.6% of 55 cases) ran in contrast to none DLL in the M. leprae group (0 of 18) (p=0.023). This result verified that M. lepromatosis is the specific cause of DLL. Additionally, 34 patients (61.8%) and eight patients were diagnosed with LL and other forms of leprosy respectively in this group.
In the M. leprae group, LL was predominant, affecting 15 (83.3%) of the 18 patients. In the dual-infection group, the clinical diagnoses included three DLL, seven LL, and four cases of borderline forms, representing features from single infections by both organisms.
Together, DLL accounted for 16 (23.2%) of the 69 patients with M. lepromatosis infection, singly or dually. The 16 patients with DLL included 12 men and four women with a median age of 49.4 years; the youngest were men aged 17, 19, and 22 years. These patients were from Michoacan (n=6), Nayarit (n=4), Guerrero (n=4), Sonora (n=1), and Queretaro (n=1). The biopsy sites for 13 (81.3%) of them were the extremities.
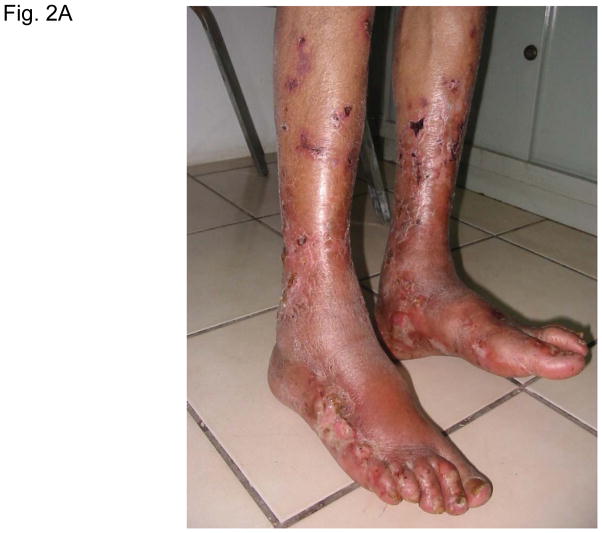
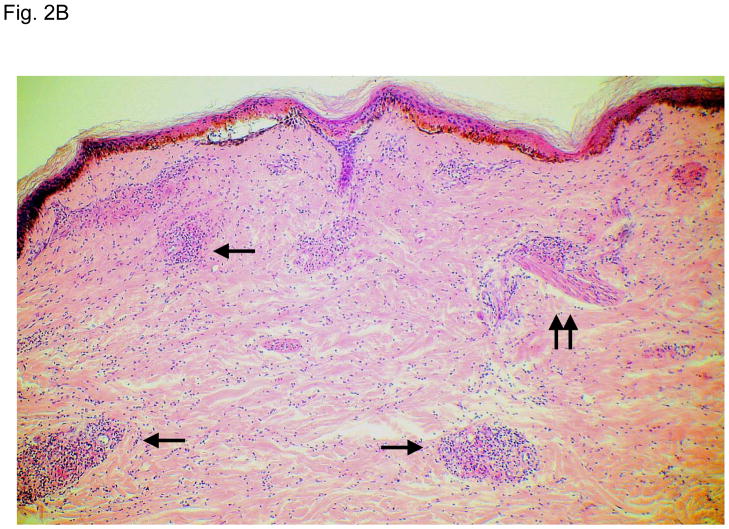
Figure 2 illustrates typical leg skin lesions (ulcers, eschars, and edema) and pathologic features of DLL with epidermal necrosis, diffuse cutaneous inflammatory infiltration, leukocytoclastic vasculitis, neuritis, invasion of the organism into endothelia, and endothelial proliferation.
Figure 2. Typical skin lesions and underlying histopathology of diffuse lepromatous leprosy caused by M. lepromatosis.
