Abstract
The altered metabolism of cancer can render cells dependent on the availability of metabolic substrates for viability. Investigating the signaling mechanisms underlying cell death in cells dependent upon glucose for survival, we demonstrate that glucose withdrawal rapidly induces supra-physiological levels of phospho-tyrosine signaling, even in cells expressing constitutively active tyrosine kinases. Using unbiased mass spectrometry-based phospho-proteomics, we show that glucose withdrawal initiates a unique signature of phospho-tyrosine activation that is associated with focal adhesions. Building upon this observation, we demonstrate that glucose withdrawal activates a positive feedback loop involving generation of reactive oxygen species (ROS) by NADPH oxidase and mitochondria, inhibition of protein tyrosine phosphatases by oxidation, and increased tyrosine kinase signaling. In cells dependent on glucose for survival, glucose withdrawal-induced ROS generation and tyrosine kinase signaling synergize to amplify ROS levels, ultimately resulting in ROS-mediated cell death. Taken together, these findings illustrate the systems-level cross-talk between metabolism and signaling in the maintenance of cancer cell homeostasis.
Keywords: cancer, metabolism, phosphatase, proteomics, reactive oxygen species
Introduction
Tumor cells exhibit an altered metabolism compared with non-transformed cells, consuming glucose and glutamine and producing lactate at prodigious rates. Building on Otto Warburg's initial observation of aerobic glycolysis in cancer tissues (Warburg, 1956), tumorigenic metabolism is thought to help satisfy the rapacious demands of highly proliferative cancer cells for biosynthetic precursors including lipids, proteins, and nucleic acids. In fact, the success of positron emission tomography imaging for non-invasive imaging of tumors using the radiolabeled glucose analog 18F-fluorodeoxyglucose relies upon the highly glycolytic tumor phenotype (Czernin and Phelps, 2002).
Additional evidence has demonstrated that many oncogenic kinases drive the high metabolic demands of cancer cells. A prime example is the serine/threonine kinase Akt, which increases aerobic glycolysis through upregulation of glucose transporter levels and suppression of fatty acid β-oxidation (Deberardinis et al, 2008). More recently, it was shown that tyrosine kinases (TKs) including FGFR1 and a constitutively active mutant of EGFR (EGFRvIII) can promote aerobic glycolysis and lipogenesis, respectively (Guo et al, 2009; Hitosugi et al, 2009).
One consequence of deregulated tumor metabolism is that cells can become dependent on the supply of metabolic substrates such as glucose or glutamine for viability (Elstrom et al, 2004; Buzzai et al, 2005; Yuneva et al, 2007; Aykin-Burns et al, 2009; Choo et al, 2010). Notably, this ‘addiction’ to metabolic substrates is influenced by signal transduction pathways. For example, expression of constitutively active Akt renders glioblastoma (GBM) cells sensitive to glucose withdrawal-mediated cell death (Elstrom et al, 2004). Alternatively, pharmacological activation or inhibition of kinases involved in energy and nutrient sensing including AMPK and mTORC1, respectively, can confer protection against glucose withdrawal-mediated cell death (Buzzai et al, 2005; Choo et al, 2010).
Based on the evidence that oncogenic signaling can impose a state of metabolic inflexibility, as well as the emerging concept that complex networks link signal transduction with metabolism (Wellen et al, 2010), we conducted a systems-level investigation into the role of signal transduction underlying cell death following metabolic perturbation in cell systems dependent on glucose for survival. Using cell lines derived from GBM, sarcoma, and melanoma, we report the unexpected observation that glucose withdrawal induces supra-physiological levels of phospho-tyrosine signaling in cells dependent on glucose for viability. Using unbiased, mass spectrometry-based phospho-tyrosine profiling, we discovered that glucose withdrawal induces a unique phospho-tyrosine signature associated with focal adhesions. Integrating this observation with the literature describing the inhibition of protein tyrosine phosphatases (PTPs) by NADPH oxidase (NOX)- and mitochondria-derived reactive oxygen species (ROS), we construct a contextual systems model describing a positive feedback loop involving ROS, PTPs, and TKs. Upon glucose deprivation, this positive feedback loop results in synergistic amplification of TK signaling and ROS levels, resulting in supra-physiological levels of tyrosine phosphorylation and ROS-mediated cell death. Taken together, our results describe a novel mechanism integrating the metabolic and signaling systems homeostasis of cancer cells.
Results
Glucose withdrawal induces supra-physiological levels of phospho-tyrosine signaling in glucose withdrawal-sensitive cells
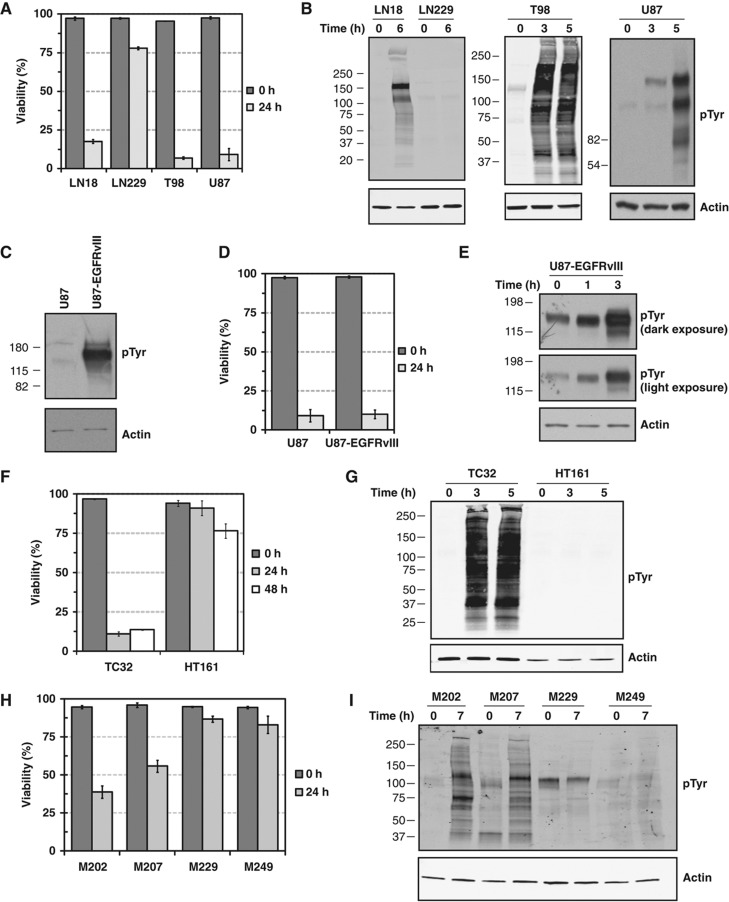
To investigate the signaling mechanisms that underlie rapid cell death following glucose withdrawal (i.e., glucose ‘addiction’), we tested the response of four GBM cell lines (LN18, LN229, T98, and U87MG) to withdrawal of glucose and the glycolytic product pyruvate, which may serve as an alternate substrate for the TCA cycle (Yang et al, 2009). Within 24 h of glucose and pyruvate withdrawal, LN18, T98, and U87MG exhibited rapid cell death, whereas LN229 cells exhibited only a minor (∼15%) loss of viability (Figure 1A), consistent with previous reports (Elstrom et al, 2004). In our systems, standard pyruvate levels (110 mg/l) did not rescue glucose starvation-induced cell death (not shown). Using an antibody that recognizes tyrosine phosphorylated proteins, western blotting demonstrated that the glucose withdrawal-sensitive cell lines (LN18, T98, and U87MG) exhibited a dramatic upregulation of phospho-tyrosine signaling within 3–6 h of glucose starvation (Figure 1B). In contrast, the glucose withdrawal-insensitive cell line LN229 showed no induction of phospho-tyrosine signaling. These data demonstrate that a severe metabolic perturbation (i.e., glucose starvation) can induce supra-physiological levels of TK signaling.
Figure 1.
Glucose withdrawal induces supra-physiological levels of tyrosine phosphorylation in cells sensitive to glucose withdrawal. (A) LN18, LN229, T98, and U87 GBM cell lines were starved of glucose and pyruvate for 24 h. Trypan blue exclusion measurements demonstrated that LN18, T98, and U87, but not LN229, show a rapid and complete loss of viability following glucose withdrawal. (B) Western blotting with a phospho-tyrosine antibody revealed that the glucose withdrawal-sensitive cell lines (LN18, T98, and U87) exhibit a dramatic induction of phospho-tyrosine signaling at the indicated times following glucose withdrawal. Conversely, the glucose withdrawal-insensitive cell line LN229 showed no increase in phospho-tyrosine levels following glucose withdrawal. Actin served as an equal loading control. (C–E) Glucose withdrawal induces supra-physiological levels of phospho-tyrosine signaling even in cells expressing a constitutively active tyrosine kinase. (C) Expression of the constitutively active EGFR mutant EGFRvIII in U87 cells induces high levels of phospho-tyrosine signaling as demonstrated by western blotting with an anti-phospho-tyrosine antibody. (D) Expression of the constitutively active EGFRvIII mutant in U87 cells (U87-EGFRvIII) does not alter the sensitivity to glucose withdrawal at 24 h. Error bars are standard deviation of the mean. (E) U87-EGFRvIII cells demonstrate a rapid induction of phospho-tyrosine signaling at the times indicated following glucose withdrawal. For phospho-tyrosine, both dark and light film exposures are shown. (F–I) Human sarcoma- and melanoma-derived cell lines also demonstrate a correlation between sensitivity to glucose withdrawal and rapid induction of supra-physiological phospho-tyrosine levels. (F) The sarcoma cell lines HT161 and TC32 were starved of glucose and pyruvate and their viability was measured by Trypan blue exclusion at the indicated times. TC32 cells show a rapid and complete loss of viability, whereas HT161 were relatively insensitive to glucose withdrawal. Error bars are standard deviation of the mean. (G) Western blotting with an anti-phospho-tyrosine antibody demonstrated that TC32, but not HT161, show induction of hyper-phosphorylation after 2 h of glucose starvation. (H) Melanoma cell lines M202, M207, M229, and M249 were starved of glucose and pyruvate for 24 h. Trypan blue exclusion measurements revealed that M202 and M207, but not M229 or M249 cells, showed significant loss of cell viability after 24 h of glucose withdrawal. Error bars are standard deviation of the mean. (I) Western blotting demonstrated that 7 h of glucose withdrawal induced supra-physiological tyrosine phosphorylation in M202 and M207, but not M229 or M249.
We next tested the specific requirements for cell death and phospho-tyrosine induction. Serum factors did not modulate cell death or the induction of phospho-tyrosine signaling following glucose withdrawal (Supplementary Figure S1). In addition, cells deprived of L-glutamine showed no decrease in cell viability and only a very modest change in tyrosine phosphorylation (Supplementary Figure S2). Comparing glucose withdrawal-sensitive cell lines (LN18 and U87) to the withdrawal-insensitive line LN229, we observed no consistent changes in either phosphorylation of the AMPK substrate pS79-acetyl coA carboxylase or stabilization of the p53 tumor suppressor (data not shown). Thus, the phospho-tyrosine induction and cell death observed following glucose withdrawal are specific to the metabolic substrate glucose and without evidence of AMPK signaling involvement.
To test whether the presence of a constitutively active TK could affect the observed response to glucose withdrawal, we next used U87 cells that overexpress a constitutively active deletion mutant of EGFR found in ∼50% of GBMs (variant III, U87-EGFRvIII) and exhibit constitutively high levels of phospho-tyrosine signaling (Figure 1C; Wang et al, 2006). Expression of EGFRvIII did not alter the rate of cell death compared with parental U87 cells (Figure 1D). Surprisingly, even in the presence of the constitutively active oncogenic kinase, glucose withdrawal induced further upregulation of tyrosine phosphorylation, suggesting that phospho-tyrosine induction is ligand independent (Figure 1E). These data demonstrate that the rapid and supra-physiological induction of TK signaling upon glucose withdrawal can occur even in cells with high basal levels of phospho-tyrosine signaling.
