Abstract
Single-molecule force spectroscopy has emerged as a powerful tool to investigate the forces and motions associated with biological molecules and enzymatic activity. The most common force spectroscopy techniques are optical tweezers, magnetic tweezers and atomic force microscopy. These techniques are described and illustrated with examples highlighting current capabilities and limitations.
Introduction
Force plays a fundamental role in biological processes. All biological motion, from cellular motility to the replication and segregation of DNA, is driven by molecular scale forces. Conversely, the elimination or reduction of motion through the binding of ligands to their cognate receptors, or through the folding of a polypeptide establishing a stable three-dimensional structure, involves the formation of bonds that overcome thermal and other forces. The ability to study these fundamental processes has been revolutionized over the last 20 years by the development of techniques that permit measurement of force and displacement generated by single molecules ranging from cells to proteins. Whereas there is an ever expanding repertoire of single-molecule manipulation techniques, including optical tweezers, magnetic tweezers, atomic force microscopy (AFM), micro-needle manipulation1, 2, biomembrane force probe3, and flow induced stretching4, 5, the first three are the most commonly employed and are the focus of this review (Table 1). I begin with an overview highlighting areas of investigation opened up by the ability to apply force and to measure displacement at the single-molecule level. This is followed by a description of the operating principles and practical implementation of optical tweezers, magnetic tweezers and the AFM. Many excellent reviews and detailed technical reports on the design, fabrication and use of these instruments are available6–12. This review is intended as an aid in choosing the technique best suited for a particular application by describing what is currently feasible with each, as well as providing details concerning strengths, limitations, and practical considerations. The enormous potential of these techniques is just beginning to be fully realized and continuing developments will provide further opportunities to probe fundamental biological processes.
Table 1.
Comparison of single-molecule force spectroscopy techniques
| Optical tweezers | Magnetic tweezersa | AFM | |
|---|---|---|---|
| Spatial resolution (nm) | 0.1–2 | 5–10 (2–10) | 0.5–1 |
| Temporal resolution (s) | 10−4 | 10−1–10−2 (10−4) | 10−3 |
| Stiffness (pN·nm−1) | 0.005–1 | 10−3–10−6 (10−4) | 10–105 |
| Force range (pN) | 0.1–100 | 10−3–102 (0.01–104) | 10–104 |
| Displacement range (nm) | 0.1–105 | 5 – 104 (5–105) | 0.5–104 |
| Probe size (μm) | 0.25–5 | 0.5–5 | 100–250 |
| Typical applications | 3-d manipulation Tethered assay Interaction assay |
Tethered assay DNA topology (3-d manipulation) |
High force pulling and interaction assays |
| Features | Low noise and drift dumbbell geometry | Force clamp Bead rotation Specific interactions |
High resolution imaging |
| Limitations | Photodamage Sample heating Non specific |
No manipulation (Force hysteresis) | Large high- stiffness probe Large minimal force Non specific |
Values for electromagnetic tweezers are shown in parenthesis
Current single-molecule manipulation spans six orders of magnitude in length (10−10–10−4 m) and force (10−14–10−8 N), ranging from the manipulation of cells (~100 μm), to the measurement of RNA polymerase advancing a single base pair (0.34 nm) along DNA13, and the mechanical disruption of covalent bonds14 (nN) and nucleic acid folding kinetics15 (~0.1 pN). The power and breadth of these techniques is highlighted by the wide variety of measurements and systems investigated. Single cells can be manipulated to probe the strength and location of receptor binding16 and adhesion or to measure traction and adhesion forces17. Viscoelastic properties can be measured on short length-scales and in small volumes18, such as within cells19. Early applications of single-molecule force and displacement measurements included the characterization of conventional motor proteins such as kinesins20, 21 and myosins22. The step size, stall-force force and processivity of these motors were established paving the way to probe fundamental questions concerning the coupling between chemical and mechanical cycles23. More generally, the application of force provides a means to selectively modify the steps in a biochemical reaction cycle that involves motion. Detailed measurements of the force extension relationship (i.e. the elasticity) of individual polymers, in particular nucleic acids1, 4, opened up the possibility of investigating unconventional nucleic-acid molecular motors that translocate or otherwise modify DNA or RNA. In parallel with these developments in single-molecule enzymology, similar techniques were developed to mechanically rupture molecular bonds24–26. The analysis of rupture force, or force spectra, provides a measure of bond energies, lifetimes, and more recently, entire energy landscapes27, 28. Force spectroscopy is employed as a tool to characterize ligand and antibody binding29 and has been extended to study the complex and multi-state unfolding of single proteins and nucleic acid structures30. Force spectroscopy in turn spurred the development of, and benefited greatly from, theoretical approaches that permit the extraction of detailed equilibrium thermodynamic parameters from inherently non-equilibrium pulling experiments27, 31, 32.
Over and above the ability to apply force and measure displacement, single-molecule techniques afford a host of additional advantages. Measuring single molecules obviates problems associated with population averaging inherent in ensemble measurements. Rare or transient phenomena that would otherwise be obscured by averaging can be resolved provided that the measurement technique has the required resolution and that the events can be captured often enough to ensure that they are not artifactual. Likewise, multi-state or multi-species distributions can be directly measured along with static and dynamic enzymatic heterogeneity33. In some instances, kinetic rates can be directly measured from single-molecule recordings or determined by analyzing the distribution of event times34–36. For the specific case of enzymes, single-molecule measurements are intrinsically synchronized and properties such as processivity are readily determined.
General considerations for single-molecule manipulation
Single-molecule measurements employing optical or magnetic tweezers or AFM have many elements in common (Table 1). Typically, one end of the molecule under study is attached to a surface and the free end is attached to a probe: an optically trapped bead, magnetic bead, or AFM tip, through which force is applied. The importance of the attachments can not be over emphasized. Ideally, the bonds at the surface and probe would specifically bind the ends of the molecule, would support infinite loads, and would not affect the mechanical or biological properties of the attached molecule. These ideals are approximated by a variety of attachment schemes ranging from non-specific adsorption to specific covalent attachments. Covalently modified nucleotides containing a carbon spacer arm terminated in a reactive moiety provide a convenient means of labeling nucleic acids at their ends with small molecules and ligand-receptor pairs such as biotin-avidin, or antibody-antigen pairs such as digoxigenin and anti-digoxigenin, are commonly used to provide specific and relatively tight binding to the surface and to the probe37. Proteins are more challenging to modify but purification tags such as biotin and hexahistidine provide a convenient means of attachment, as do reactive cysteine residues that may be naturally occurring and solvent exposed, or specifically introduced within the protein of interest. Non specific adsorption remains the simplest and most common attachment method for AFM based force spectroscopy measurements, however, more sophisticated specific attachment schemes have been developed38. Direct attachment of the molecule to the probe handle and attachment of intermediate antibodies and ligands is facilitated by commercially available functionalized polystyrene and magnetic beads, and AFM tips. The prevention of nonspecific binding interactions between the probe handle, the molecule of interest, and the surfaces of the flow cell or experimental chamber are equally if not more important than the specific binding interactions to the molecule of interest. Nonspecific binding can introduce artifact and uncertainty in the data, and in some instances can prevent the collection of any meaningful data. To prevent non-specific interactions inert proteins such as bovine serum albumen and non-ionic surfactants are used to passivate surfaces and probes.
For all three techniques, the extension of the molecule is determined from the position of the probe relative to the surface. For optical tweezers and the AFM, force is also derived from the deviation of the probe from its equilibrium position. The precision and accuracy of the measurements therefore depend critically on the ability to measure the position of the probe. Whereas the three techniques differ in the method used to measure position, they share many issues related to position measurement. The environment in which the measurements are made can profoundly influence the quality of the results. Temperature stability is the single largest concern, as the thermal expansion resulting from a 1°C temperature gradient can be hundreds of nanometers or more. Mechanical vibrations, air currents, acoustic and electrical noise are some of the other environmental factors that adversely affect single-molecule manipulation measurements. High-precision and high-stability measurements typically require instruments housed in acoustically isolated, temperature controlled environments. That said, the details of the system under study should dictate the necessary environmental conditions. For example, measurements of a fast process will be more susceptible to noise than to slow temperature drifts, whereas the opposite would hold true when measuring a slow process.
Optical tweezers
Optical tweezers (Fig. 1) or optical traps are arguably the most versatile single-molecule manipulation technique. They can exert forces in excess of 100 pN on particles ranging in size from nanometers to microns while simultaneously measuring the three-dimensional displacement of the trapped particle with sub-nanometer accuracy and sub- millisecond time resolution. These properties make them extremely well suited for the measurement of force and motion.
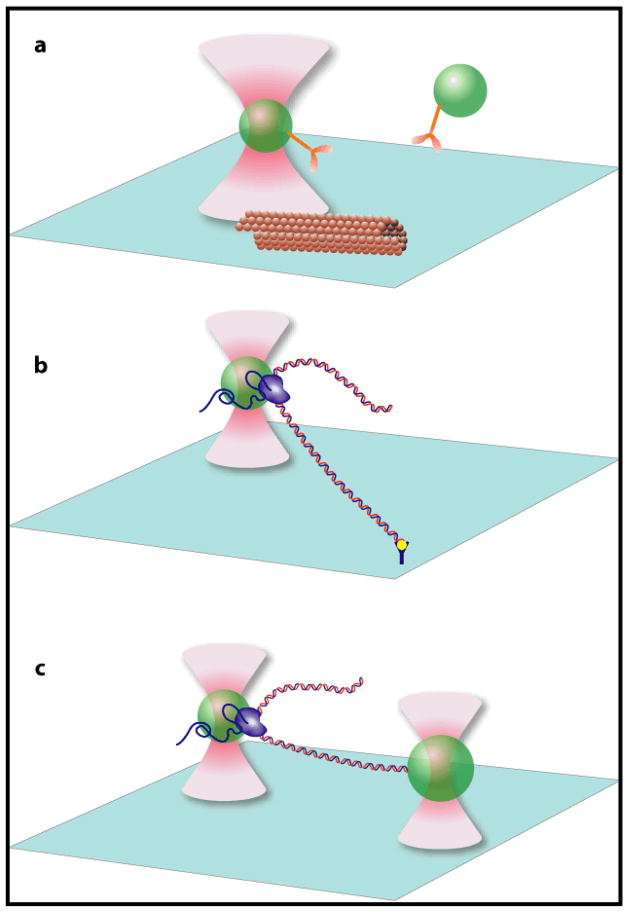
Fig. 1.
Optical tweezers based assays (not to scale). (a) Interaction assay. A low concentration of polystyrene or silica beads (green spheres) sparsely coated with kinesin molecules (yellow) are diffusing in solution. One bead is captured by the optical trap formed near the focus of an infrared laser (pink). The assay consists of bringing the trapped bead to the microtubule (brown tube) attached to the surface of the trapping chamber. The force and displacement generated by the individual kinesin molecule as it walks along the microtubule are determined from the displacement of the bead in the optical trap11, 20, 57. (b) Tethered assay. An RNA polymerase molecule (purple) is attached to an optically trapped bead (green sphere), and the free end of the DNA template (red and blue) is attached to the surface of the trapping chamber. As the DNA is transcribed, the bead is pulled along the DNA by the polymerase. By moving the stage to compensate for this motion, thereby keeping the bead at the same position in the optical trap, long transcriptional records can be obtained at a constant force63. (c) Dumbbell assay. This assay is similar to the tethered assay but the free end of the DNA is attached to a second bead, which is held in a second, independent, optical trap. The force on the bead is kept constant by moving one of the traps13, 62, 64.
An optical trap is created by focusing a laser to a diffraction-limited spot with a high numerical aperture (NA) microscope objective39. Dielectric particles in the vicinity of the focus experience a three-dimensional restoring force directed toward the focus. The dielectric particle is polarized by the optical field, and the interaction of this optically induced dipole with the steep gradient near the focus of the laser results in a force directed along the gradient. In addition to the gradient force, there is what has been termed a scattering force directed along the beam propagation direction, which results in a shift of the equilibrium trapping position slightly past the focus. To form a stable trap the gradient force along the optical axis must overcome this scattering force, which necessitates the very steep gradient obtained with a high NA objective. For small displacements (~150 nm) of the trapped object from its equilibrium position the force is linearly proportional to the displacement, and the optical trap can be well approximated as a linear spring. The spring constant, or stiffness, depends on the steepness of the optical gradient, i.e. how tightly the laser is focused, the laser power, and the polarizability of the trapped object. Particles ranging in size from ~20 nm to several microns can be stably trapped. These include single cells40, organelles within cells41, lipid vesicles42, 43 and polystyrene or silica microspheres used alone or as probes linked to a molecule of interest.