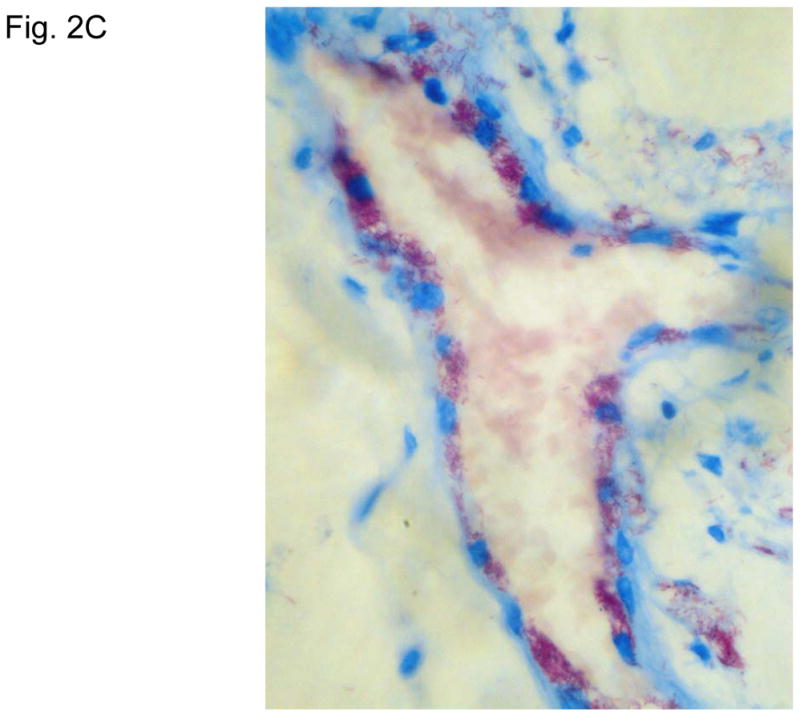
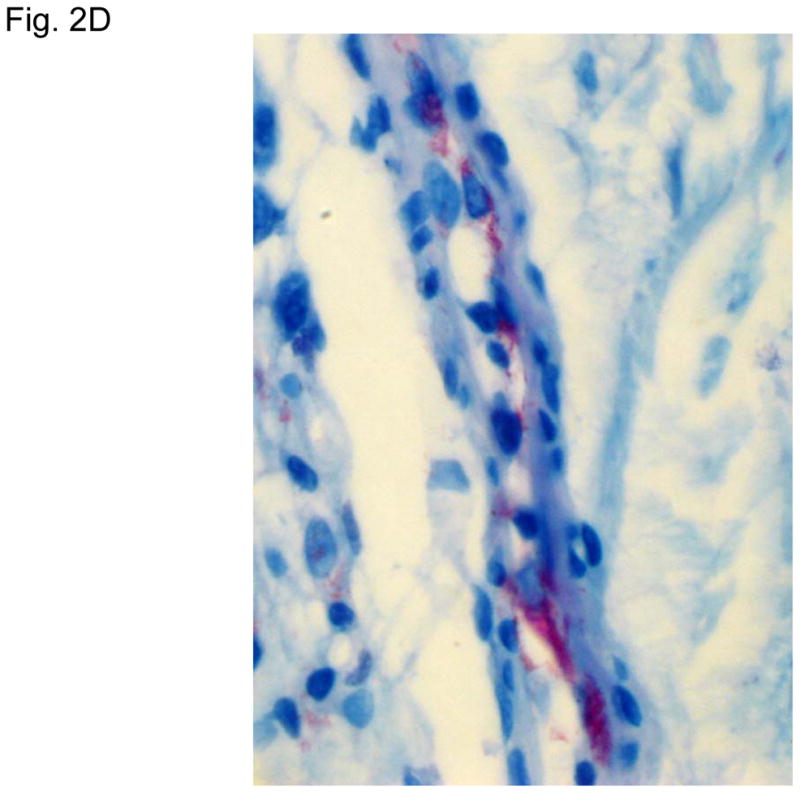
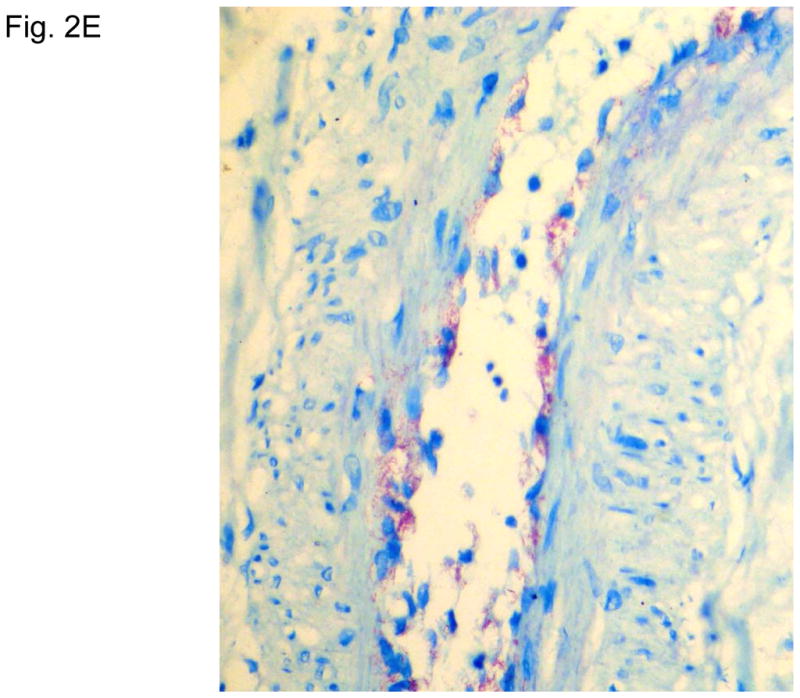
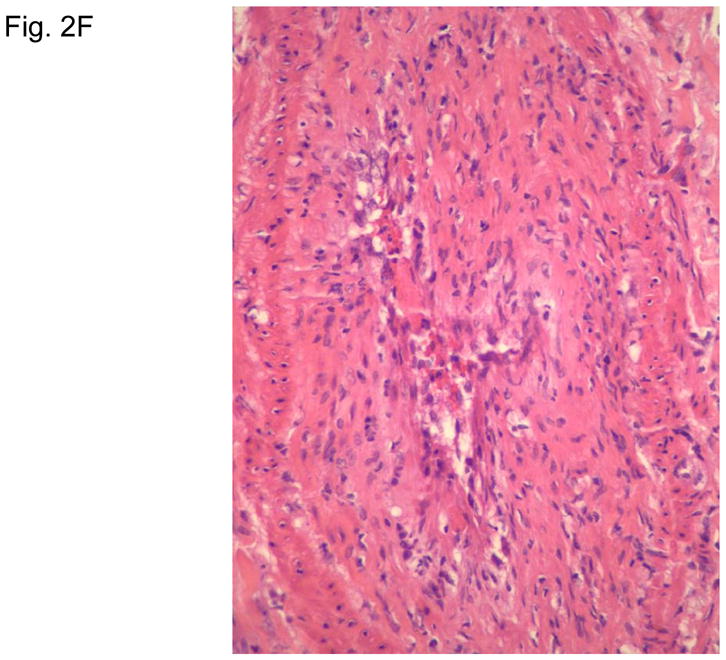
A. Advanced skin ulcers, eschars, and edema in the legs and feet. B. Epidermal necrosis and diffuse cutaneous inflammatory infiltration with leukocytoclastic vasculitis (arrows) and neuritis (dual arrows) (H & E, 35x). CDE. Invasion of the mycobacterium, as highlighted by red-colored bacilli, into the endothelia of a vein, arteriole, and medium artery respectively (Fite, 400x). E. Also endothelial proliferation. F. End stage occlusion of a medium artery (H & E, 100x).
Analysis of the PCR-negative Cases
The patients without species confirmation showed features of both the M. lepromatosis and M. leprae groups (Table 1). For instances, the geographic distribution was wide (10 of the 12 states, Fig. 1); the median age of 52 years was between the median ages of the two groups; the biopsy sites included the extremities as well as torso sites; and the clinical diagnoses were diverse (including two DLL cases). These features were not significantly different from those of the PCR-positive groups. Therefore, these PCR-negative cases were likely random misses of either species, and they should not skew the data to a particular species to render biased conclusions.