Next, we tested whether cells from cancers other than GBM exhibit a similar response to glucose withdrawal. Cell lines derived from sarcoma patient biopsies exhibited a range of sensitivities to glucose withdrawal (Figure 1F). Similarly to GBM, the glucose withdrawal-sensitive cell line TC32 demonstrated a dramatic and rapid induction of TK signaling, whereas the glucose withdrawal-insensitive HT161 line showed no increase (Figure 1G). Using cell lines derived from melanoma patient biopsies, we again found a range of sensitivities to glucose withdrawal (Figure 1H). Cells that were sensitive to glucose withdrawal (M202, M207) demonstrated a rapid induction in TK signaling, whereas glucose withdrawal-insensitive cells did not (M229 and M249; Figure 1I). Notably, the degree of glucose withdrawal-induced cell death was not correlated with cell doubling time of the melanoma cell lines (Supplementary Figure S3). Taken together, these data demonstrate a correlation between rapid cell death following glucose withdrawal (i.e., glucose ‘addiction’) and a dramatic, serum-independent induction of supra-physiological levels of TK signaling.
PTEN status influences glucose withdrawal-induced cell death and phospho-tyrosine induction
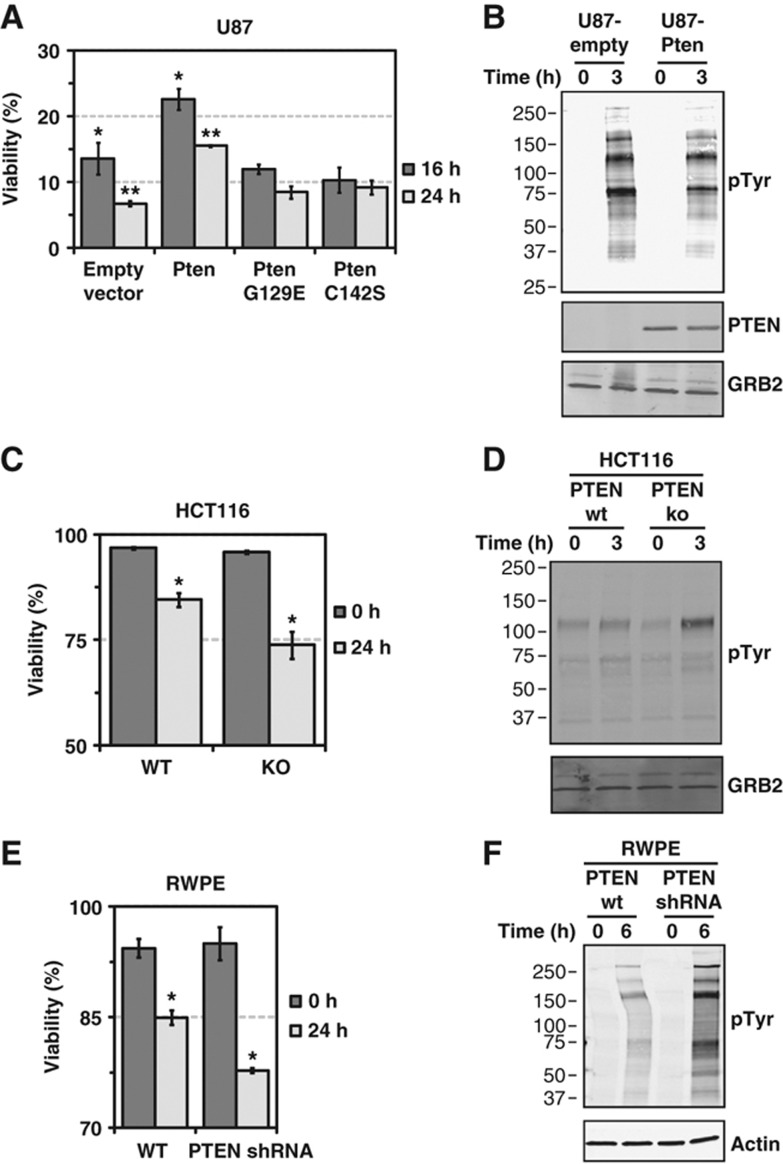
Because the expression of constitutively active Akt can render an insensitive GBM cell line (LN229) sensitive to glucose withdrawal (Elstrom et al, 2004), we next tested whether glucose withdrawal-induced TK signaling and induction of cell death were regulated by PTEN, a negative regulator of Akt signaling. To test this hypothesis, we expressed wild-type murine Pten as well as two catalytically inactive mutants in U87 cells, which are PTEN null and glucose withdrawal sensitive. Wild-type Pten-overexpressing cells demonstrated increased cell viability following glucose starvation (Figure 2A), but two catalytically inactive mutants (C124S and G129E) did not, demonstrating a requirement for the lipid phosphatase activity of Pten for partial rescue from glucose withdrawal. Furthermore, the increased resistance to glucose withdrawal-induced cell death in U87-Pten was accompanied by reduced phospho-tyrosine induction upon glucose withdrawal (Figure 2B), providing further evidence for the correlation between glucose withdrawal-induced phospho-tyrosine signaling and cell death.
Figure 2.
PTEN status regulates glucose withdrawal-induced phospho-tyrosine induction and cell death. (A, B) Murine wild-type Pten or the lipid phosphatase inactive mutants G129E and C142S were expressed in the PTEN-null GBM cell line U87. (A) U87 cells infected with an empty vector control, wild-type Pten or the lipid phosphatase inactive mutants were starved of glucose and pyruvate for 16 and 24 h, and viability was measured by Trypan blue exclusion. Expression of wild-type Pten increased survival following glucose withdrawal by roughly two-fold (P=0.04 (*) and 0.001 (**) by Student's t-test). Error bars are standard error of the mean (n=3). (B) Western blotting with an anti-phospho-tyrosine antibody demonstrated that U87-Pten cells exhibit reduced induction of phospho-tyrosine signaling after 3 h of glucose withdrawal. Pten and GRB2 served as confirmation of Pten overexpression and equal loading, respectively. (C, D) HCT116 cells with genetic knockout of PTEN show greater cell death and phospho-tyrosine induction in response to glucose withdrawal. (C) HCT116 with wild-type PTEN (WT) or genetic knockout by homologous recombination (HCT116-22, KO) were serum starved for 16 h and then starved of glucose and pyruvate for 24 h before measurement of viability by Trypan blue exclusion. PTEN knockout cells showed reduced survival at 24 h compared with wild-type cells (P-value=0.03 (*) by Student's t-test, n=4). (D) Western blotting with an antibody against phospho-tyrosine demonstrated increased phospho-tyrosine signaling in HCT116 PTEN KO cells following 3 h of glucose withdrawal. Confirmation of loss of PTEN expression in HCT116 KO cells is shown in Supplementary Figure S4. (E, F) RWPE cells with reduced PTEN expression show greater cell death in response to glucose withdrawal (P-value=0.002 (*) by Student's t-test, n=3). (E) RWPE cells with wild-type PTEN or cells infected with an shRNA targeting PTEN were starved of glucose and pyruvate for 24 h before viability measurements by Trypan blue exclusion. Confirmation of reduced PTEN expression is shown in Supplementary Figure S4. (F) Western blotting demonstrated that RWPE cells with reduced PTEN expression demonstrate greater induction of phospho-tyrosine signaling following glucose withdrawal.
We next tested whether downregulation of endogenous PTEN expression could similarly regulate the twin phenomena of TK induction and cell death following glucose withdrawal. We first tested HCT116 colon carcinoma cells with and without homologous recombination-mediated deletion of endogenous PTEN (Supplementary Figure S4). Cells lacking PTEN expression exhibited greater cell death and induction of phospho-tyrosine signaling than parental wild-type cells (Figure 2C and D). We next tested RWPE prostate epithelial cells with and without shRNA-mediated knockdown of PTEN expression and found that PTEN knockdown cells exhibited greater cell death and greater phospho-tyrosine induction than parental cells (Figure 2E and F). Notably, knockdown of PTEN expression in an already glucose withdrawal-sensitive cell line (SF268) did not further increase sensitivity to glucose withdrawal (Supplementary Figure S5).
Taken together, these data demonstrate that PTEN, a negative regulator of Akt signaling, can decrease sensitivity to glucose withdrawal-induced TK induction and cell death. However, it is clear that PTEN is not a master regulator of these phenotypes, as demonstrated by the robust induction of phospho-tyrosine signaling following glucose withdrawal even in U87 cells overexpressing PTEN (Figure 2B).
Glucose withdrawal activates multiple TKs and selected intracellular signaling pathways
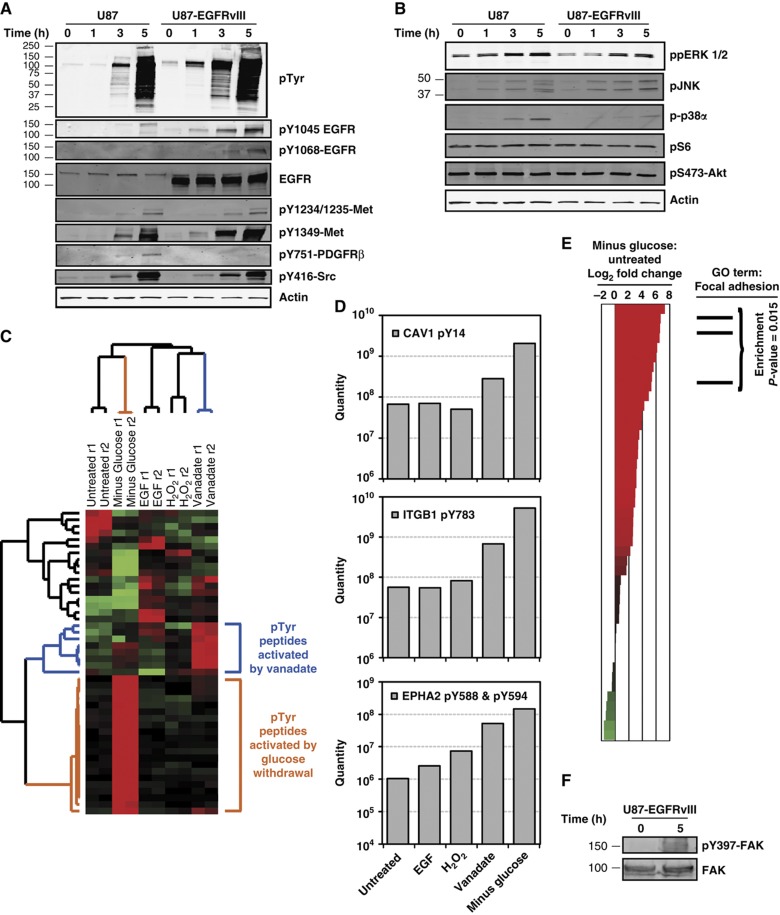
Having established that PTEN influences but does not fully control the sensitivity to glucose withdrawal, we sought to gain a more detailed perspective on which TKs were activated by glucose withdrawal. Using the glucose withdrawal-sensitive cell lines U87 and U87-EGFRvIII, we found that glucose withdrawal induced phosphorylation of EGFR on residues Y1068, indicating enzymatic activation, and Y1045, a docking site for c-Cbl (Figure 3A). The receptor TK (RTK) Met also showed strong glucose withdrawal-induced phosphorylation of residues in the kinase activation loop (Y1234/Y1235) and in the carboxy-terminal tail (Y1349). Additionally, we found that phosphorylation of PDGFRβ Y751, which regulates enzymatic activation, was increased by glucose withdrawal. Finally, we tested the non-RTK SRC and observed strong glucose withdrawal-induced phosphorylation of the enzymatic activation site in the TK domain (Y416). Thus, these data demonstrate that glucose withdrawal induces activation of multiple receptor and non-RTKs.