Technical requirements
Optical tweezers run the gamut from simple manipulation to sophisticated custom built instruments with sensitive position detectors and dynamic position control. The core elements shared by all optical tweezers are a trapping laser and a high NA microscope objective. Trapping lasers should have a Gaussian output intensity profile, or equivalently, an M2 value near unity, to achieve the smallest focal spot producing the largest optical gradient. High precision optical trapping measurements require a trapping laser with superior pointing and power stability. Fluctuations in beam pointing result in spurious motions of the optical trap, while power fluctuations result in force fluctuations, both of which increase measurement noise. Trapping lasers operating in the near infrared (800–1100 nm) minimize optically induced damage in biological specimens40. For most biological applications the trapping laser of choice is the diode pumped neodymium yttrium aluminum garnet (Nd:YAG) with a wavelength of 1064 nm. These lasers display exceptional power and pointing stability and are available with output powers in excess of 10 W. Single-mode diode lasers and high power near-infrared fiber lasers44 have also been used for optical trapping.
The high NA objective used to focus the laser is the second most important component of the optical trapping instrument. The NA of the trapping objective should be at least 1.2 to achieve the steep focus needed to create a stable trap. This necessitates the use of a water or oil immersion objective. Whereas oil immersion objectives offer the highest NA, they introduce spherical aberrations that degrade optical trap performance deep in solution45, 46. Water immersion objectives do not suffer from spherical aberrations and permit trapping deep in solution. The other factor to consider in the choice of microscope objective is the optical transmission characteristics at the trapping wavelength. High NA objectives are typically optimized for use with visible light, and their transmission can vary greatly in the near infrared40. Some objective manufacturers provide transmission curves in the near infrared but if these are not available, or if the transmission has to be accurately known, it can be measured by the dual objective technique47, 48. In our experience, fluorescence objectives and objectives specifically designed for use in the near infrared have the highest near infrared transmission, whereas highly corrected objectives have the lowest transmission.
A basic optical tweezers provides a means of non-invasively manipulating objects in solution. Adding position detection of the trapped particle permits the simultaneous measurement of displacement and force, greatly enhancing the capabilities of the instrument. Whereas several methods of position detection have been developed, the most versatile and sensitive is back focal plane (BFP) interferometry49, 50, which relies on the interference between light scattered by the trapped bead and un-scattered light to measure the three-dimensional position of the bead relative to its equilibrium position. The interference is measured with a quadrant photodiode (QPD), or position sensitive detector (PSD) placed in a plane optically conjugate to the BFP of the condenser, i.e. the BFP of the condenser is imaged onto the QPD or PSD. The detectors are sensitive to minute intensity asymmetries in the interference pattern. A trapped bead at its equilibrium position produces a symmetric interference pattern and a null detector signal. Displacement of the bead results in an asymmetric interference profile, which generates a detector signal proportional to the bead displacement. Axial motion of the trapped particle also changes the interference pattern resulting in changes in the total intensity at the detector. Sub-nanometer spatial resolution and bandwidths in excess of 100 kHz have been achieved with this detection scheme49, 51. The most straightforward implementation of BFP interferometry relies on the interference from the scattered trapping laser, however it is sometimes more convenient to employ a second low power detection laser11. By splitting one laser into two beams based on polarization, or using two different detection lasers, it is possible to track simultaneously the position of two trapped beads52. Recently, BFP detection has been implemented with light backscattered from the trapped bead53. This scheme simplifies position detection and will likely result in improved measurement stability and noise performance.
The full potential of an optical tweezers instrument can be realized with the addition of dynamic control of the trap position in the specimen plane and the position of the trapping chamber. Dynamic position control can be incorporated into a feedback loop to maintain a constant force on the trapped bead11, or to actively compensate for thermal drift54. Moving the trap by deflecting the laser beam with galvanometer or piezoelectric (piezo) actuated mirrors, or an acousto-optic deflector (AOD) can be very fast (~10 μs for an AOD), but the range of motion is typically limited to a few microns in a single axial plane. Alternatively, a piezo stage can be used to move the trapping chamber with nm accuracy while keeping the trap position fixed. Although this approach is slower (~10 ms response time), full three-dimensional control is possible over a large range of motion (~100 μm).
Optical tweezers calibration
Position and force calibration for all three techniques reviewed is accomplished by similar means: The probe is treated as a linear spring, the Brownian motion of which is related to the spring stiffness (See Box 1), and force is determined from Hooke’s law (F = −kx). Position calibration is typically accomplished by moving the probe through a known distance while recording the position signal. To provide an illustration of the calibration process, I present a detailed consideration of the techniques used to calibrate optical tweezers. Less detailed descriptions, highlighting differences or additional considerations, will be presented for the other techniques.
Box 1. Fundamental resolution limits of displacement, force and time for single-molecule force spectroscopy.
Single-molecule force spectroscopy techniques employ a micron sized probe to apply force and measure displacement. Because of their small size, these probes are subject to thermal fluctuations that impose fundamental limits on displacement, force and time resolution. To a good approximation, the single-molecule probe can be considered to be attached to a linear spring with stiffness α. This stiffness can be the intrinsic stiffness of the probe, as is the case for an optically trapped bead or an AFM cantilever, or it may be dominated by the stiffness of the molecule to which the probe is attached, as is the case for a magnetic bead in a magnetic tweezers and in many pulling experiments regardless of the technique used to apply force. Spatial resolution is determined by the thermal noise of the probe, which is given by:
| (1) |
where δx is the magnitude of the position noise, kBT is the thermal energy, and α is the spring stiffness. From Hooke’s law (F = −αx), the corresponding force resolution is:
| (2) |
The magnitude of the noise can be reduced by filtering the position data. The effect of filtering the can be determined by considering the power spectrum of the thermal motion, which is a Lorentzian (see Fig. 4):
| (3) |
where S(f) is the power per unit frequency, expressed as displacement2·Hz−1, f is the frequency in Hz, β is the hydrodynamic drag on the probe (for a sphere of radius a in a medium of viscosity η, β = 6πηa), and f0 = α·(2π β)−1 is the characteristic roll-off frequency of the motion. The area under the power spectrum is the position variance (δx)2, which is the square of the position noise amplitude. The effect of filtering the position signal can be approximated by calculating the area under the power spectrum over the frequency range from zero Hz to the filter cut-off frequency, Δf. If the cut-off frequency is much less than the roll-off frequency (f0) of the power spectrum, the position noise amplitude is approximately:
| (4) |
Spatial resolution can therefore be improved by increasing the stiffness, reducing the bandwidth (Δf), or decreasing the drag, which is proportional to the size of the probe and the viscosity of the medium. Stiffness has the greatest impact on spatial resolution, whereas bandwidth and hydrodynamic drag have lesser effects (see Fig. 4). Filtering the signal also improves the force resolution:
| (5) |
which can only be improved by decreasing the drag or the bandwidth. Decreasing the bandwidth, however, reduces the temporal resolution of the measurement. Temporal resolution is inversely proportional to the bandwidth if the signal is filtered or to the roll-off frequency (f0) of the Lorentzian if it is not. Temporal resolution can therefore be improved by increasing the stiffness or decreasing the drag. Maximal resolution of displacement, force and time is achieved by minimizing hydrodynamic drag on the probe. This can be accomplished by reducing the size of the probe, or by reducing the viscosity of the medium.
Temporal resolution and measurement bandwidth are crucially important considerations in calibration techniques that rely on thermal motion. Both power spectrum and variance stiffness calibrations rely on measuring the entirety of the noise, and significant errors are introduced if the position signal is inappropriately filtered or if the measurement technique does not have sufficient bandwidth to capture high frequency motion. Berg-Sorensen and coworkers provide an excellent treatment of these and other issues encountered in fitting power spectra to obtain stiffness161.
Accurate determination of displacement and force requires calibrating the position detector and measuring the stiffness of the optical trap. Position detector calibration is achieved by moving the trapped bead through a known distance while recording the position signal. A bead, stuck to the surface of the trapping chamber, is moved with a calibrated piezo stage, or by scanning the trapping laser faster than the trapped bead can respond (see Box 1), thereby moving the laser with respect to the effectively stationary bead55. If an independent detection laser is employed, calibration can be performed by moving the bead through the detection laser by scanning the trap position11. Due to variation among beads of nominally the same size, the best practice is to calibrate each bead prior to acquiring data.
A well calibrated position detector is helpful but not essential for calibrating trap stiffness. The simplest stiffness calibration method relies on the thermal fluctuations of the trapped bead, which is equivalent to the Brownian motion of an over-damped spring. The power spectrum of the fluctuations is a Lorentzian (see Equation 3 in Box 1) with roll-off frequency f0=α· (2πβ) −1 where α is the trap stiffness, and β is the hydrodynamic drag on the bead. The roll-off frequency can be obtained by fitting the power spectrum, which need not be calibrated, and the drag can be calculated for simple shapes. A related stiffness calibration technique makes use of the equipartition theorem, which states that each degree of freedom has ½kBT of thermal energy. Equating this with the energy associated with fluctuations of the trapped particle, ½α<x2>, where α is the trap stiffness and <x2> is the positional variance, gives an expression for the trap stiffness: α = kBT<x2>−1. Unlike the power spectrum method, the variance method requires a calibrated position detector, however neither the drag coefficient of the trapped object nor the viscosity of the medium are required. Because these two calibration methods rely on thermal fluctuations, they provide a measure of trap stiffness near the equilibrium position of the trap. Measuring the trap stiffness for greater displacements and determining the region over which the stiffness is constant requires an external force to displace the trapped bead. Rapidly moving the trapping chamber, i.e., with a piezo stage, exerts a large calculable drag force on the trapped bead and provides a measure of trap stiffness at large displacements. It is good practice to measure the stiffness as a function of laser power using all three methods, subtle problems with the trap or the position detection system can be uncovered. There are many variants7 of these stiffness calibration methods. One elegant method permits simultaneous calibration of stiffness and position by recording the power spectrum of a trapped bead while the trapping chamber is oscillated56. This technique has the advantage of not requiring knowledge of the hydrodynamic drag on the bead, thus neither the particle size nor the sample viscosity are required.
Applications of optical tweezers
The extraordinary versatility of optical tweezers is evident from the vast array of measurements this technique has enabled. In one class of optical trapping assay, the three-dimensional manipulation capabilities are exploited to impose a specific interaction between the trapped object and a fixed partner, and to measure the force and displacement resulting from the interaction (Fig. 1a). This type of assay is typified by the measurement of the force and displacement of optically trapped kinesin coated beads moving along fixed microtubules, pioneered by Block and coworkers20, 21, 57. In a similar assay, the binding probability and unbinding force was measured for virus coated beads brought into contact with erythrocytes58, and the binding strength and activation state of single fibrinogen-integrin pairs was measured on living cells16. Recently, Kessemaker and colleagues directly observed individual incorporation and shrinkage events at the dynamic end of a microtubule59. They used a pair of optical traps in a “key-hole” configuration, in which one trap applies a force on a bead attached to the microtubule, while the second trap is rapidly scanned in a line to orient the microtubule. With this novel trapping configuration, the growing end of the microtubule could be pushed against a micro-fabricated barrier and the dynamics of subunit incorporation and loss could be measured from motion of the trapped bead. Employing the same optical trapping assay, Footer and coworkers directly measured the force associated with actin polymerization60.