Discussion
Leprosy has been endemic in Mexico for at least several centuries. Thanks to many decades of sustained campaigns against the disease, however, the country achieved a national prevalence of less than 1 case per 10,000 population by 1994,30 a definition of elimination of leprosy by the World Health Organization. In 2008, only 243 new cases were reported.1 The official epidemiologic survey and a recent review 30 showed that most cases in the past several years occurred in the states included in this study. Thus, our study should reflect the leprosy picture in Mexico.
Using archived skin biopsy specimens, we have investigated the prevalence of the newly discovered M. lepromatosis as well as the well-known M. leprae in a relatively quick and cost-effective manner. The design of parallel testing for both organisms led us to draw three main conclusions: M. lepromatosis appears to be the dominant cause of leprosy in Mexico because it caused far more cases, affected patients at a younger age, and was spread through more regions of the country than M. leprae; M. lepromatosis is the specific cause of DLL but it also causes LL and other forms of leprosy, the latter being similar to M. leprae; and M. lepromatosis and M. leprae co-exist in endemic areas and may cause dual infections. These results suggest that M. lepromatosis is another significant contributing factor, unknown previously, to the puzzling clinical and geographic variations of leprosy.
The surprising prevalence of M. lepromatosis prompted us to ask why the organism was not discovered earlier. There might be several explanations: like M. leprae, M. lepromatosis has yet to be successfully cultivated; leprosy is a neglected disease of the poor; use of animal passage and molecular techniques to study leprosy in Mexico, particularly DLL, has been limited; the immune theory has explained the leprosy variations for several decades; and the known M. leprae has been the focus of studies.
The clonal patterns of the worldwide M. leprae strains suggest that leprosy spread along human migration tracks during the past 100,000 years.1,8 Leprosy in Mexico is believed to be indigenous and/or brought by the Spanish settlers of the past 500 years.30 Indigenous Mexicans (Mongoloid) came from Asia over 12,000 years ago through the Alaska Bering land bridge. Delineation of M. lepromatosis and DLL in this study lends support to the native origin of M. lepromatosis. First, the lack of description of DLL in Spain excludes a Spanish origin of the disease. Second, the dominance of M. lepromatosis and the endemic nature of DLL indicate deep roots of this species/disease in the country. Third, the endemic zone of M. lepromatosis matches the Mongoloid migration routes and settlements along the Pacific states (Figure 1), not the Gulf of Mexico states by the Spanish settlers. Fourth, the endemicity of DLL in the Caribbean 2,18 and the Brazilian DLL cases 23,24 coincide with further Mongoloid spread in Central and South Americas. And finally, the reports of DLL in Singapore 15 and Malaysia 16 are consistent with the Asian origin. Our on-going studies (Han XY et al., 2011, unpublished) also detected the presence of M. lepromatosis in Brazil, Singapore, and Myanmar. By contrast, M. leprae was likely brought to Mexico by both the original and Spanish settlers, as evidenced by its global prevalence and our findings of dual infections and only M. leprae in Tamaulipas (a Gulf state). Recently, Matsuoka et al also validly showed the presence of M. leprae only in the Gulf states, such as Tamaulipas, Nuevo Leon, Vera Cruz, and the Yucatan Peninsula, but they did not test the new agent M. lepromatosis in the Pacific and western states.31
DLL represents 15–23% of all leprosy diagnoses in Mexico, based on historic clinical data and a recent review.10,30 In our species-confirmed cases, DLL accounted for 18.4% (16 of 87) (Table 1). This consistency argues against selection and/or test biases. More important, these DLL cases were all caused by M. lepromatosis singly (13 cases) or dually with M. leprae (3 cases). This specificity (p=0.023, Table 1) thus affirms the DLL etiology to unravel the 160-year-old medical mystery. Until 2008 the etiologic agent of DLL was presumed to be M. leprae without specific microbiologic studies. Then we performed multi-gene analyses of a mycobacterium from two deaths that were misdiagnosed but showed typical clinical features of DLL and pathognomonic histopathology.5 These open-approach analyses revealed a remarkable genetic difference between the DLL organism and M. leprae, which led us to propose the new species M. lepromatosis. Here we further showed that M. lepromatosis also caused other forms of leprosy, particularly more LL than DLL (Table 1). What accounted for the different diagnoses? In view of the national leprosy control campaigns and the long history of DLL in Mexico, the diagnostic skills on the part of clinician were hardly doubted. Instead, differences in individual disease manifestation, stages of infection due to variable lead time to initial diagnosis, and/or possible co-factors were more plausible. But delineation of these aspects requires future studies. The overlapping features of LL caused by either or both species also need to be addressed.