Figure 3.
Phospho-proteomics reveals that glucose withdrawal induces a distinct signature of phospho-tyrosine signaling that is associated with focal adhesions. (A, B) Following glucose and pyruvate starvation of U87 and U87-EGFRvIII for the indicated times, western blotting revealed activation of some but not all signaling pathways following glucose withdrawal. (A) Phospho-specific antibodies against tyrosine residues demonstrated significantly increased phosphorylation of RTKs, including EGFR, Met, and PDFGRβ. The non-RTK Src also showed increased active site phosphorylation. Total EGFR and actin served as equal loading controls. (B) Phospho-specific antibodies revealed increased glucose withdrawal-induced activity of all MAPK pathways tested (ppERK 1/2, pJNK and p-p38α) but not mTOR signaling (pS235/S236-S6) or Akt signaling (pS473-Akt). (C–E) Glucose withdrawal induces a signature of hyper-phosphorylation in U87 that is associated with focal adhesions. (C) Hierarchical clustering of tyrosine phosphorylation in U87 cells reveals that glucose withdrawal induces a distinct set of phospho-events. U87 cells were treated with four stimuli known to induce tyrosine phosphorylation, including (a) EGF stimulation (10 ng/ml, 5 min), (b) vanadate treatment (1 mM, 60 min), (c) H2O2 (5 mM, 30 min) and (d) glucose and pyruvate withdrawal (3 h). Changes in phospho-tyrosine signaling were measured by quantitative, label-free mass spectrometry (Rubbi et al, 2011) and data were hierarchically clustered. Each row of the heatmap depicts an individual phosphorylation event, and each column represents a sample as labeled. In the heatmap, red and green represent normalized levels of high and low phosphorylation, respectively. Samples were measured in technical duplicate (r1 and r2). Branches of the dendrogram associated with upregulation by glucose withdrawal and vanadate treatment are colored orange and blue, respectively. See Supplementary Table 1 for quantitative phospho-peptide data. (D) Glucose withdrawal induces increased phosphorylation of proteins known to localize to focal adhesions. Tyrosine residues on integrin β 1 (ITGB1 pY783), caveolin 1 (CAV1 pY14), and ephrin 2A (EPH2A pY588 and pY594) show dramatically increased phosphorylation in response to 3 h of glucose withdrawal. (E) Phospho-events associated with focal adhesions are enriched following glucose withdrawal in U87 cells. Phospho-peptides were ranked according to the measured log2 fold change in phospho-tyrosine levels following glucose withdrawal and plotted on a waterfall plot, where red and green represent increased or decreased phosphorylation, respectively. Analysis of the phospho-peptides demonstrated an enrichment for proteins annotated with the GO term Focal Adhesion (GO:0005925) at the top of the ranked list (i.e., increased phosphorylation following glucose withdrawal) (permutation-based P-value=0.02). (F) Western blotting revealed increased FAK Y397 phosphorylation in response to glucose withdrawal. Source data is available for this figure in the Supplementary Information.
We next asked which intracellular signaling pathways downstream of RTKs were activated following glucose withdrawal. Western blotting revealed activation of three MAPK signaling pathways (ERK 1/2, JNK, and p38α) in both U87 and U87-EGFRvIII cells (Figure 3B). In contrast, we found that mTOR signaling, as measured by pS6 levels, and PI3K/Akt signaling, as measured by pSer473-Akt levels, were unaffected by glucose withdrawal. Because U87 and U87-EGFRvIII exhibit high basal levels of mTOR and Akt signaling, it is possible that these signaling pathways were saturated before glucose withdrawal. Taken together, these data indicate that the dramatic induction of supra-physiological levels of phospho-tyrosine signaling caused by glucose withdrawal is accompanied by concomitant activation of some but not all intracellular signaling pathways. This result led us to further explore the spectrum of glucose withdrawal-induced signaling using a more unbiased approach.
Phospho-proteomics reveals that glucose withdrawal induces a distinct phospho-tyrosine signature associated with focal adhesions
To obtain a more system-wide, unbiased characterization of glucose withdrawal-induced phospho-tyrosine signaling, we performed global and quantitative tyrosine phospho-profiling using label-free quantitative mass spectrometry (Skaggs et al, 2006; Rubbi et al, 2011). For both U87MG and U87MG-EGFRvIII cells, we compared glucose deprivation with three stimuli associated with activation of TK signaling: (a) EGF, a ligand for EGFR, (b) vanadate, a chemical inhibitor of PTPs, and (c) hydrogen peroxide (H2O2), a general oxidative stress.
Phospho-tyrosine profiling of U87 and U87-EGFRvIII cells identified 46 and 110 unique phosphorylation sites, respectively. Hierarchical clustering of the data revealed patterns of phospho-tyrosine peptide activation unique to all stimuli, including the two strongest perturbations, vanadate treatment and glucose withdrawal (Figure 3C; Supplementary Figure S6A). Notably, these unique patterns of phospho-tyrosine activation were preserved even when the data were normalized to overall signal strength, indicating that the observed signatures do not simply reflect increased activity of TKs (data not shown). As expected, bioinformatic analysis revealed that phospho-tyrosine sites activated by EGF stimulation were enriched for proteins annotated with the EGFR signaling pathway gene ontology term (P<0.001; Supplementary Figure S6B). Despite the presence of highly activated EGFR (Figure 3A), the EGF signaling pathway activation signature was not recapitulated by glucose withdrawal (data not shown). Instead, we noted the presence of highly phosphorylated focal adhesion-associated proteins in the glucose withdrawal cluster (Figure 3D; Supplementary Figure S6C, n=3 for U87 and 8 for U87-EGFRvIII). Indeed, bioinformatic analysis revealed that the glucose withdrawal phospho-tyrosine signature was highly enriched for peptides from focal adhesion proteins (P=0.015 and 0.004 for U87 and U87-EGFR-vIII, respectively; Figure 3E; Supplementary Figure S6D). To confirm this bioinformatic analysis, we tested whether glucose deprivation induced activation of focal adhesion kinase (FAK) and found that phosphorylation of the FAK activation site (Y397) was increased by glucose deprivation in U87-EGFRvIII (Figure 3F). Thus, unbiased phospho-proteomic profiling by LC-MS/MS revealed that glucose deprivation induces phospho-tyrosine signaling that is distinct from other phosphatase-targeted perturbations and associated with focal adhesions and activation of multiple TKs (e.g., EGFR, PDGFR, Met and Src).
Glucose withdrawal induces ROS generation in glucose withdrawal-sensitive cells
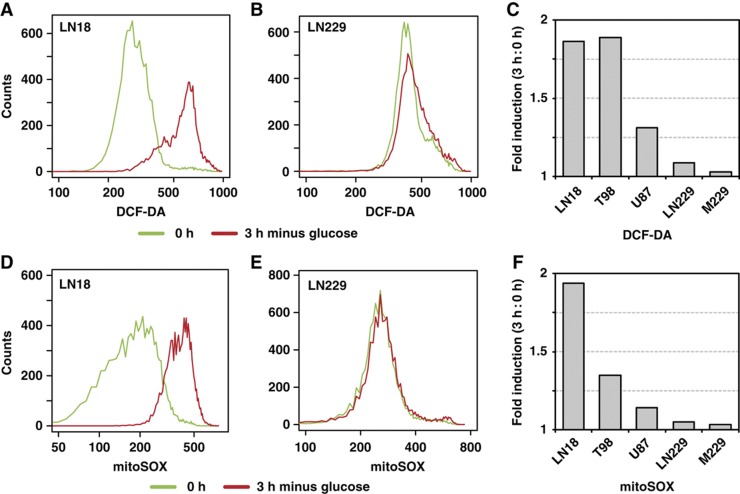
To determine how glucose withdrawal induces phospho-tyrosine signaling in glucose withdrawal-sensitive but not in insensitive cells, we tested the involvement of ROS that mediate the differential response of cancerous cells and their non-transformed counterparts to glucose withdrawal (Ahmad et al, 2005; Aykin-Burns et al, 2009). In glucose withdrawal-sensitive LN18 cells, we found that 3 h of glucose deprivation induced a nearly two-fold increase in the mean fluorescent intensity of the oxidation-dependent fluorogen DCF-DA (Figure 4A). In contrast, LN229 cells, which are insensitive to glucose withdrawal, demonstrated a negligible increase in DCF-DA signal following glucose withdrawal (Figure 4B). Similarly, we found that the glucose withdrawal-sensitive cell lines T98 and U87 exhibited increased ROS levels following glucose withdrawal, whereas the glucose withdrawal-insensitive cell line M229 exhibited no change (Figure 4C; Supplementary Figure S7A). Notably, increased ROS levels occurred at times when cells could be rescued from glucose withdrawal-induced cell death by resupplementation with glucose and pyruvate (Supplementary Figure S7B). Treatment with the H2O2 scavenger catalase abrogated the glucose withdrawal-induced increase in DCF-DA signal in both LN18 and U87 cells (Supplementary Figure S7C).
Figure 4.
Glucose withdrawal induces rapid amplification of ROS in glucose withdrawal-sensitive cells. Cells were starved of glucose and pyruvate for 0 or 3 h, stained with either the oxidation-sensitive fluorogen DCF-DA or mitoSOX, and analyzed by flow cytometry. (A, B) LN18 but not LN229 cells demonstrated increased DCF-DA fluorescence 3 h following glucose and pyruvate starvation. (C) Quantification of the fold change in DCF-DA signal (3 h:0 h) revealed that the glucose withdrawal-sensitive cell lines (LN18, T98, and U87) but not glucose withdrawal-insensitive cell lines (LN229 and M229) demonstrated increased ROS levels following glucose and pyruvate withdrawal. Histograms for all cell lines are shown in Supplementary Figure S7. (D, E) LN18 but not LN229 cells demonstrated increased mitochondrial superoxide production 3 h after glucose and pyruvate starvation. (F) Quantification of the fold change in mean mitoSOX signal (3 h:0 h) demonstrated that glucose withdrawal-sensitive cell lines (LN18, T98, and U87) but not glucose withdrawal-insensitive cell lines (LN229 and M229) demonstrated increased levels of mitochondrial ROS following glucose and pyruvate withdrawal. Histograms for all cell lines are shown in Supplementary Figure S7.
Because mitochondria can contribute to oxidative stress following glucose withdrawal (Ahmad et al, 2005), we next tested whether levels of mitochondrial ROS were increased following glucose withdrawal. Using the mitochondrial superoxide probe mitoSOX, we found that LN18 exhibited a roughly two-fold increase in mitochondrial superoxide levels (Figure 4D). In contrast, the glucose withdrawal-insensitive cell line LN229 demonstrated negligible changes following glucose withdrawal (Figure 4E). Similarly to DCF-DA, we found that other glucose withdrawal-sensitive cell lines (e.g., T98 and U87) exhibited moderate to substantial increases in mitochondrial superoxide levels upon glucose and pyruvate starvation, whereas the glucose withdrawal-insensitive cell line M229 showed little change (Figure 4F; Supplementary Figure S7D). Taken together, these data demonstrate that glucose withdrawal induces a rapid increase in ROS in cells highly dependent on glucose for survival.