In the second class of optical trapping assays, the optical trap is used as a force and displacement transducer of a probe that is attached to a substrate by the molecule of interest (Fig. 1b). This assay is typified by the measurement of translocation and force generation of individual RNA polymerase molecules as they transcribe DNA, pioneered by Block and coworkers. In this assay an immobilized RNA polymerase reels in a bead attached to the end of the transcribed DNA, which can be measured from the resulting motion of the bead in the optical trap. This single-molecule assay has revealed details of transcription including the stall force61 (~30 pN), transcriptional pausing62, 63, backtracking of the polymerase along the DNA template64, and the mechanism of translocation13, which Abbondanzieri and coworkers demonstrated is a Brownian ratchet rather than a power-stroke. These last measurements were enabled by the use of a DNA dumbbell assay in which the transcribing RNA polymerase is attached to one trapped bead, while the free end of the DNA is attached to a second optically trapped bead (Fig. 1c). Suspending both beads in solution resulted in a dramatic reduction in noise and drift, ultimately permitting observation of individual basepair steps of RNA polymerase translocation65. A similar DNA dumbbell geometry was employed by Dame and coworkers to probe the mechanical bridging of two DNA molecules by the bacterial DNA histone-like nucleoid structuring protein H-NS66. In this novel assay, two DNA molecules were extended between two pairs of optical traps and bridged by H-NS proteins. The molecular interactions between the DNA molecules and H-NS were revealed by pulling the DNA molecules apart with an unzipping (perpendicular to the DNA molecules) or a shearing (parallel to the DNA molecules) force. The mechanical unzipping of the DNA bridges revealed that individual H-NS proteins bind both DNA strands at intervals close to the helical repeat distance (~3.5 nm). The ability to manipulate multiple DNA molecules afforded by this and related single-molecule assays permit the study of inter-molecular reactions such as recombination and strand exchange.
In addition to the study of processive nucleic acid enzymes, such as the viral DNA packaging motor67, the optical trapping puling assay has been adapted to other measurements. The mechanical unfolding of proteins and later of nucleic acid structures was pioneered by the Bustamante group25, 68. Due to the larger forces required to unfold most proteins, optical trapping methods have generally given way to AFM based force spectroscopy. Conversely, the forces (~15 pN) and displacements (~nm) associated with nucleic acid folding are ideally suited for optical tweezers based measurements. Liphardt and coworkers measured the mechanical unfolding and the effect of load on the unfolding and refolding kinetics of several hairpin loops from the Tetrahymena thermophila group I ribozyme, which provided details of the folding energy landscape, including the position and height of the energy barriers, and the folding sequence of multi-hairpin structures68. The ability to manipulate and characterize nucleic acid hairpins paved the way for their use in enzymatic assays. Dumont and coworkers measured the processive opening of an RNA hairpin by the hepatitis C virus RNA helicase NS369. Since each base unwound increases the end-to-end extension of the molecule by two bases, the effective spatial resolution of the assay was increased two-fold. With this enhanced resolution individual 11 base-pair ATP-dependent unwinding events and 3.6 base-pair sub steps were observed. In a novel approach combining two heretofore disparate single-molecule approaches, Keyser and coworkers used an optical trap to directly measure the force on a single molecule of λ-DNA in a solid state nanopore as a function of applied voltage across the pore and as a function of the buffer KCL concentration70. Surprisingly, the force on the DNA in the pore remained constant at ~0.2 pN/mV independent of the salt concentration, which is consistent with ~0.5 electrons per basepair of DNA, only 25% of the full charge on the DNA70.
Although the vast majority of optical trapping configurations result in rotationally isotropic trapping potentials, work by the Rubinzstein-Dunlop group71, 72 and more recently by the Wang group73, 74, has demonstrated the ability to impose an optical torque on birefringent particles. Circularly polarized light carries spin angular momentum that can be transferred to an optical active trapped object though a process that is analogous to the transfer of linear photon momentum to a dielectric particle that is the basis of optical trapping. LaPorta and Wang used this principle to generate an optical torque clamp instrument that was capable of either spinning a birefringent particle in the optical trap, or of freezing out the thermal rotations of the particle by actively clamping the position and measuring the torque required74. In more recent work, Duefel microfabricated quartz cylinders that could be specifically labeled on one end, to which DNA would bind. This construct, in conjunction with the optical torque clamp, permitted a direct measure of the torque required to Buckle DNA as a function of applied load73.
Limitations and drawbacks of optical tweezers
The versatility and precision afforded by optical tweezers are accompanied by important limitations and drawbacks that must be carefully considered prior to and during their use.
Whereas many of the advantages afforded by optical tweezers stem from their purely optical origin, there are some important difficulties associated with using light to generate force. Since trap stiffness depends on the gradient of the optical field, optical perturbations that affect the intensity or the intensity distribution will degrade the performance of the optical tweezers. High-resolution optical trapping is therefore limited to optically homogeneous preparations and highly purified samples. Particular care must be exercised when creating multiple optical traps through polarization splitting or rapid beam scanning. Optical interference and non-ideal behavior of the beam steering optics can result in the generation of ghost traps and spurious position signals, among other artifacts. Optical tweezers also lack selectivity and exclusivity. Essentially any dielectric particle near the focus of the trapping laser will be trapped, and the number of particles that can be simultaneously trapped can be quite large. For this reason, samples in which the objects that will be trapped are freely diffusing must be kept at extremely low concentrations to prevent additional objects from being trapped once the first is captured. Moreover, trapping in cell extract or any medium containing impurities is generally precluded as trapped impurities can distort or mask the position signal, though in vivo optical trapping of lipid vesicles within eukaryotic cells43 and organelles within yeast cells41 has been successfully implemented.
The high intensity at the focus of the trapping laser that forms the optical trap, typically 109–1012 W·cm−1, results in local heating. Trapping transparent dielectric particles in water at a trapping wavelength of 1064 nm results in modest heating on the order of 1°C per 100 mW of power in the specimen plane75, 76. Trapping absorbing particles or trapping at laser wavelengths more strongly absorbed by water, can result in significantly more heating77. Local heating can influence enzymatic activity and change the local viscosity of the medium, whereas steep thermal gradients may produce convention currents that can adversely affect the measurements. Local heating in the vicinity of the optical trap can be calculated75 and several techniques have been developed to measure the temperature directly75, 77.
Optical damage induced in trapped specimens is less well understood. Trapping with laser wavelengths in the near infrared (800–1100 nm) minimizes photodamage, however an oxygen-mediated photodamage process, likely involving singlet oxygen or other reactive oxygen species, persists throughout this wavelength region40. Removing molecular oxygen from the trapping medium with an enzymatic scavenging system, or by exchanging the oxygen with an inert gas, significantly reduces photodamage40. For optical trapping involving living organisms, 830 and 970 nm trapping wavelengths have been found to minimize photodamage in E. coli40 and Chinese hamster ovary cells78.
In addition to the technical concerns with optical tweezers, there are a number of practical matters that also must be taken into account. The range of applied forces is limited to 0.1–100 pN. The low end of the range is set by the lowest stiffness ensuring trap stability. The upper limit is set by the maximum power in the specimen plane. The range of motion that can be measured with a fixed optical trap is limited to ~400 nm or less, whereas the range over which the stiffness is constant is significantly smaller (~150 nm). Range of motion can be increased almost without limit through the incorporation of dynamic position control of the trap, the stage, or both, but these necessarily increase the complexity of the optical tweezers instrument. An optical tweezers capable of applying well controlled loads and accurately measuring displacements is a complex, delicate and expensive instrument. Optical tweezers with a variety of features and capabilities are commercially available, however they don’t currently include the position detection and dynamic position control required to make sensitive single-molecule measurements of force and displacement described above. Adapting a commercially available optical tweezers, or building a custom instrument, to meet the requirements of a given set of experiments is challenging and time consuming, but it is possible, as evidenced by the increasing number of labs around the world pursing high resolution optical trapping measurements.
Magnetic tweezers
Magnetic tweezers (Fig. 2) are the most straightforward of the three techniques to implement. A basic magnetic tweezers consists of a pair of permanent magnets placed above the sample holder of an inverted microscope outfitted with a CCD camera linked to a frame grabber79, 80. Magnetic tweezers are capable of exerting forces in excess of one nN (electromagnetic tweezers), and can be used to manipulate, and importantly rotate, magnetic particles ranging in size from 0.5–5 μm. Magnetic tweezers are unique in that they afford passive, infinite bandwidth, force clamping over large displacements. These characteristics are ideally suited to the study of nucleic acid enzymes, particularly DNA topoisomerases81–83 (Fig. 2c), and the rotary motor FoF1 ATPase(references???).
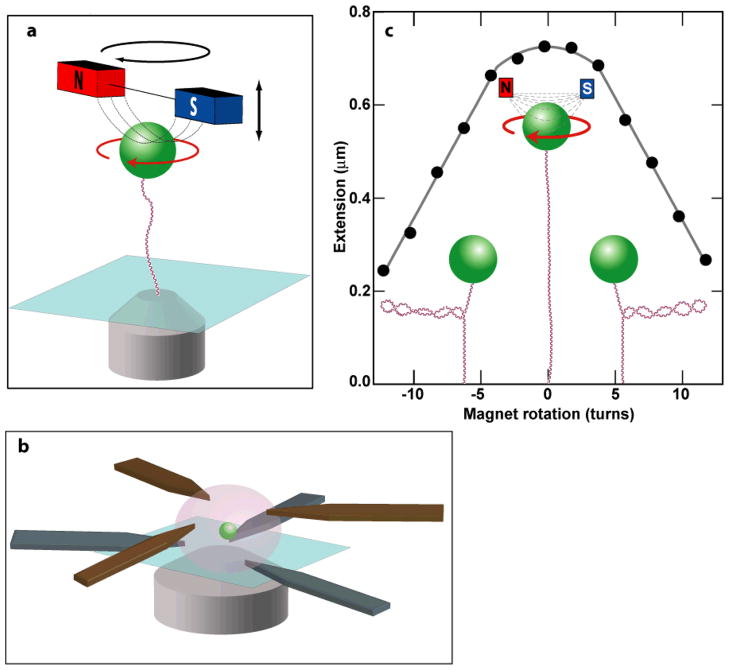
Fig. 2.
Magnetic tweezers. (a) Cartoon depicting the layout of a magnetic tweezers based on permanent magnets (not to scale). A superparamagnetic bead (green) is attached to the surface of the trapping chamber by a single molecule of DNA (red and blue). A pair of small permanent magnets (red and blue) above the trapping chamber produces a magnetic field gradient (dashed lines) along the axial direction, which results in a force on the bead directed up toward the magnets. The force is controlled by moving the magnets in the axial direction (black straight arrow). Rotation of the magnets (black circular arrow) produces rotation of the magnetic bead (red circular arrow) with a one-to-one correspondence. A microscope objective (grey) images the bead onto a CCD camera (not shown) for real time position tracking. (b) Schematic representation of an electromagnetic tweezers pole configuration permitting full three-dimensional control (adapted from reference84) (not to scale). Thin (~180 μm) pole pieces (brown and grey) are laser machined from magnetic foil. Two sets of three pole pieces are symmetrically arranged in two axial planes, which provide full three-dimensional control over the position of the bead (green sphere). The pole pieces are sandwiched between electromagnetic coils in an assembly that mounts on an inverted microscope (not shown). (c) DNA topology measured with magnetic tweezers (not to scale). The extension as a function of rotation for a 1 μm superparamagnetic bead (green) tethered to the surface by a 3 kb molecule of DNA (red and blue) under 0.4 pN of pulling force. As the DNA is over- or under-wound (supercoiled) there is a slight decrease in extension near zero turns, which is due to the accumulation of twist in the DNA molecule. At ±4 turns the DNA buckles, forming a plectoneme loop. Each subsequent turn increases the plectoneme by another loop, leading to a linear decrease in extension from 4 to 12 turns (left and right cartoons). Removal of the plectonemes by the activity of a topoisomerase can be directly observed in real time by monitoring the extension of a supercoiled DNA molecule.