DLL carries a higher mortality rate than other forms of leprosy.9,10,15–17,22 This severity is likely a result of the prominent vascular invasion by M. lepromatosis, as observed earlier by Rea and Jerskey,14 and during the discovery of this species.5 More convincingly, Vargas-Ocampo studied the histopathology of 199 cases of DLL in Mexico and noted consistent endothelial changes, with associated leukocytoclastic vasculitis and mycobacterial invasion, which ranged from early endothelial proliferation to the late stages of luminal obliteration.12 Because these changes underpin the clinical manifestation of skin purpura, ulceration, and necrosis, the author concluded that DLL is essentially a vascular disease. The natural history of DLL depicts an insidious onset but an accelerated course;10,18 this can be explained by the vascular disease once the lumen narrows to a crucial point to cause skin ischemia (purpura) and ulceration. This disease process resembles, to some extent, the progression of coronary artery heart disease. The DLL skin lesions typically start from the legs and progress to the entire body at late stage.5,10,14–16 Our finding of the extremities as the most common biopsy sites for M. lepromatosis infections, DLL in particular, is consistent with this feature. As the skin breaks down, secondary bacterial sepsis may be lethal in the pre-antibiotic era or even today in the misdiagnosed cases.5,10,15 In addition, cachexia and extensive involvement of viscera, such as the liver and/or spleen as documented clinically or on autopsy or biopsy,5,10,18,22,32 are also features of end-stage DLL.
Dual infections by M. lepromatosis and M. leprae accounted for 14 (16.1%) of all species-confirmed cases. The manifestations of these cases were more variable than those of a single infection by either species: some cases exhibited mostly features of M. lepromatosis infection, such as DLL, younger age, and the extremities as the biopsy site, whereas others exhibited mostly features of M. leprae infection, such as the chest and face/ear as biopsy sites. A number of factors may affect the manifestations, including such factors as which species infects the patient first and its duration, whether infection by the second species represents general vulnerability of the patient to these organisms or a mere chance of exposure due to common living environment, the preferred site of infection of each species, and dominance of one species over the other.
Finally, the referral and retrospective nature of this study precluded such clinical data as the lead time to diagnosis, lepra reactions, response to antimicrobial treatment, and follow-up. We were unable to review available clinical data for uniformity due to the wide geographic scope and/or long archival although these aspects bore some advantage of blindness or more objectivity for discerning disease pattern. The hit-or-miss nature of minute biopsy and the overall lower DNA quality after tissue processing (formalin fixation and paraffin embedment) and long storage also likely undermined PCR detection of the etiologic agents in a handful cases. Hence, future prospective studies should be designed to test fresh biopsy specimens and focus on clinical details and treatment outcomes caused by different species. Integration of the PCR testing into regular clinical service or national surveillance can also be considered.
Acknowledgments
This work was supported in part by a University Cancer Foundation grant from the University of Texas M. D. Anderson Cancer Center (MDACC) (to XYH) and the National Institutes of Health (NIH) grant CA16672 for M. D. Anderson’s histology core facility. The authors thank the staff of histology core facility for assistance in tissue sectioning, John Spencer PhD for helpful discussions, and Bryan Tutt for editorial assistance.
Footnotes
Conflict of interest: There is no conflict of interest to disclose for any of the authors.
Author’s contribution
XYH initiated and orchestrated the study and wrote the manuscript; XYH and KCS performed DNA experiments and data analysis and had full access to the data; SVF, LF-C, and FV-O provided tissue and clinical data of the cases; all contributed to the editing and final approval of the paper.
Dedication
Dr. Han wishes to dedicate this paper to the memory of Kurt Clement Sizer, MD who died recently of an illness.