Glucose withdrawal-induced ROS mediate TK induction and cell death
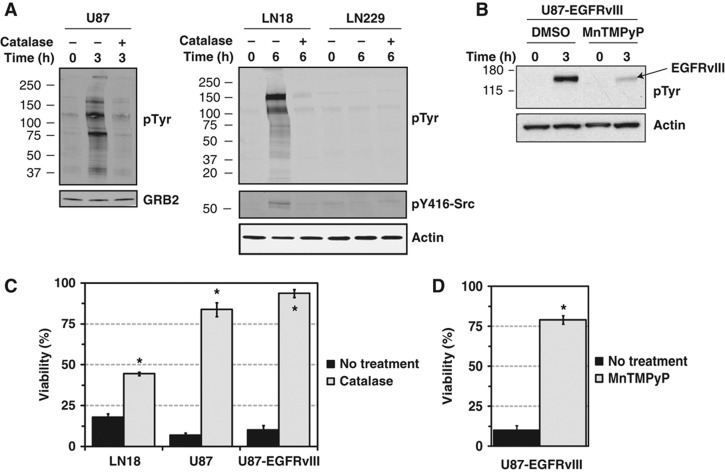
Having demonstrated that glucose withdrawal induces both supra-physiological phospho-tyrosine signaling and increased ROS levels in cells dependent on glucose for survival, we next tested the functional relationship between TK signaling and ROS induction upon glucose deprivation. In the glucose withdrawal-sensitive cell lines U87 and LN18, treatment with the H2O2 scavenger catalase ablated the induction of phospho-tyrosine signaling and activation of Src following glucose withdrawal (Figure 5A). LN229 cells, which are insensitive to glucose withdrawal, showed no change in either phospho-tyrosine signaling or Src activation in the absence and presence of catalase. Similarly to LN18, the sarcoma cell line TC32 also demonstrated reduced glucose withdrawal-induced phospho-tyrosine signaling when treated with catalase (Supplementary Figure S8A). We next tested the effects of a redox active manganic porphyrin (MnTMPyP) that can protect cells against oxidative stress and found that glucose withdrawal-induced activation of EGFRvIII phosphorylation was strongly attenuated in U87-EGFRvIII cells by MnTMPyP treatment (Figure 5B). These data indicate that ROS mediate the induction of phospho-tyrosine signaling upon glucose withdrawal.
Figure 5.
Glucose withdrawal-induced ROS mediate tyrosine kinase induction and cell death. (A) U87, LN18, and LN229 cells were starved of glucose and pyruvate for the indicated times with or without the H2O2 scavenger catalase (250 U/ml for U87, 1 kU/ml for LN18 and LN229). Western blotting demonstrated that glucose withdrawal-induced phospho-tyrosine signaling was ablated by catalase treatment in U87 and LN18. In addition, glucose withdrawal-induced activation of the Src active site (Y416) in LN18 cells required catalase-sensitive ROS. Actin served as an equal loading control. (B) U87-EGFRvIII cells were starved of glucose and pyruvate for 3 h in the presence of either DMSO or the antioxidant MnTMPyP (25 μM). Western blotting revealed that MnTMPyP treatment reduced tyrosine phosphorylation of EGFRvIII (∼155 kDa, the most prominent band in the phospho-tyrosine western blot) following glucose withdrawal. (C, D) ROS are required for cell death following glucose withdrawal. (C) The glucose withdrawal-sensitive cell lines LN18, U87, and U87-EGFRvIII were starved of glucose and pyruvate with or without catalase, and viability was measured by Trypan blue exclusion 24 h later. Catalase treatment rescued cells from glucose withdrawal-induced cell death (P-value <1 × 10−3 (*) by Student's t-test). Error bars are standard deviation of the mean (n=3–13). (D) U87-EGFRvIII cells were starved of glucose and pyruvate for 24 h in the absence or presence of the antioxidant MnTMPyP. Treatment with MnTMPyP rescued U87-EGFRvIII from glucose withdrawal-induced cell death (P-value <1 × 10−3 (*) by Student's t-test, n=13 and 3 for No treatment and MnTMPyP treatment, respectively).
Next, we tested whether ROS also mediate cell death upon glucose withdrawal. In several glucose withdrawal-sensitive GBM and sarcoma cell lines, treatment with the H2O2 scavenger catalase rescued cells from glucose withdrawal-induced cell death (Figure 5C; Supplementary Figure S8D). PEG-conjugated catalase, which can enter cells by endocytosis (Beckman et al, 1988), also protected cells against glucose withdrawal-induced cell death (data not shown). Supporting this observation, treatment with either MnTMPyP or cell-permeable reduced glutathione protected U87-EGFRvIII cells against glucose withdrawal-induced cell death (Figure 5D; Supplementary Figure S8E). Taken together, these data indicate that glucose withdrawal-induced ROS are functionally required for rapid cell death following glucose withdrawal.
NOX- and mitochondria-derived ROS contribute to glucose withdrawal-induced phospho-tyrosine signaling
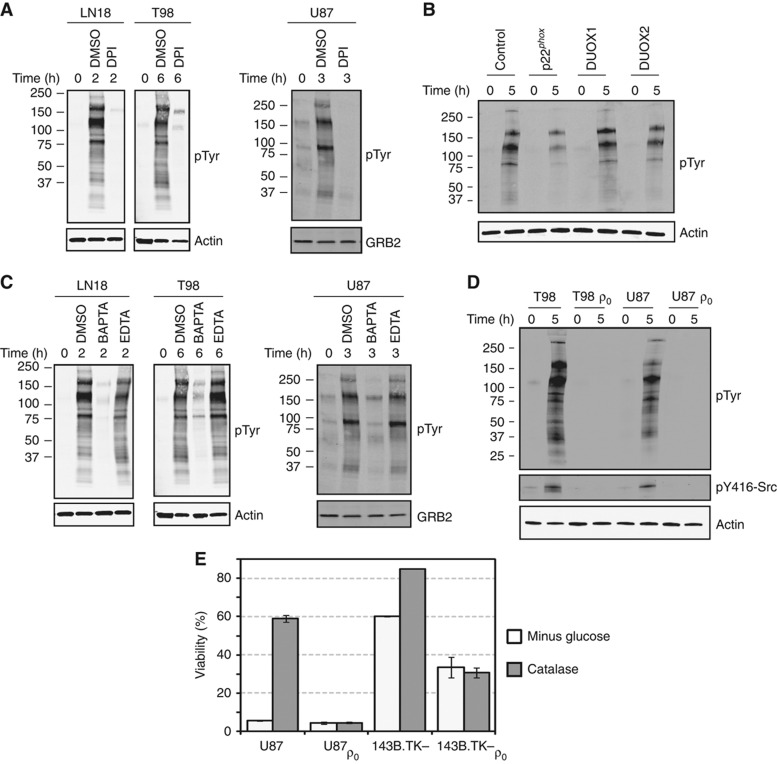
Having demonstrated that ROS contribute to phospho-tyrosine induction following glucose and pyruvate starvation, we next sought to determine the cellular source(s) of ROS that mediate glucose withdrawal-induced phospho-tyrosine signaling. We first focused on NOX, a membrane bound protein complex that generates superoxide anion because NOX is associated with focal adhesions (Ushio-Fukai, 2006) and the glucose withdrawal-induced phospho-tyrosine signature determined by our phospho-tyrosine mass spectrometry experiments is closely associated with focal adhesions (Figure 4). Treatment with DPI, a chemical inhibitor of flavo-proteins including NOX, completely abrogated glucose withdrawal-induced phospho-tyrosine signaling in LN18, T98, and U87 cells (Figure 6A). To confirm the role of NOX in glucose withdrawal-induced phospho-tyrosine signaling, we next targeted the p22phox organizer subunit that is required for the activity of NOX1–4 (Kawahara et al, 2005). Indeed, knockdown of p22phox expression by siRNA attenuated glucose withdrawal-induced phospho-tyrosine signaling (Figure 6B; Supplementary Figure S9). Notably, siRNA targeting the Ca2+-activated, p22phox-independent NOX family members DUOX1, DUOX2, and NOX5 were unable to affect glucose withdrawal-induced phospho-tyrosine signaling (Figure 6B and not shown). Thus, NOX-derived ROS contribute to the rapid upregulation of phospho-tyrosine signaling following glucose withdrawal in U87 cells.
Figure 6.
NADPH oxidase- and mitochondria-derived ROS contribute to glucose withdrawal-induced phospho-tyrosine signaling. (A) Inhibition of NOX inhibits glucose withdrawal-induced signaling. LN18, T98, and U87 cells were starved of glucose and pyruvate in the presence of DMSO or DPI (1 μM). Western blotting demonstrated that NOX activity is required for the induction of phospho-tyrosine signaling. (B) Knockdown of the NOX subunit p22phox attenuates phospho-tyrosine signaling following glucose withdrawal. U87 cells were reverse transfected with control, non-targeting siRNA or siRNA against p22phox, DUOX1 or DUOX2. Forty-eight hours later, cells were starved of glucose and pyruvate for 5 h. Western blotting demonstrated that knockdown of p22phox but not DUOX1/2 attenuated glucose withdrawal-induced phospho-tyrosine signaling. p22phox knockdown efficiency was >90% (Supplementary Figure S9). (C) LN18, T98, and U87 cells were starved of glucose and pyruvate in the presence of either DMSO or BAPTA-AM (25 μM). Western blotting with an anti-phospho-tyrosine antibody demonstrated that chelation of intracellular Ca2+ by BAPTA-AM completely abrogated glucose withdrawal-induced phospho-tyrosine signaling. Treatment with extracellular EDTA (25 μM) had no effect. Actin and GRB2 served as equal loading controls. (D) ρ0 derivatives of T98 and U87 cells do not exhibit upregulation of phospho-tyrosine signaling or activation of Src in response to glucose withdrawal. (E) Catalase rescues parental but not ρ0 cells from glucose withdrawal-induced cell death. Cells were starved of glucose and pyruvate with or without catalase (1 kU/ml), and viability was measured by Trypan blue exclusion 24 h later.
Next, we sought to determine whether mitochondrial superoxide also contribute to glucose withdrawal-induced phospho-tyrosine signaling. Because intracellular calcium flux has been linked to mitochondrial-mediated superoxide generation (Mbaya et al, 2010), we tested whether intracellular Ca2+ contributes to glucose withdrawal-induced mitochondrial superoxide production. Indeed, treatment with the cell permeable Ca2+ chelator BAPTA-AM reduced glucose withdrawal-induced mitochondrial superoxide levels (Supplementary Figure S10). Consistent with the hypothesis that mitochondrial ROS mediate glucose withdrawal-induced phospho-tyrosine signaling, treatment with BAPTA-AM but not with extracellular EDTA substantially abrogated the induction of phospho-tyrosine in the glucose withdrawal-sensitive cell lines LN18, T98, and U87 (Figure 6C).
To more directly test the role of mitochondrial superoxide generation in glucose withdrawal-induced phospho-tyrosine signaling, we obtained or derived ρ0 sublines of T98 and U87 GBM cells and the osteosarcoma line 143B.TK− (Supplementary Figure S11). Because several protein products required for the mitochondrial electron transport chain are encoded on the mitochondrial genome, these ρ0 cells, which lack mitochondrial DNA, are unable to generate mitochondrial superoxide (Hashiguchi and Zhang-Akiyama, 2009). In all three ρ0 derivatives, glucose withdrawal-induced phospho-tyrosine signaling was absent (Figure 6D; Supplementary Figure S11C). Glucose withdrawal did induce cell death in the mitochondria-deficient U87 ρ0 and 143B.TK− ρ0 cells, but this death was not rescued by the H2O2 scavenger catalase (Figure 6E). Together, these data demonstrate that the rapid upregulation of phospho-tyrosine signaling following glucose and pyruvate starvation involves both NOX- and mitochondria-mediated ROS generation.