Magnetic tweezers are similar in concept to optical tweezers; a magnetic particle in an external magnetic field experiences a force proportional to the gradient of the square of the magnetic field. High forces can be achieved with relatively small magnetic field strengths provided a very steep field gradient can be generated. The fields generated by sharp electromagnetic tips84, or small permanent magnets85, have been used to apply forces in excess of 200 pN on micron sized magnetic particles. Due to the steep gradient however, the force falls off rapidly with displacement away from the magnet. Consequently, appreciable force can only be applied on a particle in close proximity to the magnet, and the force is not constant for small displacements of the magnetic particle in the vicinity of the magnet. Larger magnets provide a higher magnetic field strength and a shallower field gradient, resulting in forces that vary more slowly over a larger area. A single magnet can be used to supply an attractive pulling force on a magnetic particle, but a minimum of two magnets are required to generate torque and apply force. For the most common case in which a superparamagnetic bead is manipulated, the external field induces a magnetic moment in the bead, which experiences a force in the direction of, and proportional to, the field gradient12. Superparamagnetic beads are available from a number of suppliers (Dynal – Invitrogen, Bangs Laboratories, New England Biolabs, and Polysciences) with a variety of chemical and ligand modifications, as they are commonly used in bio-separation applications. Superparamagnetic beads ranging in size from ~500 nm to 4.5 μm are typically composed of ~10–20 nm magnetite particles embedded in a porous matrix sphere enclosed in a protective polymer shell. In the absence of an external magnetic field, the magnetic domains are thermally disordered and there is no residual magnetization, which prevents aggregation. The application of an external magnetic field orients the magnetic domains resulting in a large magnetic moment aligned with the field. In practice, the alignment is not perfect as some of the domains are constrained within the bead resulting in a fixed preferential polarization axis with respect to the bead12. In an external field the bead experiences a torque aligning this preferential axis along the direction of the field. Rotating the external field therefore results in bead rotation. Estimates of the applied torque for a one micron magnetic bead are in excess of 103 pN·nm, orders of magnitude larger than biologically relevant torques. Magnetic tweezers based on permanent magnets and electromagnets have been developed and used for single-molecule force spectroscopy.
Permanent magnet configuration: Technical requirements and calibration
The simplest magnetic tweezers employ a pair of permanent rare earth magnets to generate the magnetic field (Fig. 2a). Neodymium iron boron (Nd2Fe14B) magnets, also called NIB or simply neodymium magnets, are the strongest available permanent magnets with magnetic fields in excess of 1.3 Tesla. They are classified by the magnetic energy density in units of megagauss-oersteds. The highest energy density magnets available are in the range of N45–N50. Typically the magnets are several mm on a side and are configured with the north pole of one magnet facing the south pole of the other, separated by a ~1 mm gap. An iron ring surrounding the two magnets provides a return path for the magnetic flux and reduces stray fields. In this configuration the magnetic field strength decreases roughly exponentially with a characteristic length scale comparable to the separation between the magnets. Consequently, the force on the magnetic particle changes in proportion to displacement with a characteristic length scale of one mm, resulting in an effective stiffness on the order of 10−6 pN·nm−1. As a result of this negligible stiffness, magnetic tweezers afford an infinite bandwidth, passive force clamp. The change in force on a magnetic particle that moves a full ten μm is only 0.01 pN. It is possible to perform experiments at constant force with optical tweezers or AFMs, but it requires sophisticated active feedback, or for the recently described passive optical force clamp, is limited to a relatively small range of motion (~50 nm)52. As a consequence of the force-clamp properties of the magnetic tweezers, they are insensitive to drift and noise in the position of the magnets, which considerably relaxes the design constraints on the magnet translation and rotation mechanisms. Magnetic tweezers based on permanent magnets are well suited for constant force experiments, but they are unable to manipulate magnetic particles in three dimensions. Instead, they provide a constant one-dimensional pulling force without a local minimum. Experiments must therefore be carried out with magnetic particles attached to a surface of the microscope chamber, most commonly by a single molecule of DNA. In a configuration developed by Bensimon and Croquette, the microscope chamber is mounted on an inverted microscope and the magnets are held above the chamber, providing an upward pulling force on the magnetic beads79 (Fig. 2a). Rotation of the magnets, accomplished by coupling them to a rotary motor under computer control, permits rotating tethered superparamagnetic beads. The sample is illuminated through the gap in the magnets with a collimated light emitting diode. Interference between light scattered off the bead and unscattered light produces a well defined pattern of concentric rings around the image of the bead captured on a CCD camera. The lateral position of the bead is determined from the centroid of the interference pattern, and the axial position of the bead is determined from the intensity distribution of the interference pattern, which is a strong function of the position of the bead with respect to the focus of the objective86. Calibration of the axial interference signal involves recording the interference pattern at a series of focal positions, accomplished by moving the objective with a piezo focusing element. Axial motion of the bead can then be determined in real-time by correlating the instantaneous diffraction pattern to the calibration pattern. This tracking technique, related to digital holography87, permits real-time three-dimensional video based tracking with an accuracy of ~2–4 nm laterally and 10 nm in the axial dimension86. Sample drift artifacts can be reduced by tracking the position of the mobile bead relative to a reference bead stuck to the surface of the microscope chamber. The force on the magnetic beads is controlled by adjusting the position of the magnets above the microscope chamber, usually with a motorized platform under computer control. Calibration of the force as a function of magnet distance relies on a variance based equipartition method similar to that used for the calibration of optical tweezers. A magnetic bead tethered to the surface of the microscope chamber by a DNA molecule and subjected to an upward force can be treated as an inverted pendulum with lateral stiffness αx = Fz·L−1, where Fz is the axial force and L is the length of the tether. The lateral stiffness is determined from the transverse fluctuations through the equipartition relation: ½αx<x2> = ½kBT, and the tether length is measured directly. The maximum force depends on the magnetic moment of the beads, which scales with the volume and on how close the magnets approach the sample. By using thin sample chambers (~100 μm), thereby reducing the minimum magnet separation, forces in excess of 20 pN can be applied on 1 μm superparamagnetic beads88. This configuration of magnetic tweezers has been used extensively to measure the topology of individual DNA molecules80, 89–91, and to study topoisomerases81, 92–94.
Electromagnet configuration: Technical requirements and calibration
Magnetic tweezers have also been designed with electromagnets84, 86 (Fig. 2b). Electromagnets have the advantage that force and rotation can be controlled by changing the current rather than moving the magnets, which permits faster and simpler control over these parameters. However, to increase the magnetic field the electromagnet coils are typically wrapped around soft iron or Mu-metal pole pieces95, which introduce significant hysteresis in the magnetic field as a function of current. Moreover, the high current required to generate significant force also generates significant heat, necessitating the implementation of active cooling in some electromagnetic configurations. Generally speaking, electromagnets produce lower magnetic fields and hence lower forces when configured in a similar manner as permanent magnets86. However, since both force and rotation can be controlled by adjusting the amplitude or phase of the coil current rather than mechanically moving the magnets, electromagnets can be placed in close proximity to the magnetic beads. Simple, single-pole electromagnetic tweezers have been developed that can apply up to 10 nN of pulling force on a 4.5μm magnetic bead ~10μm away from the pole piece96. More recently, Fisher and coworkers developed a microfabricated electromagnetic system in which they employed six thin foil electromagnetic pole pieces to achieve three-dimensional-position control (Fig 2b), combined with laser-based particle tracking to achieve position feedback with 100 kHz bandwidth84. High forces and multi-dimensional control have been achieved with other microfabricated or micromachined electromagnetic tweezers97. Unlike optical tweezers, electromagnet tweezers do not create a stable three-dimensional trapping potential, rather an effective potential is created through active control of the applied force. As a result, properties of the feedback loop determine the effective stiffness86, which can nevertheless be measured with the same techniques used to determine the stiffness of optical tweezers. For laser based tracking, position calibration is accomplished in a similar manner to optical tweezers84, whereas for video based position tracking, calibration techniques developed for the permanent magnetic configuration are employed86.
Applications of magnetic tweezers
Magnetic tweezers offer some advantages over other force spectroscopy techniques, and are particularly well suited for certain measurements. They do not suffer from the problems of sample heating and photodamage that plague optical tweezers. Moreover, magnetic manipulation is exquisitely selective for the magnetic beads used as probes, and is generally insensitive to the sample and microscope chamber preparation. These features permit non-invasive force and displacement measurements in complex, heterogeneous environments, including the interior of cells98 and within entangled biopolymer networks99. Due to the properties of the magnetic field used to impose force, magnetic tweezers offer the prospect of highly parallel single-molecule measurements, which would be difficult or impossible to achieve with other single-molecule force spectroscopy techniques100. Permanent magnet configurations are relatively simple to assemble, and they combine force clamp properties with the ability to impose rotation. This combination makes magnetic tweezers ideally suited for the study of DNA topology and topoisomerases79, 81, 82, 101. Tethering the DNA molecule to the sample chamber and to the bead through multiple bonds produces rotationally constrained attachments that permit the DNA molecule to be over-or under-wound by rotating the magnets (Fig. 2b). Unprecedented control over the topological state of the DNA can be achieved with magnetic tweezers permitting sensitive measurements of topoisomerase activity at the single-molecule level. For example, Koster and coworkers probed the details of the process by which topoisomerase IB relaxes supercoils. By measuring the relaxation of supercoiled DNA by topoisomerase IB as a function of load, they demonstrated that rotation of the DNA around the single strand of DNA across from the transiently generated nick is hindered by rotational friction within the enzyme102. In follow up experiments, they measured the activity of topoisomerase IB bound to camptothecin, a potent anti-tumor chemotherapy agent. Their results indicate that in the presence of the drug, supercoil relaxation becomes asymmetric, which they were able to demonstrate has in vivo implications103. In another series of experiments, Revyakin and coworkers used magnetic tweezers to dissect the dependence of transcription initiation on DNA supercoiling density104, and more recently, were able to demonstrate that RNA polymerase “scrunches” the DNA template during abortive initiation105.
Limitations and drawbacks
Despite their many unique features, magnetic tweezers are not nearly as versatile as optical tweezers or the AFM. The robust permanent magnet configuration lacks the manipulation ability of other techniques. Although, the ability to rotate magnetic beads has proved useful, the large applied torque precludes the direct measure of rotation or torque generation. Furthermore, the bandwidth and sensitivity are limited by the video based detection, which prevents the direct measurement of very fast or very small displacements. Electromagnetic tweezers permit full three-dimensional manipulation, however this requires cumbersome feedback control in addition to sophisticated custom machined pole pieces, and has not yet achieved the sensitivity of some other force spectroscopy techniques. Moreover, producing the large magnetic field and field gradient requires high current electromagnets that can produce significant heating, or small, closely spaced, pole pieces that no longer preserve the constant-force benefit of magnetic tweezers. Electromagnetic manipulation has a great deal of potential, which is being realized through continuing technological and theoretical developments.
Atomic Force Microscopy
The atomic force microscope (AFM, Fig. 3A) is perhaps the most familiar of the three force spectroscopy techniques covered in this review, and is by far the simplest in concept. The AFM106, 107 is a version of the scanning probe microscope in which the properties of the surface are investigated with a proximal probe. This technique allows mapping of the surface characteristics at sub-nanometer resolution. The AFM was initially developed to overcome limitations of the scanning tunneling microscope in imaging non-conductive samples108, 109. However, the possibility of modifying the surface and manipulating individual molecules made AFM an ideal tool for biological applications110–114. One significant advantage of the technique is simple and rapid sample preparation. Another important feature of the AFM is the ability to conduct measurements of biological samples under near-physiological conditions115, 116.
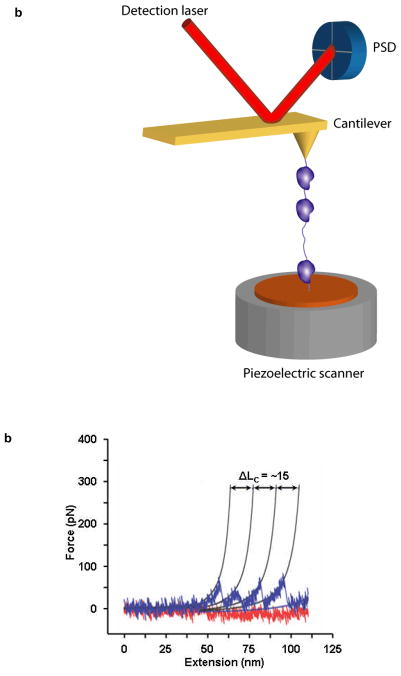
Fig. 3.