References
- 1.Monot M, Honoré N, Garnier T, et al. On the origin of leprosy. Science. 2005;308:1040–2. doi: 10.1126/science/1109759. [DOI] [PubMed] [Google Scholar]
- 2.Gelber RH. Leprosy (Hansen’s Disease) In: Kasper DL, Braunwald E, Fauci AS, Hauser SL, Longo DL, Jameson JL, editors. Harrison’s Principles of Internal Medicine. 16. New York: McGraw-Hill; 2005. pp. 966–972. [Google Scholar]
- 3.World Health Organization. Weekly Epidemiological Record. 2008 Aug 15;83(33):293–300. [Google Scholar]
- 4.Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis. 1966 Jul-Sep;34(3):255–73. [PubMed] [Google Scholar]
- 5.Han XY, Seo Y-H, Sizer KC, Taylor S, May GS, Spencer JS, Li W, Nair RG. A new Mycobacterium species causing diffuse lepromatous leprosy. Am J Clin Pathol. 2008;130:856–864. doi: 10.1309/AJCPP72FJZZRRVMM. [DOI] [PubMed] [Google Scholar]
- 6.Han XY, Sizer KC, Thompson EJ, Kabanja J, Li J, Hu P, Gómez-Valero L, Silva FJ. Comparative sequence analysis of Mycobacterium leprae and the new leprosy-causing Mycobacterium lepromatosis. J Bacteriol. 2009;191 (19):6067–74. doi: 10.1128/JB.00762-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole ST, Eiglmeier K, Parkhill J, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–11. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 8.Monot M, Honoré N, Garnier T, Zidane N, et al. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet. 2009;41:1282–9. doi: 10.1038/ng.477. [DOI] [PubMed] [Google Scholar]
- 9.Lucio R, Alvarado Y. Opusculo sobre el mal de San Lazaro o elefanciasis de los Griegos. Mexico: M. Murguia y Cia; 1852. p. 53. [Google Scholar]
- 10.Latapi F, Chevez-Zamora A. The “spotted” leprosy of Lucio: an introduction to its clinical and histological study. Int J Lepr. 1948;16:421–37. [Google Scholar]
- 11.Frenken JH. Diffuse leprosy of Lucio and Latapi. Oranjestad, Aruba, Netherlands Antilles: De Wit; 1963. [Google Scholar]
- 12.Vargas-Ocampo F. Diffuse leprosy of Lucio and Latapí: a histologic study. Lepr Rev. 2007;78(3):248–60. [PubMed] [Google Scholar]
- 13.Rea TH. Lucio’s phenomenon: an overview. Lepr Rev. 1979;50:107–12. doi: 10.5935/0305-7518.19790016. [DOI] [PubMed] [Google Scholar]
- 14.Rea TH, Jerskey RS. Clinical and histologic variations among thirty patients with Lucio’s phenomenon and pure and primitive diffuse lepromatosis (Latapi’s lepromatosis) Int J Lepr Other Mycobact Dis. 2005;73(3):169–88. [PubMed] [Google Scholar]
- 15.Ang P, Tay YK, Ng SK, Seow CS. Fatal Lucio’s phenomenon in two patients with previously undiagnosed leprosy. J Am Acad Dermatol. 2003;48:958–61. doi: 10.1067/mjd.2003.326. [DOI] [PubMed] [Google Scholar]
- 16.Choon SE, Tey KE. Lucio’s phenomenon: a report of three cases seen in Johor, Malaysia. Int J Dermatol. 2009;48:984–88. doi: 10.1111/j.1365-4632.2009.04078.x. [DOI] [PubMed] [Google Scholar]
- 17.Kumari R, Thappa DM, Basu D. A fatal case of Lucio phenomenon from India. Dermatol Online J. 2008 Feb 28;14(2):10. [PubMed] [Google Scholar]
- 18.Romero A, Ibarra AB, Fallas M. Clinical study of lepromatous leprosy in Costa Rica. Int J lepr. 1949;17:27–33. [PubMed] [Google Scholar]