Glucose withdrawal-induced ROS oxidize and inhibit PTPs
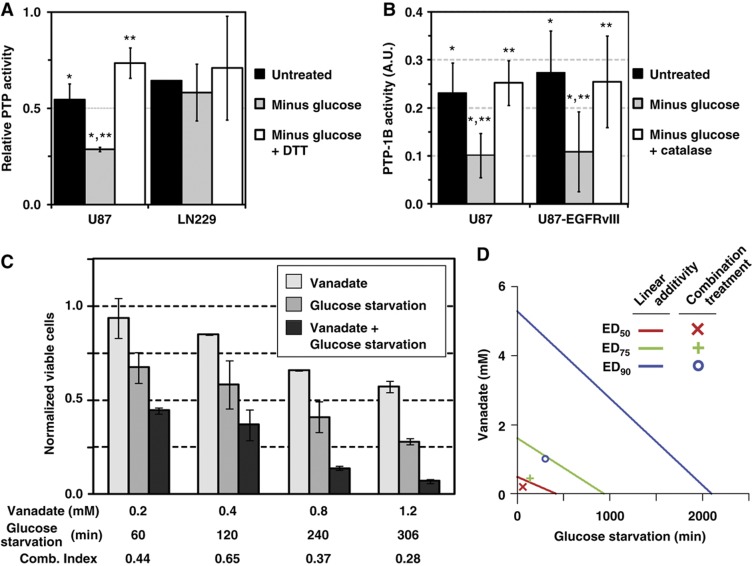
Because ROS are required for glucose withdrawal-induced phospho-tyrosine signaling and ROS can inhibit PTPs by oxidation of the catalytic cysteine residue (Lee et al, 1998a; Meng et al, 2002), we hypothesized that supra-physiological levels of phospho-tyrosine signaling following glucose withdrawal were mediated by oxidative inhibition of PTPs. We first measured total cellular PTP activity in U87 and LN229 cell lysates using a phospho-substrate dephosphorylation assay. Under standard culture conditions, lysates from both cell lines had detectable levels of PTP activity. Following glucose withdrawal, PTP activity of the glucose withdrawal-sensitive cell line U87 was reduced by roughly two-fold whereas the PTP activity of the insensitive cell line LN229 showed no change (Figure 7A; Supplementary Figure S12A). Confirming that PTP activity was inhibited by oxidation, the reducing agent DTT rescued the PTP activity of U87 cell lysates following glucose withdrawal.
Figure 7.
Glucose withdrawal-induced ROS mediate oxidative inhibition of protein tyrosine phosphatases. (A) Glucose withdrawal causes oxidative inhibition of PTP activity in glucose withdrawal-sensitive cells. U87 and LN229 cells were starved of glucose for 0 or 3 h, and the ability of cell lysates to dephosphorylate a phospho-substrate was measured by quantitative western blotting. Addition of DTT to the dephosphorylation reaction reduced oxidized PTPs. Data were normalized to a dephosphorylation reaction with the PTP inhibitor vanadate added (P=0.04 (*) and 0.01 (**) by Student's t-test (n=3 for U87). A representative blot is shown in Supplementary Figure S12A. (B) Glucose withdrawal inhibits PTP-1B activity by oxidation. U87 and U87-EGFRvIII cells were starved of glucose and pyruvate for 1.5 h with or without catalase. PTP-1B was then immunoprecipitated from cell lysates under anaerobic conditions and incubated with the colorimetric phosphatase substrate pNPP. PTP-1B activity was normalized to a no-antibody control and expressed in arbitrary units (AU). For both cell lines, glucose withdrawal reduced PTP-1B activity by two-fold and catalase treatment rescued PTP-1B activity from the effects of glucose withdrawal (P<0.01 (*) and 0.02 (**) by combined Fisher's method of paired Student's t-test for U87 and U87-EGFRvIII, n=4 for U87, n=8 for U87-EGFRvIII). (C) Glucose and pyruvate starvation and vanadate treatment synergistically kill U87 cells. U87 cells were exposed to increasing doses of vanadate for 1.5 h and then starved of glucose and pyruvate for the indicated times. The number of viable cells was measured by Trypan blue exclusion 24 h later. To assess the degree of synergy, the combination index (Comb. Index) was calculated using the method of Chou and Talalay (1984). Values less than one indicate positive synergy. Error bars are the standard deviation of the mean (n=2). (D) Isobologram plot of the effect of glucose and pyruvate starvation combined with vanadate treatment demonstrates positive synergy. The effective doses (ED) of glucose and pyruvate starvation (min) and vanadate treatment (mM) are plotted on the x and y axis, respectively. Lines of linear additivity connect the ED for ED50, ED75, and ED100 for individual treatments. Because the experimentally measured responses to combinations of glucose starvation and vanadate treatment (combination treatments) lie to the left of the linear lines of additivity, glucose starvation and vanadate treatment interact with positive synergy. Source data is available for this figure in the Supplementary Information.
To measure the effect of glucose withdrawal on a specific PTP, we focused on PTP-1B because (a) PTP-1B can be inhibited by oxidation of the catalytic cysteine (Lee et al, 1998a; Mahadev et al, 2001; Salmeen et al, 2003; Lou et al, 2008) and (b) our phospho-proteomic data revealed a 15-fold increase in phosphorylation of PTP-1B Y20 following glucose starvation (Supplementary Figure S12B). In both U87 and U87-EGFRvIII cells, glucose withdrawal reduced PTP-1B activity by roughly two-fold (Figure 7B). Confirming the functional significance of glucose withdrawal-induced ROS, treatment with the H2O2 scavenger catalase rescued PTP-1B activity. Further confirming PTP-1B inhibitory oxidation, treatment of immunoprecipitates with DTT also increased phosphatase activity (data not shown). Taken together, these results show that glucose withdrawal inhibits PTP activity through oxidation by ROS.
Having established that glucose withdrawal-generated ROS inhibit cellular PTP activity, we next tested whether chemical inhibition of PTPs can induce ROS generation. Using the general oxidation probe DCF-DA, we found that vanadate treatment induced oxidative stress in LN18, T98, and U87 GBM cell lines (Supplementary Figure S13). The observation of both PTP inhibition-induced ROS and ROS-mediated PTP inhibition suggested the existence of a positive feedback loop. We next sought to determine if initiating this positive feedback loop at two points would result in synergistic amplification. We measured the viability of U87 cells after exposure to different dose and time combinations of vanadate inhibition of PTPs and glucose withdrawal (Figure 7C). We found that combinations of vanadate treatment and glucose deprivation killed U87 cells more effectively than either treatment alone. Using the method of Chou and Talalay (1984), we calculated that co-treatment with glucose starvation and vanadate gave combination indices of less than one, indicative of positive synergy. Plotting the linear additive isoboles further supported that combinations of glucose starvation and vanadate exhibited positive synergy (Figure 7D). These data demonstrate that increased TK signaling can promote sensitivity to glucose withdrawal, supporting the existence of a positive feedback loop between glucose withdrawal-induced ROS generation, PTP inhibition by ROS-mediated oxidation, increased TK signaling, and further ROS generation.
Discussion
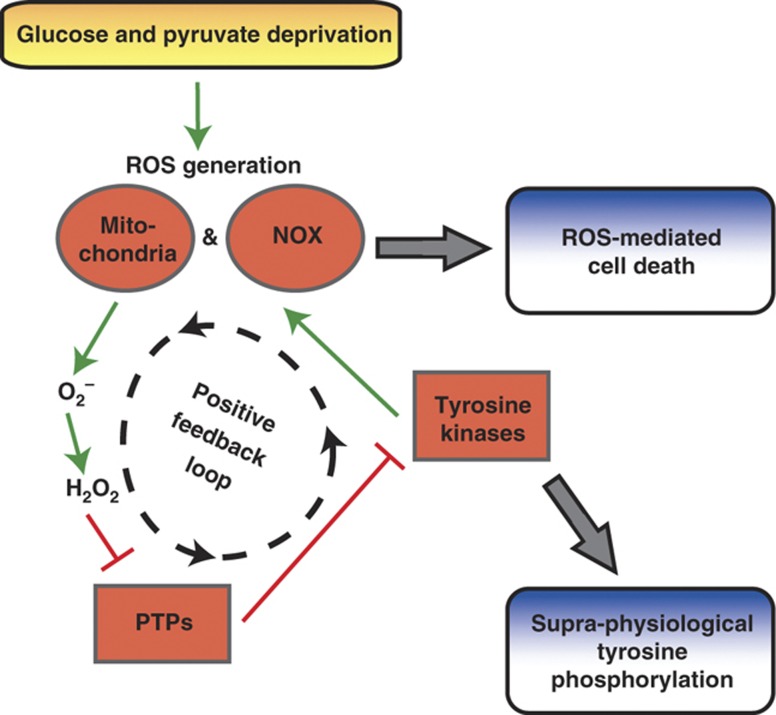
The reprogramming of cellular metabolism during oncogenesis has attracted considerable recent attention, even being named one of the ‘emerging’ hallmarks of cancer (Hanahan and Weinberg, 2011). The metabolic reprogramming that helps satisfy the voracious appetite of tumor cells for biosynthetic precursors can also render cells exquisitely sensitive to nutrient deprivation (e.g., glucose and glutamine; Elstrom et al, 2004; Yuneva et al, 2007; Aykin-Burns et al, 2009; Yang et al, 2009). Here, we extend our understanding of this phenomenon with the phospho-tyrosine proteomic-based discovery that glucose deprivation provokes a systems-level positive feedback loop between ROS generation, PTPs, and TK signaling in cells dependent on glucose for survival (Figure 8). Building on the unexpected observation that glucose withdrawal induces supra-physiological levels of phospho-tyrosine signaling, our systems-level feedback amplification loop model integrates the observations that (a) glucose withdrawal induces oxidative stress (Spitz et al, 2000; Aykin-Burns et al, 2009) and can activate diverse intracellular kinases including ERK, JNK, and Lyn (Lee et al, 1998b, 2000; Blackburn et al, 1999), (b) focal adhesions and RTKs serve as sites of NOX-mediated ROS generation (Lee et al, 1998a; Wu et al, 2005; Diaz et al, 2009), and (c) ROS can inhibit PTPs, inducing further TK signaling (Lee et al, 1998a; Mahadev et al, 2001; Meng et al, 2002). Taken together, this systems perspective reveals that glucose deprivation activates a positive feedback amplification loop, as indicated by supra-physiological levels of phospho-tyrosine signaling, until ROS accumulate above a toxicity threshold resulting in cell death.
Figure 8.
Glucose withdrawal activates a positive feedback loop resulting in supra-physiological phospho-tyrosine signaling and ROS-mediated cell death. In cells dependent on glucose for survival, glucose and pyruvate deprivation induces oxidative stress driven by NOX and mitochondria. This oxidative stress provokes a positive feedback loop in which NOX and mitochondria generate superoxide anion (O2−), which dismutes to hydrogen peroxide (H2O2) and inhibits PTPs by oxidation (e.g., PTP-1B and PTEN). Without the negative regulation of PTPs, TKs including EGFR and Src activate NOX at focal adhesions, further amplifying ROS generation. This glucose withdrawal-induced positive feedback loop results in supra-physiological levels of tyrosine phosphorylation and ROS-mediated cell death.
Notably, the systems-wide positive feedback loop described here also functions in localized, subcellular contexts. A similar feedback amplification loop involving signaling, ROS, and PTPs occurs physically proximal to RTKs where EGF (Lee et al, 1998a) and insulin (Mahadev et al, 2001) induce oxidative inhibition of PTP-1B to promote signaling. Related positive feedback loops also occur at T-cell receptor signaling complexes (Kwon et al, 2010) and focal complexes during endothelial cell migration (Wu et al, 2005) and the formation of invadopodia (Diaz et al, 2009). Here, we have demonstrated that a metabolic perturbation (i.e., glucose withdrawal) initiates a positive feedback amplification loop driven by NOX- and mitochondria-derived ROS generation resulting in cell-wide consequences on phospho-tyrosine signaling (Figure 3) and PTP activity (Figure 7), ultimately resulting in cell death. In spatially localized contexts such as invadopodia or T-cell receptor signaling complexes, positive feedback is quickly dampened by reduction and re-activation of oxidized PTPs. However, in the case of metabolic deficiency, cells are unable to maintain redox homeostasis, perhaps due to depletion of cellular pools of NADPH (Ahmad et al, 2005), which drives the amplification of ROS until a cellular toxicity threshold is breached and cells undergo ROS-mediated cell death. Thus, the ROS-PTP-TK positive feedback amplification loop set in motion by glucose withdrawal resembles positive feedback loops that are quickly dampened under normal, nutrient-rich conditions.