(a). Cartoon of the atomic force microscope (not to scale). The AFM consists of a cantilever with a sharp tip (yellow) held above a piezo scanning stage (grey). Deflection of the AFM cantilever is measured from the displacement of a low power laser (red beam) reflected off the cantilever on a position sensitive device (PSD) (blue disc). A typical AFM pulling experiment is displayed in which a poly-protein molecule (purple) is attached to the sample surface (copper) and the AFM cantilever tip. The piezo stage is retracted along the axial direction, increasing the separation between the cantilever and the sample surface. The force on the molecule is provided by the cantilever defection, and the extension of the molecule is equal to the separation between the AFM tip and the sample surface. (b) MFP stretching curve displays the force-induced unfolding of individual domain repeats of a hypothetical tetramer protein. Thin solid lines are the WLC fits to the data with a persistence length PL of ~ 0.8 nm and the contour length increment ΔLC of 15 nm.
Although the AFM is primarily an imaging tool, it also allows measurement of inter- and intramolecular interaction forces with pN resolution. This specialized version of AFM is called a molecular force probe (MFP) or 1D AFM, and is fundamentally different from an imaging AFM. When used in an imaging mode, the AFM cantilever scans the surface of the specimen, line after line, whereas for a MFP it is moved only in the vertical direction, perpendicular to the specimen plane26, 117. The vertical motion of the cantilever is controlled by piezo-electric actuators affording sub-nanometer resolution. The displacement of the cantilever is monitored directly with either a capacitor or a linear voltage differential transformer. As a result, high-resolution force versus extension curves of single molecules can be recorded using the MFP.
Technical requirements and calibration
With MFP measurements two types of characteristic data can be obtained while pulling on the protein or other molecule of interest: (1) force and (2) extension. Proteins can be described as springs that generate a restoring force when mechanically stretched. The extension is the distance between the anchoring points at which the two ends the protein are attached, such as the surface of a coverslip and the cantilever tip. It is important to note that neither force nor extension data are recorded directly, but rather through ‘handles’ that can be manipulated using a MFP.
Force is generally calculated from the bending of the cantilever of known spring constant. The cantilever stiffness depends on the material properties and shape of the cantilever with typical values from 10 to 105 pN/nm. To obtain precise force data, each cantilever must be properly calibrated before use. Cantilevers from the same lot manufactured from the same source may have very different spring constants, especially if they have a sophisticated shape or if they are coated with functionalized or reflecting layers to improve their performance. The most commonly used calibration methods can be divided into four principal groups118. The stiffness of the cantilever can be obtained: (1) through comparison with a reference cantilever of known stiffness, (2) through calibration using thermal vibrations, (3) by the method of added particle masses, or (4) by combining measurements of the resonant frequency with the cantilever physical dimensions and material properties. In general, if the cantilever is compared with a reference cantilever the stiffness accuracy will be ~10%, but the positioning and the calibration of the load can be troublesome and potentially destructive. The accuracy of calibrating cantilevers with the thermal fluctuation method varies between 10 and 20%, but precise control of thermal fluctuations is essential. This method is commonly used to calibrate soft cantilevers, and it requires mathematical analysis of the resonance curve acquired for each cantilever. Calibration by the method of static deflection with added mass is conceptually straightforward, however the positioning and exact determination of the additional load is challenging. Furthermore, the calibration process can potentially destroy the cantilever. The accuracy of this method is ~15%. Cantilever stiffness is most accurately determined with the method of scaling from the resonance frequency, which gives results with 5 to 10% error. It is important to mention that accurate measurements of the cantilever dimensions and determination of effective mass is crucial when this method of calibration is applied118.
The extension data is obtained by measuring the change in displacement between the handles that are attached at the ends of the protein or other molecule of interest. The accuracy of this data is determined by the quality of the piezo-stage. Piezo-stages used in MFPs provide angstrom level resolution and with the advent of closed-loop position feedback control, these devices are accurate and not susceptible to the drift and hysteresis commonly associated with piezo-electric actuators. Data must be corrected for the deflection of the cantilever associated with the measured force. The exact extension of the stretched protein can be obtained only if the attachment points can be precisely located, e.g., by using microfabricated cantilevers. The exact position of the tip of the soft cantilever, relative to the fixed surface, such as a coverslip, can also be determined directly with the use of a calibrated evanescent field119.
To facilitate mechanically strong and specific interactions between the sample molecule and the handles (i.e. cantilever tips and surfaces) for stretching measurements, the ends of the biomolecule needs to be attached specifically. In the MFP both the cantilever and the surface, between which the molecule is tethered, can be chemically modified in order to form specific bonds. The attachment of the molecules to the surfaces can be achieved with several different methods. Non-specific binding is simplest way to attach molecules to the specimen surface such as glass, mica, or gold26. However, non-specific absorption results in large uncertainty in the location of the attachments points on the molecule, which complicates data interpretation. The molecule of interest can be specifically attached by its extremities using antibodies120, 121, but the contribution of the antibodies to the elastic response of the total system must be taken into consideration. The same concern applies with molecular handles formed by streptavidin/biotin bonds122 or by avidin/biotin bonds123 are used to hold the molecules. Highly specific and mechanically strong interactions like hexahistidine/Ni-NTA124 or gold/SH-group125 can be utilized, but these bonds are very sensitive to reducing agents. Photoactivated cross-linkers121 and polyethylene-glycol (PEG) handles126 are perhaps the most versatile methods for specific attachment of proteins.
Handles also can be used as a tool to alleviate the problem of multiple proteins binding between the cantilever and the surface. Even the sharpest available cantilever is significantly larger than the molecule under study, so the possibility of more than one molecule being attached to the tip of the cantilever is a significant concern. The problem of pulling an ensemble of molecules instead of a single molecule can be addressed in several different ways (dilution, using a tip with suitable geometry or statistical analysis of the data), but perhaps the most direct method is the use of handles with special mechanical characteristics. In this method termed ‘mechanical fingerprinting’122 the protein of interest is serially linked, most commonly with DNA, or with a well -characterized oligomer of Titin’s I27 domain. During the stretch, the DNA attached to the molecule of interest, undergoes a characteristic transition at 65 pN. If this 65 pN transition is present on stretching curves, one can be sure that only one molecule is being manipulated. Similarly, in the case of the I27 oligomer linkage, the characteristic sawtooth pattern in stretching curves due to the unfolding of individual Titan domains ensures that a single molecule is being stretched127–129. A recent approach for specific attachment of single molecules exploits the properties of extremely sharp functionalized carbon nanotubes130.
Stretching curves; measurements of elasticity and force
Single-molecule force-extension measurements are obtained by mechanically perturbing the structure of a molecule. The stretch-release curves, also known as the force-extension relationships, provide valuable information on the structure, the folding and unfolding processes and even the activity of the molecule. Measurement of the stretching curve is relatively straightforward. The MFP tip is lowered towards the surface, or the sample is lifted toward the cantilever tip, by piezo-electric actuators. After the initial contact with the surface, the cantilever is pressed into the surface, on which the sample is deposited, with a predetermined constant force. The tip is then retracted. Attachment of the sample (RNA, DNA, protein, etc.) will tether the cantilever tip to the surface resulting in the bending of the cantilever toward the surface due to the increasing force upon further retraction. The value of this force is obtained using Hooke’s Law:
where d is the cantilever deflection and k is the cantilever spring constant. In most cases the force versus extension curve displays non-linear behavior indicative of an entropic spring, which is well described by the worm like chain (WLC) entropic elasticity model131 (Fig. 3B). Multidomain proteins, such as homo-oligomers of Titin’s I27 domain or ankyrin oligomers display characteristic sawtooth patterns upon stretching. For these homo-oligomers, individual domain unfolding events may be resolved (Fig. 3B). Between two unfolding events the molecule behaves like a non-linear elastic spring. These regions of the stretching curve can be fit with the WLC model to obtain the persistence length and contour length of the unfolded regions of the molecule, as shown in Figure 3B. The change in contour length associated with unfolding events is given by the difference in contour lengths computed prior to and after a rupture event.
Applications of AFM
Commercial AFMs are available from several sources, though custom built AFMs offer greater flexibility and experimental control. Although pulling assays are relatively straightforward to set up, care must be taken in the preparation of samples to minimize background and artifactual signals. Even under well-controlled sample conditions, distinguishing between binding the molecule of interest and nonspecific binding to the surface can be challenging. Distinguishing molecules of interest from nonspecific interactions is facilitated by working with natural or engineered multidomain peptides that have a characteristic force unfolding fingerprint. Functionalizing the AFM tip with antibodies or other ligands specific for the molecule of interest is an alternative means of improving specificity 38.
AFM based force spectroscopy has emerged as a powerful tool to study the rupture of molecular bonds, ranging from covalent bonds14, to the unfolding of proteins132 and nucleic acids107. Schwaiger and coworkers discovered unfolding intermediates with MFP by mechanical perturbation of the actin-crosslinking protein, filamin133. The effect of the direction of pulling on the unfolding pathway has been clearly demonstrated in the case of ubiquitin134 and a (protein containing only beta sheets)n all-beta protein, E2lip3135. Dietz and coworkers have developed a technique to apply force along different directions of a protein, rather than the typical single axis defined by the N- and C-termini of the protein under study136. By introducing pairs of specific cysteine mutations in the green fluorescent protein (GFP), the monomers in a poly-GFP peptide could be linked in a well defined geometry through disulfide bonds between the cysteine residues. The unfolding pathways for five different pulling directions varied dramatically, and there was a six-fold variation in rupture force. Parallel unfolding pathways have also been demonstrated using MFP. Employing mutants of the I27 domain of Titin, Wright and coworkers demonstrated changes in the flux between different transition states on parallel folding pathways137.
Force-clamp spectroscopy is an emerging technique for studying mechanochemistry at the single-molecule level. Recently, AFM pulling experiments have been harnessed to probe the effects of force and conformation on enzyme function. By measuring the rate of disulfide reduction by E. coli thioredoxin as a function of force on an engineered cysteine bond in a polypeptide chain, Wiita and coworkers were able to deduce sub-angstrom level conformation changes in the substrate during enzyme catalysis138. In similar experiments, Szoszkiewicz and coworkers examined the kinetics disulfide bond reduction using MFP combined with force clamp spectroscopy139. Another noteworthy achievement in the field of mechanochemistry is the work of Greene and coworkers140 who showed that the protein kinase domains of Titin unfold in a step-wise manner, and speculated that kinases in muscle cells may be activated in response to the forces generated during each contraction/relaxation cycle, i.e. may act as a force-sensor.
MFP can also be used to investigate the structure of supramolecular assemblies. Brown and coworkers used atomic force microscopy to study the forced unfolding of engineered linear oligomers of fibrinogen to elucidate the elastic properties of blood clots141. More recently, Lim and coworkers showed that the elastic properties of blood clots are mainly determined by the coiled-coil helices of fibrinogen142. In another example, Kellermayer and coworkers described the structure and self assembly mechanism of amyloid β-fibrils143 and myosin thick filaments144.
An interesting new development is the combination of AFM imaging with force mapping and spectroscopy143. This technique allows to investigator to perform an image scan of the surface and subsequently target specific topological structures for stretching measurements with subnanometer accuracy. Oesterhelt and coworkers measured the unbinding forces and force spectrum for the removal of bacteriorhodopsin from patches of purple membrane145. The membrane patches were initially imaged with the AFM to find the location of an individual bacteriorhodopsin molecule, which was then extracted with the same AFM tip. These experiments highlight the unique capability of AFM instruments to both image the surface topology at high resolution and measure the unbinding or unfolding forces at well defined locations.
Force spectroscopy measurements of ligand binding, antibody-antigen interactions, and protein unfolding are well established and widely employed. MFP is now routinely used to probe the affinity, mechanics and recognition properties of biomolecular interactions such as ligand/receptor pairs146, 147, antibody/antigen interactions148, 149, protein/DNA150 and DNA/DNA interactions107.
Drawback, limitations and future perspectives
The main drawbacks and limitations of the AFM stem from the large size and relatively high stiffness of the cantilevers, which impose a lower bound on the useful force range and a reduced bandwidth, particularly in aqueous conditions (see Box 1). The forces associated with many biological processes and structures are therefore difficult to study with the AFM. Specificity is a second major concern in many AFM pulling experiments. It can be difficult to discriminate between interactions of the AFM tip with the molecule of interest from non-specific interactions, or inappropriate contacts with the molecule of interest, such as binding at an intermediate position rather than at one of the ends.