- 19.Kaur C, Thami GP, Mohan H. Lucio phenomenon and Lucio leprosy. Clin Exp Dermatol. 2005;30:525–27. doi: 10.1111/j.1365-2230.2005.01860.x. [DOI] [PubMed] [Google Scholar]
- 20.Golchai J, Zargari O, Maboodi A, Maboodi A, Granmayeh S. Lepromatous leprosy with extensive unusual ulcerations and cachexia. Is it the first case of Lucio’s phenomenon from Iran? Int J Lepr Other Mycobact Dis. 2004;72:56–9. doi: 10.1489/1544-581X(2004)072<0056:LLWEUU>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 21.Ezzedine K, Malvy D, Beylot C, Longy-Boursier M. Autochthonous leprosy in metropolitan France presenting with a diffuse infiltration of the face and febrile illness. Int J Dermatol. 2009;48:69–72. doi: 10.1111/j.1365-4632.2009.03831.x. [DOI] [PubMed] [Google Scholar]
- 22.Derbes VJ, Samuels M, Williams OP, Walsh JJ. Diffuse leprosy; case in a Louisiana Negro. Arch Dermatol. 1960 Feb;81:210–24. doi: 10.1001/archderm.1960.03730020046009. [DOI] [PubMed] [Google Scholar]
- 23.Costa IM, Kawano LB, Pereira CP, Nogueira LS. Lucio’s phenomenon: a case report and review of the literature. Int J Dermatol. 2005;44:566–71. doi: 10.1111/j.1365-4632.2005.02566.x. [DOI] [PubMed] [Google Scholar]
- 24.Azulay-Abulafia L, Pereira Spinelli L, Hardmann D, et al. Lucio phenomenon. Vasculitis or occlusive vasculopathy? Hautarzt. 2006;57:1101–5. doi: 10.1007/s00105-005-1086-3. [in German] [DOI] [PubMed] [Google Scholar]
- 25.Fenniche S, Benmously R, Sfia M, et al. Late-occurring cutaneous vasculitis after successful treatment of diffuse lepromatous leprosy: Lucio’s phenomenon. Med Trop (Mars) 2007;67:65–8. [in French] [PubMed] [Google Scholar]
- 26.Han XY, Pham AS, Tarrand JJ, Sood PK, Luthra R. Rapid and accurate identification of mycobacteria by sequencing hypervariable regions of the 16S ribosomal RNA gene. Am J Clin Pathol. 2002;118:796–801. doi: 10.1309/HN44-XQYM-JMAQ-2EDL. [DOI] [PubMed] [Google Scholar]
- 27.Liesack W, Pitulle C, Sela S, Stackebrandt E. Nucleotide sequence of the 16S rRNA from Mycobacterium leprae. Nucleic Acids Res. 1990;18:5558. doi: 10.1093/nar/18.18.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox RA, Kempsell K, Fairclough L, Colston MJ. The 16S ribosomal RNA of Mycobacterium leprae contains a unique sequence which can be used for identification by the polymerase chain reaction. J Med Microbiol. 1991;35:284–90. doi: 10.1099/00222615-35-5-284. [DOI] [PubMed] [Google Scholar]
- 29.Plikaytis BB, Gelber RH, Shinnick TM. Rapid and sensitive detection of Mycobacterium leprae using a nested-primer gene amplification assay. J Clin Microbiol. 1990;28(9):1913–7. doi: 10.1128/jcm.28.9.1913-1917.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez Roa RI, Morris MF. Leprosy in Mexico. Jpn J Lepr. 2006;75(1):51–8. doi: 10.5025/hansen.75.51. [DOI] [PubMed] [Google Scholar]
- 31.Matsuoka M, Gonzalez AV, Estrada I, Carreno-Martinez, Fafutis-Morris M. Various genotypes of Mycobacterium leprae from Mexico reveal distinct geographic distribution. Lepr Rev. 2009;80:322–326. [PubMed] [Google Scholar]
- 32.Rea TH, Bevans L, Taylor CR. The histopathology of the spleen from a patient with lepromatous leprosy. Int J Lepr Other Mycobact Dis. 1980 Sep;48(3):285–90. [PubMed] [Google Scholar]