This work highlights the emerging concept of systems integration between oncogenic signaling networks and metabolism. For example, constitutively active oncogenic kinases, including myristoylated Akt and the activated EGFR mutant EGFRvIII, can promote aerobic glycolysis and lipogenesis, respectively (Elstrom et al, 2004; Guo et al, 2009). Conversely, glucose metabolism can influence signaling, such as the requirement of the hexosamine biosynthetic pathway for signaling through IL3-RA (Wellen et al, 2010). Here, our model demonstrates the bidirectional interactions between signaling and metabolism. Our model predicts that either reduced TK signaling or increased PTP activity at sites of ROS generation (e.g., NOX complexes and mitochondria) should protect cells from glucose withdrawal-induced cell death. However, our data and model suggest that inhibition of a single kinase or overexpression of a single PTP is likely insufficient to rescue cells from glucose withdrawal-induced cell death precisely because cell death is controlled by a systems-level positive feedback loop that simultaneously activates and inhibits multiple TKs and PTPs, respectively. Indeed, overexpression of PTEN, which functions as both a lipid and PTP (Myers et al, 1997) and can undergo oxidative inactivation by RTK-induced NOX1 activity (Kwon et al, 2004; Boivin et al, 2008), only partially dampened the positive feedback loop in U87 cells (Figure 2). The diminished activity of PTPs following glucose starvation (Figure 7) is also consistent with the observation by us and others that serine/threonine MAPK signaling is induced by glucose withdrawal (Figure 3B; Lee et al, 1998b, 2000; Blackburn et al, 1999), as dual-specificity phosphatases, which dephosphorylate MAPK enzymes, can be inhibited by oxidation of the catalytic cysteine residue (Kamata et al, 2005). Our observations thus add to the growing evidence supporting the systems-level integration of metabolism and signaling homeostasis.
The integrated nature of the ROS-PTP-TK positive feedback loop described here offers an opportunity for therapeutic intervention. Indeed, combinatorial activation of the ROS-PTP-TK positive feedback loop with glucose deprivation and PTP inhibition exhibited synergistic killing of U87 cells (Figure 7C and D). Because many types of cancer cells exhibit increased levels of ROS and weakened redox buffering compared with normal cells (Szatrowski and Nathan, 1991; Toyokuni et al, 1995), ROS-promoting small molecule drugs can be selectively toxic to cancer cells (Trachootham et al, 2006; Raj et al, 2011; Shaw et al, 2011), Similarly, therapeutics targeting the metabolic inflexibility of cancer are being pursued for selective toxicity to tumor cells (Yuneva, 2008; Simons et al, 2009). Our results highlight the possibility of unifying these two concepts through the judicious selection of therapeutic cocktails targeted against redox homeostasis and the metabolic inflexibility of cancer.
In support of this hypothesis, combinations of the glycolytic inhibitor 2-deoxyglucose and redox modulators (e.g., bulthione sulfoximine and antimycin A) demonstrate enhanced cytotoxicity compared with either agent alone (Andringa et al, 2006; Fath et al, 2009). Alternatively, it may be possible to alter the redox balance of tumor cells using physiological signals such as fasting, which can slow the growth of tumor xenografts (Kalaany and Sabatini, 2009) and enhance the efficacy of high-dose chemotherapy in mouse models (Lee et al, 2012). In light of our data demonstrating that glucose withdrawal-induced phospho-tyrosine signaling is driven by focal adhesions, it is interesting to note that normal epithelial cells exhibit loss of glucose transporters and oxidative stress following detachment from extracellular matrix (Schafer et al, 2009). Thus, the integrated nature of metabolism, redox homeostasis, and signaling may permit ‘synthetic lethal’ therapeutic approaches with selective toxicity toward tumor cells.
Finally, we speculate that this glucose withdrawal-initiated positive feedback loop may regulate the survival of metabolically altered tumor cells and/or select for metabolic phenotypes in nutrient-limited environments in vivo. Recently, it was demonstrated that low glucose media can select for colorectal cancer cells with mutated KRAS and increased GLUT1 expression (Yun et al, 2009). In a manner similar to the hypoxic selection of cells with increased resistance to apoptosis (Graeber et al, 1996), intermittent glucose deprivation could select against cells dependent on glucose for survival. Additionally, because hypoxia selects for cells with increased glycolysis (Kim et al, 2007), it may be possible that hypoxic and therefore glycolytic tumors are preferentially sensitive to metabolic inhibition.
In summary, cellular redox homeostasis is maintained by the balance between ROS generation by required metabolic functions and ROS elimination. Likewise, signaling homeostasis is controlled by balancing kinase and phosphatase activity. Here, we demonstrate that the cellular microenvironment (i.e., nutrient availability) can alter the cellular redox balance, provoking a signaling-based positive feedback loop that amplifies ROS levels above a toxicity threshold resulting in cell death. This positive feedback loop demonstrates the complex, systems-level integration of homeostatic control mechanisms for metabolism (e.g., redox balance) and TK signaling (e.g., PTPs). Furthermore, this systems integration offers a scaffold for synergistic combinations of therapeutics targeting signaling, metabolism, and redox homeostasis.
Materials and methods
Cell culture
All cell lines were cultured in high-glucose DMEM (4.5 g/l glucose, 110 mM pyruvate; Mediatech) supplemented with 10% (v/v) FBS (Omega Scientific) plus 1% (v/v) SPF (Invitrogen). For glucose starvation, cells were washed twice with PBS and then incubated in DMEM without glucose and pyruvate (0 g/l glucose, 0 mM pyruvate; Invitrogen) supplemented with 10% dialyzed FBS (Omega Scientific) plus 1% SPF. GBM cell lines LN18, LN229, T98, and U87 were purchased from American Tissue Culture Collection (ATCC). U87-EGFRvIII (Wang et al, 2006) and the melanoma cell lines M202, M207, M229, and M249 (Nazarian et al, 2010; Sondergaard et al, 2010) have been described previously. HT161 and TC32 sarcoma cell lines were generously donated by Christopher Denny (UCLA). U87-Pten wild-type, C124S, and G129E were constructed by amplification of murine Pten constructs (Liliental et al, 2000) and cloning into the retroviral backbone pDS-FB-hygro using Gateway technology (Invitrogen). Stable overexpression of wild-type Pten and mutants was achieved by retroviral transduction of U87 cells using retrovirus produced in 293T as described elsewhere (Rubbi et al, 2011) followed by selection with hygromycin. HCT116 PTEN knockout cells were derived and provided by T Waldman (Georgetown University). RWPE-1 and SF268 sublines with stable PTEN knockdown were generated by retroviral transduction using a PTEN-targeted human specific shRNA (pSIREN-RetroQ-PTEN; Vivanco et al, 2007). Viruses were generated by transfecting the hairpin construct into an amphotropic packaging cell line. Infected cells were selected with 3 μg/ml puromycin. To generate ρ0 cells lacking mitochondrial DNA, cells were cultured for 4–6 weeks in media supplemented with dialyzed serum, 50 ng/ml ethidium bromide, and 50 μg/ml uridine (Hashiguchi and Zhang-Akiyama, 2009). To confirm the loss of mitochondrial DNA, genomic and mitochondrial DNA was harvested using a DNeasy kit (Qiagen), and the presence of the mitochondrially encoded tRNA-Leu was assessed by PCR (forward primer, 5′-GATGGCAGAGCCCGGTAATCGC-3′; reverse primer, 5′-TAAGCATTAGGAATGCCATTGCG-3′). Presence of the nuclear genome was confirmed by PCR against a region of the X chromosome (forward primer, 5′-GAAGGTGAAGGTCGGAGTC-3′; reverse primer, 5′-GAAGATGGTGATGGGATTTC-3′). To functionally confirm the loss of the mitochondrial genome, ρ0 cells were tested for uridine auxotrophy (Hashiguchi and Zhang-Akiyama, 2009).
Antibodies and reagents
Antibodies used for western blotting and immunoprecipitation included: anti-phospho-tyrosine (clone 4G10) from Upstate; anti-phospho-ERK1/2, anti-phospho-JNK, anti-phospho-p38alpha, anti-phospho-S235/236-S6, anti-phospho-S473-Akt, anti-phospho-Y1045-EGFR, anti-phospho-Y1068-EGFR, anti-total EGFR, anti-phospho-Y1349 Met, anti-phospho-Y1234/Y1235-Met, anti-phospho-751-PDGFRβ, anti-phospho-Y416-SRC, and anti-phospho-Y397-FAK from Cell Signaling Technology; anti-PTEN from Cascade Biosciences; anti-total FAK from BD Biosciences; anti-total GRB2 and anti-total PTP-1B from Santa Cruz Biotechnology. Catalase and pNPP were purchased from Sigma. DPI, MnTMPyP, and glutathione monoethyl ester (GSH-MEE) were from Calbiochem. CM-H2-DCF-DA and mitoSOX were from Invitrogen.
Cell viability analysis
Viability was measured by Trypan blue exclusion using a Vi-Cell XR 2.03 viability analyzer (Becton-Dickinson) using optimized, cell type-specific imaging parameters to distinguish live and dead cells.
Western blotting
Cells were lysed in modified RIPA buffer (50 mM Tris–HCl (pH 7.5), 150 NaCl, 10 mM β-glycerophosphate, 1% NP-40, 0.25% sodium deoxycholate, 10 mM sodium pyrophosphate, 30 mM sodium fluoride, 1 mM EDTA, 1 mM vanadate, 20 μg/ml aprotinin, 20 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride). Whole cell lysates were resolved by SDS–PAGE on 4–15% gradient gels and blotted onto nitrocellulose membranes (Bio-Rad). Membranes were blocked overnight and then incubated sequentially with primary and either HRP-conjugated (Pierce) or IRDye-conjugated secondary antibodies (Li-Cor). Blots were imaged using either Amersham ECL Western Blotting Detection Reagents (GE Healthcare) or the Odyssey Infrared Imaging System (Li-Cor).
Flow cytometry
Cells were incubated with either 2.5 μM CM-H2DCFDA or mitoSOX (Invitrogen) for 30 min before analysis using a Becton Dickinson FACScan analytic flow cytometer in the UCLA Jonsson Comprehensive Cancer Center and Center for AIDS Research Flow Cytometry Core Facility. Cells were gated using forward scatter and side scatter to remove debris and dead cells, and 10 000 live cell events were recorded. To quantify changes in DCF-DA or mitoSOX signal, mean fluorescent intensity after gating was used.
Knockdown of p22phox subunit of NOX
siRNA constructs targeting p22phox (M-011020) and a non-targeting control siRNA (D-001206) were purchased from Thermo Scientific. Cells were reverse transfected with 30 pM siRNA. To assess knockdown efficiency, total RNA was extracted using the RNeasy kit (Qiagen) and reverse transcribed using the SuperScript VILO cDNA kit (Invitrogen). The levels of p22phox cDNA were quantified by qPCR and normalized to expression of GAPDH using the following primers: p22phox forward 5′-GGCGCTTCACCCAGTGGTACTTTGG-3′ and reverse 5′-TAGGTAGATGCCGCTCGCAATGGC-3′; GAPDH forward 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse 5′-GAAGATGGTGATGGGATTTC-3′.