Current cutting-edge AFM techniques have developed a high level of sophistication. Significant steps have been made toward technological improvements of data acquisition speed. One of the most interesting and scientifically potent improvements to the field is the high-speed AFM151, which makes it possible to follow molecular events in real time. Another promising direction of development is the combination of different single-molecule techniques. Some of these techniques already exist, like the combination of AFM imaging with force mapping and spectroscopy, AFM combined with fluorescent imaging or combination of MFP with TIRF-based technique, which allows (using a suitable sample) following the process of protein unfolding or refolding via changes in force and emission of a fluorophore in the sample.
Prospects and outlook
Over the past two decades there have been remarkable developments and refinements in single-molecule force spectroscopy techniques that have opened up new avenues of biophysical and biological research. The power and success of these techniques are illustrated by the rapidly growing number of applications and systems to which they are being applied, and by the fact that these once cutting-edge techniques are becoming more commonplace, some even commercialized. With this strong foundation to build on, the field will be able to address harder problems and bigger challenges. Primary among these is the need to develop single-molecule force spectroscopy techniques that can be applied within living cells. With a few notable exceptions41, 97, single-molecule manipulation has been primarily concerned with in vitro measurements of purified proteins, or with probes attached to cell membranes. Performing the same or similar experiments in vivo will provide an unparalleled view of how individual enzymes function in their native environment and how cells generate and respond to forces at the molecular level. Although these measurements are beginning to be undertaken, a great deal of work remains to overcome the many difficulties associated with attempting to bring single-molecule techniques to the interior of the cell. Improvements in specific and efficient labeling of intracellular proteins with appropriate manipulation “handles” will be required, in addition to less damaging and less invasive manipulation techniques that are insensitive to the crowded, inhomogeneous conditions found within a cell. Along the same lines, improvements in single-molecule force spectroscopy methods are required to permit the study of macromolecular protein machines152, and multi-enzyme complexes153–155. The combination of force-spectroscopy techniques with other single-molecule methods, in particular single-molecule fluorescence detection modalities156–158, is an important development that will grow in importance as the complexity of the questions addressed increases. At the other extreme, increasing parallelism159 and through-put in single-molecule force spectroscopy measurements will be important developments. Finally, the ability to manipulate and control unlabeled proteins has recently been described160 but this and other emerging techniques have yet to be fully realized. In addition to these developments, new technologies will enable the continuing development of novel single-molecule force spectroscopy techniques.
Concluding remarks
The single-molecule force spectroscopy techniques covered in this review constitute powerful versatile tools to study motion and force associated with biomolecules. Many factors influence the decision as to which, if any, of these techniques is suited for a particular measurement or biological system of interest. Determining the relevant range of forces and displacements and the required temporal and spatial resolution is relatively straightforward. Other factors to consider include the compatibility of the system under study with the different techniques, and the trade-offs between the cost, complexity and convenience of the techniques. Finally, as the rapid progress in the field demonstrates, current techniques can be modified, improved or extended to satisfy the requirements of the desired measurement and system under study. Meeting these challenges will ensure the continuing refinement of current techniques and the development of novel approaches.
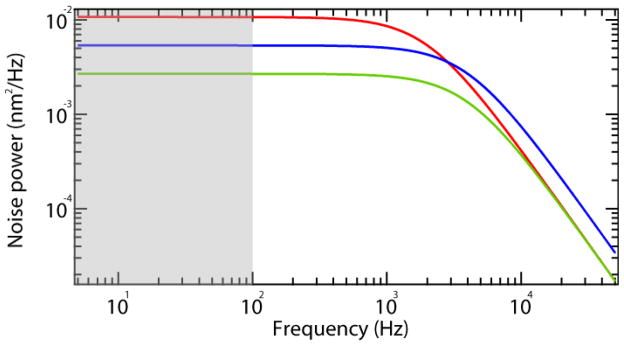
Fig. 4.
Influence of probe size, stiffness and measurement bandwidth on spatial resolution. The power spectrum (Eq. 3) of the fluctuations of a probe particle attached to a spring undergoing Brownian motion. The noise power amplitude expressed as nm2·Hz−1 is plotted as a function of frequency on a double logarithmic scale. The power spectrum for a one micron particle with a roll-off frequency of 2 kHz is shown in red. The power spectrum for the same particle with half the drag coefficient is shown in blue, and the power spectrum for a two-fold increase in stiffness is shown in green. Both of these changes increase the roll-off frequency while simultaneously decreasing the low frequency amplitude of the power spectrum. However, the area under the curve, and hence the position noise, is reduced only by increasing the stiffness (Eq. 1). The decrease in low frequency amplitude is compensated by the extended roll-off frequency when the drag is halved (blue trace), consequently the noise is equivalent for the red and blue curves. This is no longer the case if the signals are filtered. The reduction in noise achieved by filtering is illustrated by the shaded grey region that corresponds to a frequency bandwidth of 100 Hz. By limiting the measurement bandwidth to 100 Hz, the noise is reduced to the area under the curves in the shaded region, and there is a large difference in noise among the red, blue and green curves (see Eq. 4).
Acknowledgments
KCN and AN are supported by the Intramural Program of the National Heart Lung and Blood Institute, National Institutes of Health. We would like to thank Dr. Grace Liou, Dr. Richard Neuman, and Dr. Yasuharu Takagi for critical reading of the manuscript. KCN would also like to thank his mentors David Bensimon and Vincent Croquette at the Ecole Normale Supérieure, Paris, France, and Steven Block at Stanford University, in addition to members of their labs for making this work possible.
References
- 1.Cluzel P, et al. DNA: An extensible molecule. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 2.Kishino A, Yanagida T. Force measurements by micromanipulation of a single actin filament by glass needles. Nature. 1988;334:74–76. doi: 10.1038/334074a0. [DOI] [PubMed] [Google Scholar]
- 3.Evans E, Ritchie K, Merkel R. Sensitive force technique to probe molecular adhesion and structural linkages at biological interfaces. Biophys J. 1995;68:2580–2587. doi: 10.1016/S0006-3495(95)80441-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith SB, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–6. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 5.Kim SJ, Blainey PC, Schroeder CM, Xie XS. Multiplexed single-molecule assay for enzymatic activity on flow-stretched DNA. Nat Methods. 2007;4:397–399. doi: 10.1038/nmeth1037. [DOI] [PubMed] [Google Scholar]
- 6.Greenleaf WJ, Woodside MT, Block SM. High-resolution, single-molecule measurements of biomolecular motion. Annu Rev Biophys Biomol Struct. 2007;36:171–90. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neuman KC, Block SM. Optical trapping. Rev Sci Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. This is a detailed and thorough technical review of optical trapping. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanase M, Biais N, Sheetz M. Magnetic tweezers in cell biology. Methods Cell Biol. 2007;83:473–93. doi: 10.1016/S0091-679X(07)83020-2. [DOI] [PubMed] [Google Scholar]
- 9.Zlatanova J, Lindsay SM, Leuba SH. Single molecule force spectroscopy in biology using the atomic force microscope. Prog Biophys Mol Biol. 2000;74:37–61. doi: 10.1016/s0079-6107(00)00014-6. [DOI] [PubMed] [Google Scholar]
- 10.Lee CK, Wang YM, Huang LS, Lin S. Atomic force microscopy: determination of unbinding force, off rate and energy barrier for protein-ligand interaction. Micron. 2007;38:446–61. doi: 10.1016/j.micron.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Lang MJ, Asbury CL, Shaevitz JW, Block SM. An automated two-dimensional optical force clamp for single molecule studies. Biophys J. 2002;83:491–501. doi: 10.1016/S0006-3495(02)75185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neuman KC, Lionnet T, Allemand JF. Single-molecule micromanipulation techniques. Ann Rev Mater Res. 2007;37:33–67. [Google Scholar]
- 13.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. This is an experimental tour de force in which individual 0.34 nm base-pair steps of transcribing RNA polymerases were directly measured with an optical tweezers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grandbois M, Beyer M, Rief M, Clausen-Schaumann H, Gaub HE. How strong is a covalent bond? Science. 1999;283:1727–1730. doi: 10.1126/science.283.5408.1727. [DOI] [PubMed] [Google Scholar]
- 15.Hohng S, et al. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the holliday junction. Science. 2007;318:279–83. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Litvinov RI, Shuman H, Bennett JS, Weisel JW. Binding strength and activation state of single fibrinogen-integrin pairs on living cells. Proc Natl Acad Sci U S A. 2002;99:7426–31. doi: 10.1073/pnas.112194999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prass M, Jacobson K, Mogilner A, Radmacher M. Direct measurement of the lamellipodial protrusive force in a migrating cell. J Cell Biol. 2006;174:767–72. doi: 10.1083/jcb.200601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacKintosh FC, Schmidt CF. Microrheology. Curr Opin Colloid Interface Sci. 1999;4:300–307. [Google Scholar]
- 19.Daniels BR, Masi BC, Wirtz D. Probing single-cell micromechanics in vivo: The microrheology of C-elegans developing embryos. Biophys J. 2006;90:4712–4719. doi: 10.1529/biophysj.105.080606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Block SM, Goldstein LS, Schnapp BJ. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–52. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 21.Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–84. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 22.Simmons RM, et al. Force on single actin filaments in a motility assay measured with an optical trap. Adv Exp Med Biol. 1993;332:331–6. doi: 10.1007/978-1-4615-2872-2_32. [DOI] [PubMed] [Google Scholar]
- 23.Tinoco I, Jr, Bustamante C. The effect of force on thermodynamics and kinetics of single molecule reactions. Biophys Chem. 2002;101–102:513–33. doi: 10.1016/s0301-4622(02)00177-1. This paper provides a detailed and comprehensive treatment of the effects of force on single-molecule reactions. The concepts and analysis presented form the underpinning of single-molecule force spectroscopy. [DOI] [PubMed] [Google Scholar]
- 24.Tskhovrebova L, Trinick J, Sleep JA, Simmons RM. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 1997;387:308–12. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 25.Kellermayer MSZ, Smith SB, Granzier HL, Bustamante C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 26.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. This paper and the preceding paper (#25) appeared together and present the first demonstrations of the mechanical unfolding of proteins. [DOI] [PubMed] [Google Scholar]
- 27.Hummer G, Szabo A. Free energy surfaces from single-molecule force spectroscopy. Acc Chem Res. 2005;38:504–13. doi: 10.1021/ar040148d. [DOI] [PubMed] [Google Scholar]
- 28.Woodside MT, et al. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc Natl Acad Sci U S A. 2006;103:6190–6195. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim CT, Zhou EH, Li A, Vedula SRK, Fu HX. Experimental techniques for single cell and single molecule biomechanics. Mater Sci Eng C-Biomimetic Supramol Syst. 2006;26:1278–1288. [Google Scholar]
- 30.Zhuang XW, Rief M. Single-molecule folding. Curr Opin Struct Biol. 2003;13:88–97. doi: 10.1016/s0959-440x(03)00011-3. [DOI] [PubMed] [Google Scholar]
- 31.Evans E. Probing the relation between force--lifetime--and chemistry in single molecular bonds. Annu Rev Biophys Biomol Struct. 2001;30:105–28. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 32.Jarzynski C. Nonequilibrium equality for free energy differences. Phys Rev Lett. 1997;78:2690–2693. This seminal paper presents the remarkable Jarzynski equality that relates the equilibrium free energy difference to non-equilibrium measurements of work. The Jarzynski equality and subsequent relations based on the equality permit the extraction of folding free energies from out-of equilibrium mechanical unfolding experiments. [Google Scholar]
- 33.Lu HP, Xun L, Xie XS. Single-molecule enzymatic dynamics. Science. 1998;282:1877–82. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 34.Xie XS, Lu HP. Single-molecule enzymology. J Biol Chem. 1999;274:15967–70. doi: 10.1074/jbc.274.23.15967. [DOI] [PubMed] [Google Scholar]
- 35.Hua W, Young EC, Fleming ML, Gelles J. Coupling of kinesin steps to ATP hydrolysis. Nature. 1997;388:390–3. doi: 10.1038/41118. [DOI] [PubMed] [Google Scholar]
- 36.Schnitzer MJ, Block SM. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386–90. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 37.Hermanson G. Bioconjugate Techniques. Academic Press; 1996. [Google Scholar]
- 38.Hinterdorfer P, Dufrene YF. Detection and localization of single molecular recognition events using atomic force microscopy. Nat Methods. 2006;3:347–55. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 39.Ashkin A, Dziedzic JM, Bjorkholm JE, Chu S. Observation of a single-beam gradient force optical trap for dielectric particles. Opt Lett. 1986;11:288–290. doi: 10.1364/ol.11.000288. This classic paper is the first experimental demonstration of the single-beam gradient trap, or optical tweezers. [DOI] [PubMed] [Google Scholar]
- 40.Neuman KC, Chadd EH, Liou GF, Bergman K, Block SM. Characterization of photodamage to escherichia coli in optical traps. Biophys J. 1999;77:2856–63. doi: 10.1016/S0006-3495(99)77117-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sacconi L, Tolic-Norrelykke IM, Stringari C, Antolini R, Pavone FS. Optical micromanipulations inside yeast cells. Appl Optics. 2005;44:2001–2007. doi: 10.1364/ao.44.002001. [DOI] [PubMed] [Google Scholar]
- 42.Cherney DP, Bridges TE, Harris JM. Optical trapping of unilamellar phospholipid vesicles: Investigation of the effect of optical forces on the lipid membrane shape by confocal-raman microscopy. Anal Chem. 2004;76:4920–4928. doi: 10.1021/ac0492620. [DOI] [PubMed] [Google Scholar]
- 43.Gross SP. Application of optical traps in vivo. Methods Enzymol. 2003;361:162–74. doi: 10.1016/s0076-6879(03)61010-4. [DOI] [PubMed] [Google Scholar]
- 44.Lee WM, Reece PJ, Marchington RF, Metzger NK, Dholakia K. Construction and calibration of an optical trap on a fluorescence optical microscope. Nat Protoc. 2007;2:3226–38. doi: 10.1038/nprot.2007.446. [DOI] [PubMed] [Google Scholar]
- 45.Neuman KC, Abbondanzieri EA, Block SM. Measurement of the effective focal shift in an optical trap. Opt Lett. 2005;30:1318–20. doi: 10.1364/ol.30.001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fallman E, Axner O. Influence of a glass-water interface on the on-axis trapping of micrometer-sized spherical objects by optical tweezers. Appl Opt. 2003;42:3915–26. doi: 10.1364/ao.42.003915. [DOI] [PubMed] [Google Scholar]
- 47.Viana NB, Rocha MS, Mesquita ON, Mazolli A, Neto PAM. Characterization of objective transmittance for optical tweezers. Appl Optics. 2006;45:4263–4269. doi: 10.1364/ao.45.004263. [DOI] [PubMed] [Google Scholar]
- 48.Misawa H, Koshioka M, Sasaki K, Kitamura N, Masuhara H. Three-dimensional optical trapping and laser ablation of a single polymer latex particle in water. J Appl Phys. 1991;70:3829–3836. [Google Scholar]
- 49.Rohrbach A, Stelzer EHK. Three-dimensional position detection of optically trapped dielectric particles. J Appl Phys. 2002;91:5474–5488. [Google Scholar]
- 50.Gittes F, Schmidt CF. Interference model for back-focal-plane displacement detection in optical tweezers. Opt Lett. 1998;23:7–9. doi: 10.1364/ol.23.000007. [DOI] [PubMed] [Google Scholar]
- 51.Peterman EJG, van Dijk MA, Kapitein LC, Schmidt CF. Extending the bandwidth of optical-tweezers interferometry. Rev Sci Instrum. 2003;74:3246–3249. [Google Scholar]
- 52.Greenleaf WJ, Woodside MT, Abbondanzieri EA, Block SM. Passive all-optical force clamp for high-resolution laser trapping. Phys Rev Lett. 2005;95 doi: 10.1103/PhysRevLett.95.208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carter AR, King GM, Perkins TT. Back-scattered detection provides atomic-scale localization precision, stability, and registration in 3D. Opt Express. 2007;15:13434–13445. doi: 10.1364/oe.15.013434. [DOI] [PubMed] [Google Scholar]
- 54.Carter AR, et al. Stabilization of an optical microscope to 0.1 nm in three dimensions. Appl Opt. 2007;46:421–7. doi: 10.1364/ao.46.000421. [DOI] [PubMed] [Google Scholar]
- 55.Vermeulen KC, et al. Calibrating bead displacements in optical tweezers using acousto-optic deflectors. Rev Sci Instrum. 2006;77 [Google Scholar]
- 56.Tolic-Norrelykke SF, et al. Calibration of optical tweezers with positional detection in the back focal plane. Rev Sci Instrum. 2006;77 [Google Scholar]
- 57.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–7. doi: 10.1038/365721a0. This landmark paper reports the first direct measurement of the steps taken by individual kinesin. [DOI] [PubMed] [Google Scholar]
- 58.Mammen M, et al. Optically controlled collisions of biological objects to evaluate potent polyvalent inhibitors of virus-cell adhesion. Chem Biol. 1996;3:757–763. doi: 10.1016/s1074-5521(96)90252-5. [DOI] [PubMed] [Google Scholar]
- 59.Kerssemakers JWJ, et al. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 60.Footer MJ, Kerssemakers JW, Theriot JA, Dogterom M. Direct measurement of force generation by actin filament polymerization using an optical trap. Proc Natl Acad Sci U S A. 2007;104:2181–6. doi: 10.1073/pnas.0607052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang MD, et al. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–7. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 62.Herbert KM, et al. Sequence-resolved detecton of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–47. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 64.Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003 doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moffitt JR, Chemla YR, Izhaky D, Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc Natl Acad Sci U S A. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–90. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 67.Smith DE, et al. The bacteriophage phi 29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 68.Liphardt J, Onoa B, Smith SB, Tinoco I, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. This paper was the first to show the mechanical unfolding of RNA structures and it laid the groundwork for subsequent measurements of enzymatic unfolding of nucleic acid structures. [DOI] [PubMed] [Google Scholar]
- 69.Dumont S, et al. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–8. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keyser UF, et al. Direct force measurements on DNA in a solid-state nanopore. Nature Physics. 2006;2:473–477. [Google Scholar]
- 71.Bayoudh S, Mehta M, Rubinsztein-Dunlop H, Heckenberg NR, Critchley C. Micromanipulation of chloroplasts using optical tweezers. J Microsc. 2001;203:214–22. doi: 10.1046/j.1365-2818.2001.00843.x. [DOI] [PubMed] [Google Scholar]
- 72.Friese MEJ, Nieminen TA, Heckenberg NR, Rubinsztein-Dunlop H. Optical alignment and spinning of laser-trapped microscopic particles. Nature. 1998;394:348–50. [Google Scholar]
- 73.Deufel C, Forth S, Simmons CR, Dejgosha S, Wang MD. Nanofabricated quartz cylinders for angular trapping: DNA supercoiling torque detection. Nat Methods. 2007;4:223–5. doi: 10.1038/nmeth1013. [DOI] [PubMed] [Google Scholar]
- 74.La Porta A, Wang MD. Optical torque wrench: angular trapping, rotation, and torque detection of quartz microparticles. Phys Rev Lett. 2004;92:190801. doi: 10.1103/PhysRevLett.92.190801. [DOI] [PubMed] [Google Scholar]
- 75.Peterman EJG, Gittes F, Schmidt CF. Laser-induced heating in optical traps. Biophys J. 2003;84:1308–1316. doi: 10.1016/S0006-3495(03)74946-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abbondanzieri EA, Shaevitz JW, Block SM. Picocalorimetry of transcription by RNA polymerase. Biophysical Journal. 2005;89:L61–L63. doi: 10.1529/biophysj.105.074195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seol Y, Carpenter AE, Perkins TT. Gold nanoparticles: enhanced optical trapping and sensitivity coupled with significant heating. Opt Lett. 2006;31:2429–2431. doi: 10.1364/ol.31.002429. [DOI] [PubMed] [Google Scholar]
- 78.Liang H, et al. Wavelength dependence of cell cloning efficiency after optical trapping. Biophys J. 1996;70:1529–33. doi: 10.1016/S0006-3495(96)79716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Strick T, Allemand J, Croquette V, Bensimon D. Twisting and stretching single DNA molecules. Prog Biophys Mol Biol. 2000;74:115–40. doi: 10.1016/s0079-6107(00)00018-3. [DOI] [PubMed] [Google Scholar]
- 80.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–7. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 81.Charvin G, Strick TR, Bensimon D, Croquette V. Tracking topoisomerase activity at the single-molecule level. Annu Rev Biophys Biomol Struct. 2005;34:201–19. doi: 10.1146/annurev.biophys.34.040204.144433. [DOI] [PubMed] [Google Scholar]
- 82.Strick TR, Croquette V, Bensimon D. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature. 2000;404:901–4. doi: 10.1038/35009144. This was the first single-molecule measurement of topoisomerase activity with magnetic tweezers. [DOI] [PubMed] [Google Scholar]
- 83.Gore J, et al. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature. 2006;439:100–4. doi: 10.1038/nature04319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fisher JK, et al. Thin-foil magnetic force system for high-numerical-aperture microscopy. Rev Sci Instrum. 2006;77 doi: 10.1063/1.2166509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yan J, Skoko D, Marko JF. Near-field-magnetic-tweezer manipulation of single DNA molecules. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70:011905. doi: 10.1103/PhysRevE.70.011905. [DOI] [PubMed] [Google Scholar]
- 86.Gosse C, Croquette V. Magnetic tweezers: Micromanipulation and force measurement at the molecular level. Biophys J. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee SH, et al. Characterizing and tracking single colloidal particles with video holographic microscopy. Opt Express. 2007;15:18275–18282. doi: 10.1364/oe.15.018275. [DOI] [PubMed] [Google Scholar]
- 88.Lionnet T, Joubaud S, Lavery R, Bensimon D, Croquette V. Wringing out DNA. Phys Rev Lett. 2006;96:178102. doi: 10.1103/PhysRevLett.96.178102. [DOI] [PubMed] [Google Scholar]
- 89.Allemand JF, Bensimon D, Lavery R, Croquette V. Stretched and overwound DNA forms a Pauling-like structure with exposed bases. Proc Natl Acad Sci U S A. 1998;95:14152–7. doi: 10.1073/pnas.95.24.14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Strick TR, Croquette V, Bensimon D. Homologous pairing in stretched supercoiled DNA. Proc Natl Acad Sci U S A. 1998;95:10579–83. doi: 10.1073/pnas.95.18.10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strick TR, Allemand JF, Bensimon D, Croquette V. Behavior of supercoiled DNA. Biophys J. 1998;74:2016–28. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dekker NH, et al. The mechanism of type IA topoisomerases. Proc Natl Acad Sci U S A. 2002;99:12126–31. doi: 10.1073/pnas.132378799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dekker NH, et al. Thermophilic topoisomerase I on a single DNA molecule. J Mol Biol. 2003;329:271–82. doi: 10.1016/s0022-2836(03)00320-6. [DOI] [PubMed] [Google Scholar]
- 94.Charvin G, Bensimon D, Croquette V. Single-molecule study of DNA unlinking by eukaryotic and prokaryotic type-II topoisomerases. Proc Natl Acad Sci U S A. 2003;100:9820–5. doi: 10.1073/pnas.1631550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Conroy RS. In: Handbook of Molecular Force Spectroscopy. Noy A, editor. Springer; US: 2008. pp. 23–96. [Google Scholar]
- 96.Bausch AR, Ziemann F, Boulbitch AA, Jacobson K, Sackmann E. Local measurements of viscoelastic parameters of adherent cell surfaces by magnetic bead microrheometry. Biophys J. 1998;75:2038–2049. doi: 10.1016/S0006-3495(98)77646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.de Vries AH, Krenn BE, van Driel R, Kanger JS. Micro magnetic tweezers for nanomanipulation inside live cells. Biophys J. 2005;88:2137–44. doi: 10.1529/biophysj.104.052035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bausch AR, Moller W, Sackmann E. Measurement of local viscoelasticity and forces in living cells by magnetic tweezers. Biophys J. 1999;76:573–579. doi: 10.1016/S0006-3495(99)77225-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Keller M, Schilling J, Sackmann E. Oscillatory magnetic bead rheometer for complex fluid microrheometry. Rev Sci Instrum. 2001;72:3626–3634. [Google Scholar]