Phospho-substrate dephosphorylation assay
Phospho-substrate was isolated via immunoprecipitation from U87-EGFRvIII cells treated with vanadate (1 mM) for 1 h. Cells were washed with degassed PBS, snap frozen in liquid nitrogen, introduced into an anaerobic chamber, lysed in dephosphorylation assay buffer (1% (v/v) Triton X-100, 50 mM Tris (pH 7.5), 250 mM NaCl, 3 mM EDTA, 20 μg/ml aprotinin, 20 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride) supplemented with ROS scavengers (250 U/ml catalase and 125 U/ml superoxide dismutase; Calbiochem), and phospho-substrate (i.e., phospho-EGFRvIII) was immunoprecipitated from using a total EGFR antibody. To test the effects of glucose starvation on PTP activity, U87 and LN229 cells were starved of glucose for 0 or 3 h and lysed in dephosphorylation assay buffer as above. Endogenous EGFR was immunodepleted from cell lysates, and the U87 or LN229 supernatant was incubated with phospho-substrate for 30 min at 37oC in the presence or absence of vanadate (1 mM) or DTT (2 mM). The remaining phospho-substrate was assessed by quantitative western blotting and normalized to total amount of substrate. Phosphatase activity was normalized to the vanadate control and expressed as the fraction of phosphorylation removed during the in vitro reaction.
Vanadate and glucose deprivation synergy assay
Cells were treated with increasing concentrations of vanadate for 90 min and then starved of glucose and pyruvate for 15–360 min. After starvation, DMEM containing glucose and pyruvate was added back to the cells. Twenty-four hours after the initial vanadate treatment, the number of viable cells was quantified by Trypan blue staining on a Vi-Cell XR 2.03 viability analyzer (Becton-Dickinson). To determine synergistic, additive, or antagonistic effects of vanadate and glucose deprivation, we used the combination index method of Chou and Talalay (1984) using CalcuSyn (Biosoft). This method takes into account both potency (median dose (Dm) or IC50) and the shape of the dose-effect curve (the m value) to calculate the combination index (CI). A CI >1 indicates an antagonistic effect; a CI <1 indicates positive synergy.
PTP-1B phosphatase activity assay
PTP-1B activity was measured using a protocol modified from Pani et al (2000). Following treatment, cells were washed once with degassed PBS and snap frozen in liquid nitrogen. Cells were introduced into an anaerobic hood supplemented with nitrogen and lysed in degassed Triton-only lysis buffer without PTP inhibitors (1% (v/v) Triton X-100, 50 mM Tris (pH 7.5), 150 mM NaCl, 10 mM sodium pyrophosphate, 30 mM sodium fluoride, 2 mM EDTA, 20 μg/ml aprotinin, 20 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride) supplemented with ROS scavengers (250 U/ml catalase and 125 U/ml superoxide dismutase; Calbiochem). Equal amounts of total protein were then immunoprecipitated with PTP-1B antibody under anaerobic conditions at 4oC. Immunoprecipitates were incubated with 10 mM p-nitrophenyl phosphate (Sigma) in HEPES buffer (62 mM HEPES (pH 7), 6.25 mM EDTA) for 2 h at 37oC, quenched with NaOH, and absorbance at 405 nm was measured. Data were normalized to a control without PTP-1B antibody.
Quantitative, label-free phospho-tyrosine peptide mass spectrometry
U87 and U87-EGFRvIII cells were grown, processed, and analyzed by MS independently. Phospho-tyrosine peptide immunoprecipitation was performed with a pan-specific anti-phospho-tyrosine antibody (clone 4G10; Millipore) using 2 × 108 cells (30 mg of total protein) as previously described (Skaggs et al, 2006; Rubbi et al, 2011). Direct quantitative comparison of our label-free approach with a label-based approach (SILAC) showed high concordance in quantitation and standard error (r=0.95; Supplementary Figure S14 of Rubbi et al, 2011). Phosphorylated peptides were analyzed by LC-MS/MS with an Eksigent autosampler coupled with a Nano2DLC pump (Eksigent) and LTQ-Orbitrap (Thermo Fisher Scientific). The samples were loaded onto an analytical column (10 cm, 75 mm inside diameter) packed with 5 mm Integrafit Proteopep2 300 Å C18 (New Objective). Peptides were eluted into the mass spectrometer with a high-performance liquid chromatography (HPLC) gradient of 5–40% buffer B in 45 min followed by a gradient of 40–90% buffer B in 10 min, where buffer A contained 0.1% formic acid in water and buffer B contained 0.1% formic acid in acetonitrile. All HPLC solvents were Ultima Gold quality (Fisher Scientific). Mass spectra were collected in positive ion mode with the Orbitrap for parent mass determination and with the LTQ for data-dependent MS/MS acquisition of the top five most abundant peptides. Each sample was analyzed twice (technical duplicate runs). MS/MS fragmentation spectra were searched with SEQUEST (Version v.27, rev. 12; Thermo Fisher Scientific) against a database containing the combined human-mouse International Protein Index (IPI) protein database (downloaded December 2006 from ftp.ebi.ac.uk). Search parameters included carbamidomethyl cysteine (*C) as a static modification and phosphorylated tyrosine and oxidized methionine (*M) as dynamic modifications. Results derived from database searching were filtered using the following criteria: Xcorr >1.0(+1), 1.5(+2), 2(+3); peptide probability score <0.001; ΔCn >0.1; and mass accuracy 25 p.p.m. with Bioworks version 3.2 (Thermo Electron Corp.). Mass spectra have been deposited in the PRIDE database (http://www.ebi.ac.uk/pride/, accession number 19835–19854).
We estimate the false-positive rate of sequence assignments at 0.5% on the basis of a composite target-reversed decoy database search strategy. Ascore was used to more accurately localize the phosphate on the peptide (Beausoleil et al, 2006). As is common in data-dependent MS2 fragmentation sequencing, some peptides identified by sequencing in one sample may not be sequenced or identified in another sample even if the peak is present. Peptide peaks sequenced in some samples but not in others were located in the remaining samples by aligning the chromatogram elution profiles by means of a dynamic time warping algorithm (Prakash et al, 2006). An extended explanation of the strategy used in this work, and example performance results, can be found in the supporting information of Zimman et al (2010) and Rubbi et al (2011). Relative amounts of the same phospho-peptide across samples run together were determined with custom software from our laboratory to integrate the area under the unfragmented (MS1) monoisotopic peptide peak. All peaks corresponding to phospho-sites discussed in the text were inspected manually, and any errors in the automated quantitation were corrected. Quantitative phospho-peptide values can be found in Supplementary Tables S1 and S2.
The number of unique phosphorylation sites identified in our experiments was determined by collapsing multiple phospho-peptide ions representing the same phosphorylation site. Multiple detections of the same phospho-site include phospho-peptides of different ion charge state, modification (e.g., oxidized methionine), and miscleavage by trypsin. Multiple detections were compared with ensure no disagreement in trend, and the MS ion with the highest intensity across the samples was kept as representative for subsequent data analysis. The residue numbers listed for phospho-sites correspond to the indicated IPI accession number. Peptide quantities were unit normalized and hierarchically clustered using the Pearson correlation in Cluster 3.0 (Eisen et al, 1998).
Enrichment analysis for functional signaling classes within phospho-tyrosine signatures
Phospho-tyrosine peptides were ranked by the log2-transformed fold change observed for a given perturbation (e.g., glucose withdrawal compared with untreated U87 cells). Peptides from proteins belonging to a specified gene ontology term (e.g., Focal Adhesion GO:0005925) were annotated using AMIGO (http://amigo.geneontology.org), and we then calculated a Kolmogorov-Smirnov statistic against the expected distribution. The statistical significance of enrichment was then determined by permutation analysis. Briefly, permutation analysis was performed by randomly shuffling the peptide ranked list, followed by calculation of the Kolmogorov-Smirnov statistic for this permutation. After 1000 permutations, the fraction of randomly ranked lists resulting in a Kolmogorov-Smirnov statistic greater than or equal to the observed value was defined as the permutation-based frequency of random occurrence (i.e., the permutation-based P-value).
Supplementary Material
Supplementary Figures S1–13
Acknowledgments
NAG is a postdoctoral trainee supported by the UCLA Scholars in Oncologic Molecular Imaging (SOMI) program, NIH grant R25T CA098010 and the UCLA Tumor Biology Program United States Health and Human Services Ruth L. Kirschstein Institutional National Research Service Award T32 CA009056. TGG received support from the Jonsson Comprehensive Cancer Center (JCCC) and The California Institute of Technology–University of California, Los Angeles Joint Center for Translational Medicine (JCTM). The UCLA JCCC and Center for AIDS Research Flow Cytometry Core Facility is supported by National Institutes of Health awards CA-16042 and AI-28697, and by the JCCC, the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA. We thank Christopher Denny (UCLA) for generously donating the HT161 and TC32 sarcoma cell lines. We also thank Björn Titz (UCLA), Harvey Herschman (UCLA), and Heather Christofk (UCLA) for helpful discussions and critical reading of the manuscript. We thank Mohammad Atefi (UCLA) for assistance with CalcuSyn.