- 100.Danilowicz C, Greenfield D, Prentiss M. Dissociation of ligand-receptor complexes using magnetic tweezers. Anal Chem. 2005;77:3023–8. doi: 10.1021/ac050057+. [DOI] [PubMed] [Google Scholar]
- 101.Charvin G, Allemand JF, Strick TR, Bensimon D, Croquette V. Twisting DNA: single molecule studies. Contemp Phys. 2004;45:383–403. [Google Scholar]
- 102.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–4. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 103.Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–7. doi: 10.1038/nature05938. This is an elegant paper in which the mechanism of action of a type I topoisomerase inhibitor measured at the single-molecule level is shown to be directly related to its effects in vivo. [DOI] [PubMed] [Google Scholar]
- 104.Revyakin A, Ebright RH, Strick TR. Single-molecule DNA nanomanipulation: improved resolution through use of shorter DNA fragments. Nat Methods. 2005;2:127–38. doi: 10.1038/nmeth0205-127. [DOI] [PubMed] [Google Scholar]
- 105.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–43. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys Rev Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 107.Lee GU, Chrisey LA, Colton RJ. Direct measurement of the forces between complementary strands of DNA. Science. 1994;266:771–3. doi: 10.1126/science.7973628. [DOI] [PubMed] [Google Scholar]
- 108.Dai P, et al. X-ray-diffraction and scanning-tunneling-microscopy studies of a liquid-crystal film adsorbed on single-crystal graphite. Phys Rev B Condens Matter. 1993;47:7401–7407. doi: 10.1103/physrevb.47.7401. [DOI] [PubMed] [Google Scholar]
- 109.Binnig G, Garcia N, Rohrer H. Conductivity sensitivity of inelastic scanning tunneling microscopy. Phys Rev B Condens Matter. 1985;32:1336–1338. doi: 10.1103/physrevb.32.1336. [DOI] [PubMed] [Google Scholar]
- 110.Marti O, et al. Scanning probe microscopy of biological samples and other surfaces. J Microsc. 1988;152:803–9. doi: 10.1111/j.1365-2818.1988.tb01452.x. [DOI] [PubMed] [Google Scholar]
- 111.Engel A. Biological applications of scanning probe microscopes. Annu Rev Biophys Biophys Chem. 1991;20:79–108. doi: 10.1146/annurev.bb.20.060191.000455. [DOI] [PubMed] [Google Scholar]
- 112.Lindsay SM. Biological scanning probe microscopy comes of age. Biophys J. 1994;67:2134–5. doi: 10.1016/S0006-3495(94)80702-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bustamante JO, Liepins A, Prendergast RA, Hanover JA, Oberleithner H. Patch clamp and atomic force microscopy demonstrate TATA-binding protein (TBP) interactions with the nuclear pore complex. J Membr Biol. 1995;146:263–72. doi: 10.1007/BF00233946. [DOI] [PubMed] [Google Scholar]
- 114.Shao Z, Yang J, Somlyo AP. Biological atomic force microscopy: from microns to nanometers and beyond. Annu Rev Cell Dev Biol. 1995;11:241–65. doi: 10.1146/annurev.cb.11.110195.001325. [DOI] [PubMed] [Google Scholar]
- 115.Drake B, et al. Imaging crystals, polymers, and processes in water with the atomic force microscope. Science. 1989;243:1586–9. doi: 10.1126/science.2928794. [DOI] [PubMed] [Google Scholar]
- 116.Bustamante C, Rivetti C, Keller DJ. Scanning force microscopy under aqueous solutions. Curr Opin Struct Biol. 1997;7:709–16. doi: 10.1016/s0959-440x(97)80082-6. [DOI] [PubMed] [Google Scholar]
- 117.Rief M, Oesterhelt F, Heymann B, Gaub HE. Single Molecule Force Spectroscopy on Polysaccharides by Atomic Force Microscopy. Science. 1997;275:1295–7. doi: 10.1126/science.275.5304.1295. [DOI] [PubMed] [Google Scholar]
- 118.Cumpson PJ, Zhdan P, Hedley J. Calibration of AFM cantilever stiffness: a microfabricated array of reflective springs. Ultramicroscopy. 2004;100:241–51. doi: 10.1016/j.ultramic.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 119.Sarkar A, Robertson RB, Fernandez JM. Simultaneous atomic force microscope and fluorescence measurements of protein unfolding using a calibrated evanescent wave. Proc Natl Acad Sci U S A. 2004;101:12882–6. doi: 10.1073/pnas.0403534101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Leake MC, Wilson D, Gautel M, Simmons RM. The elasticity of single titin molecules using a two-bead optical tweezers assay. Biophys J. 2004;87:1112–35. doi: 10.1529/biophysj.103.033571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kellermayer MS. Delayed dissociation of in vitro moving actin filaments from heavy meromyosin induced by low concentrations of Triton X-100. Biophys Chem. 1997;67:199–210. doi: 10.1016/s0301-4622(97)00044-6. [DOI] [PubMed] [Google Scholar]
- 122.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–9. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 123.Florin EL, Moy VT, Gaub HE. Adhesion forces between individual ligand-receptor pairs. Science. 1994;264:415–7. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 124.Yasuda R, Noji H, Kinosita K, Jr, Yoshida M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell. 1998;93:1117–24. doi: 10.1016/s0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]
- 125.Rief M, Gautel M, Schemmel A, Gaub HE. The mechanical stability of immunoglobulin and fibronectin III domains in the muscle protein titin measured by atomic force microscopy. Biophys J. 1998;75:3008–14. doi: 10.1016/S0006-3495(98)77741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Baumgartner W, et al. Cadherin interaction probed by atomic force microscopy. Proc Natl Acad Sci U S A. 2000;97:4005–10. doi: 10.1073/pnas.070052697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Marszalek PE, et al. Mechanical unfolding intermediates in titin modules. Nature. 1999;402:100–3. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- 128.Carrion-Vazquez M, et al. Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering. Prog Biophys Mol Biol. 2000;74:63–91. doi: 10.1016/s0079-6107(00)00017-1. [DOI] [PubMed] [Google Scholar]
- 129.Steward A, Toca-Herrera JL, Clarke J. Versatile cloning system for construction of multimeric proteins for use in atomic force microscopy. Protein Sci. 2002;11:2179–83. doi: 10.1110/ps.0212702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Katz E, Willner I. Biomolecule-functionalized carbon nanotubes: applications in nanobioelectronics. Chemphyschem. 2004;5:1084–104. doi: 10.1002/cphc.200400193. [DOI] [PubMed] [Google Scholar]
- 131.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–600. doi: 10.1126/science.8079175. In this work the non-linear elasticity of DNA was measured and an fit with an analytical expression. These results underpin all of the subsequent single-molecule force spectroscopy measurements of DNA processing enzymes. [DOI] [PubMed] [Google Scholar]
- 132.Fernandez JM, Li HB. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science. 2004;303:1674–1678. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
- 133.Schwaiger I, Kardinal A, Schleicher M, Noegel AA, Rief M. A mechanical unfolding intermediate in an actin-crosslinking protein. Nat Struct Mol Biol. 2004;11:81–5. doi: 10.1038/nsmb705. [DOI] [PubMed] [Google Scholar]
- 134.Carrion-Vazquez M, et al. The mechanical stability of ubiquitin is linkage dependent. Nat Struct Biol. 2003;10:738–43. doi: 10.1038/nsb965. [DOI] [PubMed] [Google Scholar]
- 135.Brockwell DJ, et al. Pulling geometry defines the mechanical resistance of a beta-sheet protein. Nat Struct Biol. 2003;10:731–7. doi: 10.1038/nsb968. [DOI] [PubMed] [Google Scholar]
- 136.Dietz H, Berkemeier F, Bertz M, Rief M. Anisotropic deformation response of single protein molecules. Proc Natl Acad Sci U S A. 2006;103:12724–8. doi: 10.1073/pnas.0602995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wright CF, Lindorff-Larsen K, Randles LG, Clarke J. Parallel protein-unfolding pathways revealed and mapped. Nat Struct Biol. 2003;10:658–62. doi: 10.1038/nsb947. [DOI] [PubMed] [Google Scholar]
- 138.Wiita AP, et al. Probing the chemistry of thioredoxin catalysis with force. Nature. 2007;450:124. doi: 10.1038/nature06231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Szoszkiewicz R, et al. Dwell time analysis of a single-molecule mechanochemical reaction. Langmuir. 2008;24:1356–64. doi: 10.1021/la702368b. [DOI] [PubMed] [Google Scholar]
- 140.Greene D, et al. Single-molecule force spectroscopy reveals a stepwise unfolding of C. elegans giant protein kinase domains. Biophys J. 2008 doi: 10.1529/biophysj.108.130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Brown AE, Litvinov RI, Discher DE, Weisel JW. Forced unfolding of coiled-coils in fibrinogen by single-molecule AFM. Biophys J. 2007;92:L39–41. doi: 10.1529/biophysj.106.101261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lim BB, Lee EH, Sotomayor M, Schulten K. Molecular basis of fibrin clot elasticity. Structure. 2008;16:449–59. doi: 10.1016/j.str.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 143.Kellermayer MS, et al. Reversible mechanical unzipping of amyloid beta-fibrils. J Biol Chem. 2005;280:8464–70. doi: 10.1074/jbc.M411556200. [DOI] [PubMed] [Google Scholar]
- 144.Decker B, Kellermayer MS. Periodically arranged interactions within the myosin filament backbone revealed by mechanical unzipping. J Mol Biol. 2008;377:307–10. doi: 10.1016/j.jmb.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 145.Oesterhelt F, et al. Unfolding pathways of individual bacteriorhodopsins. Science. 2000;288:143–6. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 146.Yu J, et al. Single-molecule force spectroscopy study of interaction between transforming growth factor beta1 and its receptor in living cells. J Phys Chem B. 2007;111:13619–25. doi: 10.1021/jp0758667. [DOI] [PubMed] [Google Scholar]
- 147.Moy VT, Florin EL, Gaub HE. Intermolecular forces and energies between ligands and receptors. Science. 1994;266:257–9. doi: 10.1126/science.7939660. [DOI] [PubMed] [Google Scholar]
- 148.Hinterdorfer P, Baumgartner W, Gruber HJ, Schilcher K, Schindler H. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc Natl Acad Sci U S A. 1996;93:3477–81. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Allen S, et al. Detection of antigen-antibody binding events with the atomic force microscope. Biochemistry. 1997;36:7457–63. doi: 10.1021/bi962531z. [DOI] [PubMed] [Google Scholar]
- 150.Yu J, Jiang Y, Ma X, Lin Y, Fang X. Energy landscape of aptamer/protein complexes studied by single-molecule force spectroscopy. Chem Asian J. 2007;2:284–9. doi: 10.1002/asia.200600230. [DOI] [PubMed] [Google Scholar]
- 151.Ando T, et al. A high-speed atomic force microscope for studying biological macromolecules. Proc Natl Acad Sci U S A. 2001;98:12468–72. doi: 10.1073/pnas.211400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wen JD, et al. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tanner NA, et al. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat Struct Mol Biol. 2008;15:170–6. doi: 10.1038/nsmb.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hamdan SM, et al. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27:539–49. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 155.Lee JB, et al. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–4. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 156.Ishijima A, et al. Simultaneous observation of individual ATPase and mechanical events by a single myosin molecule during interaction with actin. Cell. 1998;92:161–71. doi: 10.1016/s0092-8674(00)80911-3. [DOI] [PubMed] [Google Scholar]
- 157.Lang MJ, Fordyce PM, Engh AM, Neuman KC, Block SM. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat Methods. 2004;1:133–9. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li PT, Bustamante C, Tinoco I., Jr Real-time control of the energy landscape by force directs the folding of RNA molecules. Proc Natl Acad Sci U S A. 2007;104:7039–44. doi: 10.1073/pnas.0702137104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chiou PY, Ohta AT, Wu MC. Massively parallel manipulation of single cells and microparticles using optical images. Nature. 2005;436:370–372. doi: 10.1038/nature03831. [DOI] [PubMed] [Google Scholar]
- 160.Cohen AE, Moerner WE. Suppressing Brownian motion of individual biomolecules in solution. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4362–4365. doi: 10.1073/pnas.0509976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Berg-Sorensen K, Flyvbjerg H. Power spectrum analysis for optical tweezers. Rev Sci Instrum. 2004;75:594–612. [Google Scholar]