Author contributions: NG and TG conceived the project and designed the experiments; NG, MT, BK, MZ, and IV performed the experiments; EK and TG wrote the analytical pipeline for label-free phospho-peptide quantitation; NG and TG performed statistical analysis and data interpretation; HW, MAT, AR, RL, IM, and PM provided reagents and interpreted the data; NG and TG wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahmad IM, Aykin-Burns N, Sim JE, Walsh SA, Higashikubo R, Buettner GR, Venkataraman S, Mackey MA, Flanagan SW, Oberley LW, Spitz DR (2005) Mitochondrial O2*- and H2O2 mediate glucose deprivation-induced stress in human cancer cells. J Biol Chem 280: 4254–4263 [DOI] [PubMed] [Google Scholar]
- Andringa KK, Coleman MC, Aykin-Burns N, Hitchler MJ, Walsh SA, Domann FE, Spitz DR (2006) Inhibition of glutamate cysteine ligase activity sensitizes human breast cancer cells to the toxicity of 2-deoxy-D-glucose. Cancer Res 66: 1605–1610 [DOI] [PubMed] [Google Scholar]
- Aykin-Burns N, Ahmad IM, Zhu Y, Oberley LW, Spitz DR (2009) Increased levels of superoxide and H2O2 mediate the differential susceptibility of cancer cells versus normal cells to glucose deprivation. Biochem J 418: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol 24: 1285–1292 [DOI] [PubMed] [Google Scholar]
- Beckman JS, Minor RL Jr., White CW, Repine JE, Rosen GM, Freeman BA (1988) Superoxide dismutase and catalase conjugated to polyethylene glycol increases endothelial enzyme activity and oxidant resistance. J Biol Chem 263: 6884–6892 [PubMed] [Google Scholar]
- Blackburn RV, Spitz DR, Liu X, Galoforo SS, Sim JE, Ridnour LA, Chen JC, Davis BH, Corry PM, Lee YJ (1999) Metabolic oxidative stress activates signal transduction and gene expression during glucose deprivation in human tumor cells. Free Radic Biol Med 26: 419–430 [DOI] [PubMed] [Google Scholar]
- Boivin B, Zhang S, Arbiser JL, Zhang ZY, Tonks NK (2008) A modified cysteinyl-labeling assay reveals reversible oxidation of protein tyrosine phosphatases in angiomyolipoma cells. Proc Natl Acad Sci USA 105: 9959–9964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzai M, Bauer DE, Jones RG, Deberardinis RJ, Hatzivassiliou G, Elstrom RL, Thompson CB (2005) The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid beta-oxidation. Oncogene 24: 4165–4173 [DOI] [PubMed] [Google Scholar]
- Choo AY, Kim SG, Vander Heiden MG, Mahoney SJ, Vu H, Yoon SO, Cantley LC, Blenis J (2010) Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol Cell 38: 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC, Talalay P (1984) Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 22: 27–55 [DOI] [PubMed] [Google Scholar]
- Czernin J, Phelps ME (2002) Positron emission tomography scanning: current and future applications. Annu Rev Med 53: 89–112 [DOI] [PubMed] [Google Scholar]
- Deberardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB (2008) The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7: 11–20 [DOI] [PubMed] [Google Scholar]
- Diaz B, Shani G, Pass I, Anderson D, Quintavalle M, Courtneidge SA (2009) Tks5-dependent, nox-mediated generation of reactive oxygen species is necessary for invadopodia formation. Sci Signal 2: ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, Thompson CB (2004) Akt stimulates aerobic glycolysis in cancer cells. Cancer Res 64: 3892–3899 [DOI] [PubMed] [Google Scholar]
- Fath MA, Diers AR, Aykin-Burns N, Simons AL, Hua L, Spitz DR (2009) Mitochondrial electron transport chain blockers enhance 2-deoxy-D-glucose induced oxidative stress and cell killing in human colon carcinoma cells. Cancer Biol Ther 8: 1228–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, Giaccia AJ (1996) Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature 379: 88–91 [DOI] [PubMed] [Google Scholar]
- Guo D, Hildebrandt IJ, Prins RM, Soto H, Mazzotta MM, Dang J, Czernin J, Shyy JY, Watson AD, Phelps M, Radu CG, Cloughesy TF, Mischel PS (2009) The AMPK agonist AICAR inhibits the growth of EGFRvIII-expressing glioblastomas by inhibiting lipogenesis. Proc Natl Acad Sci USA 106: 12932–12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Hashiguchi K, Zhang-Akiyama QM (2009) Establishment of human cell lines lacking mitochondrial DNA. Methods Mol Biol 554: 383–391 [DOI] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, Xie J, Gu TL, Polakiewicz RD, Roesel JL, Boggon TJ, Khuri FR, Gilliland DG, Cantley LC, Kaufman J, Chen J (2009) Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal 2: ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaany NY, Sabatini DM (2009) Tumours with PI3K activation are resistant to dietary restriction. Nature 458: 725–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata H, Honda S, Maeda S, Chang L, Hirata H, Karin M (2005) Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell 120: 649–661 [DOI] [PubMed] [Google Scholar]
- Kawahara T, Ritsick D, Cheng G, Lambeth JD (2005) Point mutations in the proline-rich region of p22phox are dominant inhibitors of Nox1- and Nox2-dependent reactive oxygen generation. J Biol Chem 280: 31859–31869 [DOI] [PubMed] [Google Scholar]
- Kim JW, Gao P, Dang CV (2007) Effects of hypoxia on tumor metabolism. Cancer Metastasis Rev 26: 291–298 [DOI] [PubMed] [Google Scholar]
- Kwon J, Lee SR, Yang KS, Ahn Y, Kim YJ, Stadtman ER, Rhee SG (2004) Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc Natl Acad Sci USA 101: 16419–16424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J, Shatynski KE, Chen H, Morand S, de Deken X, Miot F, Leto TL, Williams MS (2010) The nonphagocytic NADPH oxidase Duox1 mediates a positive feedback loop during T cell receptor signaling. Sci Signal 3: ra59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Raffaghello L, Brandhorst S, Safdie FM, Bianchi G, Martin-Montalvo A, Pistoia V, Wei M, Hwang S, Merlino A, Emionite L, de Cabo R, Longo VD (2012) Fasting cycles retard growth of tumors and sensitize a range of cancer cell types to chemotherapy. Sci Transl Med 4: 124ra127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Kwon KS, Kim SR, Rhee SG (1998a) Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J Biol Chem 273: 15366–15372 [DOI] [PubMed] [Google Scholar]
- Lee YJ, Galoforo SS, Berns CM, Chen JC, Davis BH, Sim JE, Corry PM, Spitz DR (1998b) Glucose deprivation-induced cytotoxicity and alterations in mitogen-activated protein kinase activation are mediated by oxidative stress in multidrug-resistant human breast carcinoma cells. J Biol Chem 273: 5294–5299 [DOI] [PubMed] [Google Scholar]
- Lee YJ, Galoforo SS, Sim JE, Ridnour LA, Choi J, Forman HJ, Corry PM, Spitz DR (2000) Dominant-negative Jun N-terminal protein kinase (JNK-1) inhibits metabolic oxidative stress during glucose deprivation in a human breast carcinoma cell line. Free Radic Biol Med 28: 575–584 [DOI] [PubMed] [Google Scholar]
- Liliental J, Moon SY, Lesche R, Mamillapalli R, Li D, Zheng Y, Sun H, Wu H (2000) Genetic deletion of the Pten tumor suppressor gene promotes cell motility by activation of Rac1 and Cdc42 GTPases. Curr Biol 10: 401–404 [DOI] [PubMed] [Google Scholar]
- Lou YW, Chen YY, Hsu SF, Chen RK, Lee CL, Khoo KH, Tonks NK, Meng TC (2008) Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. FEBS J 275: 69–88 [DOI] [PubMed] [Google Scholar]
- Mahadev K, Zilbering A, Zhu L, Goldstein BJ (2001) Insulin-stimulated hydrogen peroxide reversibly inhibits protein-tyrosine phosphatase 1b in vivo and enhances the early insulin action cascade. J Biol Chem 276: 21938–21942 [DOI] [PubMed] [Google Scholar]
- Mbaya E, Oules B, Caspersen C, Tacine R, Massinet H, Pennuto M, Chretien D, Munnich A, Rotig A, Rizzuto R, Rutter GA, Paterlini-Brechot P, Chami M (2010) Calcium signalling-dependent mitochondrial dysfunction and bioenergetics regulation in respiratory chain Complex II deficiency. Cell Death Differ 17: 1855–1866 [DOI] [PubMed] [Google Scholar]
- Meng TC, Fukada T, Tonks NK (2002) Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol Cell 9: 387–399 [DOI] [PubMed] [Google Scholar]
- Myers MP, Stolarov JP, Eng C, Li J, Wang SI, Wigler MH, Parsons R, Tonks NK (1997) P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc Natl Acad Sci USA 94: 9052–9057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R, Shi HB, Wang Q, Kong XJ, Koya RC, Lee H, Chen ZG, Lee MK, Attar N, Sazegar H, Chodon T, Nelson SF, McArthur G, Sosman JA, Ribas A, Lo RS (2010) Melanomas acquire resistance toB-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468: 973–U377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani G, Colavitti R, Bedogni B, Anzevino R, Borrello S, Galeotti T (2000) A redox signaling mechanism for density-dependent inhibition of cell growth. J Biol Chem 275: 38891–38899 [DOI] [PubMed] [Google Scholar]
- Prakash A, Mallick P, Whiteaker J, Zhang HD, Paulovich A, Flory M, Lee H, Aebersold R, Schwikowski B (2006) Signal maps for mass spectrometry-based comparative proteomics. Mol Cell Proteomics 5: 423–432 [DOI] [PubMed] [Google Scholar]
- Raj L, Ide T, Gurkar AU, Foley M, Schenone M, Li X, Tolliday NJ, Golub TR, Carr SA, Shamji AF, Stern AM, Mandinova A, Schreiber SL, Lee SW (2011) Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature 475: 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rubbi L, Titz B, Brown L, Galvan E, Komisopoulou E, Chen SS, Low T, Tahmasian M, Skaggs B, Muschen M, Pellegrini M, Graeber TG (2011) Global phosphoproteomics reveals crosstalk between Bcr-Abl and negative feedback mechanisms controlling Src signaling. Sci Signal 4: ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D (2003) Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 423: 769–773 [DOI] [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS (2009) Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Winslow MM, Magendantz M, Ouyang C, Dowdle J, Subramanian A, Lewis TA, Maglathin RL, Tolliday N, Jacks T (2011) Selective killing of K-ras mutant cancer cells by small molecule inducers of oxidative stress. Proc Natl Acad Sci USA 108: 8773–8778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons AL, Mattson DM, Dornfeld K, Spitz DR (2009) Glucose deprivation-induced metabolic oxidative stress and cancer therapy. J Cancer Res Ther 5Suppl 1: S2–S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaggs BJ, Gorre ME, Ryvkin A, Burgess MR, Xie Y, Han Y, Komisopoulou E, Brown LM, Loo JA, Landaw EM, Sawyers CL, Graeber TG (2006) Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR-ABL mutants. Proc Natl Acad Sci USA 103: 19466–19471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondergaard JN, Nazarian R, Wang Q, Guo D, Hsueh T, Mok S, Sazegar H, MacConaill LE, Barretina JG, Kehoe SM, Attar N, von Euw E, Zuckerman JE, Chmielowski B, Comin-Anduix B, Koya RC, Mischel PS, Lo RS, Ribas A (2010) Differential sensitivity of melanoma cell lines with BRAFV600E mutation to the specific Raf inhibitor PLX4032. J Transl Med 8: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz DR, Sim JE, Ridnour LA, Galoforo SS, Lee YJ (2000) Glucose deprivation-induced oxidative stress in human tumor cells. A fundamental defect in metabolism? Ann NY Acad Sci 899: 349–362 [DOI] [PubMed] [Google Scholar]
- Szatrowski TP, Nathan CF (1991) Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res 51: 794–798 [PubMed] [Google Scholar]
- Toyokuni S, Okamoto K, Yodoi J, Hiai H (1995) Persistent oxidative stress in cancer. FEBS Lett 358: 1–3 [DOI] [PubMed] [Google Scholar]
- Trachootham D, Zhou Y, Zhang H, Demizu Y, Chen Z, Pelicano H, Chiao PJ, Achanta G, Arlinghaus RB, Liu J, Huang P (2006) Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell 10: 241–252 [DOI] [PubMed] [Google Scholar]
- Ushio-Fukai M (2006) Localizing NADPH oxidase-derived ROS. Sci STKE 2006: re8. [DOI] [PubMed] [Google Scholar]
- Vivanco I, Palaskas N, Tran C, Finn SP, Getz G, Kennedy NJ, Jiao J, Rose J, Xie W, Loda M, Golub T, Mellinghoff IK, Davis RJ, Wu H, Sawyers CL (2007) Identification of the JNK signaling pathway as a functional target of the tumor suppressor PTEN. Cancer Cell 11: 555–569 [DOI] [PubMed] [Google Scholar]
- Wang MY, Lu KV, Zhu S, Dia EQ, Vivanco I, Shackleford GM, Cavenee WK, Mellinghoff IK, Cloughesy TF, Sawyers CL, Mischel PS (2006) Mammalian target of rapamycin inhibition promotes response to epidermal growth factor receptor kinase inhibitors in PTEN-deficient and PTEN-intact glioblastoma cells. Cancer Res 66: 7864–7869 [DOI] [PubMed] [Google Scholar]
- Warburg O (1956) On respiratory impairment in cancer cells. Science 124: 269–270 [PubMed] [Google Scholar]
- Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, Rabinowitz JD, Coller HA, Thompson CB (2010) The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev 24: 2784–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RF, Xu YC, Ma Z, Nwariaku FE, Sarosi GA Jr., Terada LS (2005) Subcellular targeting of oxidants during endothelial cell migration. J Cell Biol 171: 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Sudderth J, Dang T, Bachoo RG, McDonald JG, DeBerardinis RJ (2009) Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res 69: 7986–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Wilson JK, Markowitz S, Zhou S, Diaz LA Jr., Velculescu V, Lengauer C, Kinzler KW, Vogelstein B, Papadopoulos N (2009) Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science 325: 1555–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M (2008) Finding an ‘Achilles' heel’ of cancer: the role of glucose and glutamine metabolism in the survival of transformed cells. Cell Cycle 7: 2083–2089 [DOI] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y (2007) Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol 178: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimman A, Chen SS, Komisopoulou E, Titz B, Martinez-Pinna R, Kafi A, Berliner JA, Graeber TG (2010) Activation of aortic endothelial cells by oxidized phospholipids: a phosphoproteomic analysis. J Proteome Res 9: 2812–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–13