Abstract
The phenotype of an organism is determined by its genotype and environment. An interaction between these two arises from the differential effect of the environment on gene expression in distinct genotypes; however, the genomic properties identifying these are not well understood. Here we analyze the transcriptomes of five C. elegans strains (genotype) cultivated in five growth conditions (environment), and find that highly regulated genes, as distinguished by intergenic lengths, motif concentration, and expression levels, are particularly biased toward genotype–environment interactions. Sequencing these strains, we find that genes with expression variation across genotypes are enriched for promoter single-nucleotide polymorphisms (SNPs), as expected. However, genes with genotype–environment interactions do not significantly differ from background in terms of their promoter SNPs. Collectively, these results indicate that the highly regulated nature of particular genes predispose them for exhibiting genotype–environment interaction as a consequence of changes to upstream regulators. This observation may provide a deeper understanding into the origin of the extraordinary gene expression diversity present in even closely related species.
Keywords: C. elegans , genome regulation, genomic sequencing, genotype-environment interaction, transcriptomics
Introduction
A genotype–environment interaction occurs when the effect of a genetic locus is different in magnitude or direction across environments (Mackay et al, 2009). For example, consider a gene induced by temperature in one geographical isolate but uniformly expressed regardless of temperature in another isolate (Figure 1A). Intuitively, the interaction arises as the environmental expression profile across genotypes is not different by a global factor, but rather different for particular environments. While genomic sequences are now readily available, and transcriptomic expression data are available across many different strains, species, and environmental perturbations (Gasch et al, 2000; Rifkin et al, 2003; Yanai et al, 2004; Tirosh et al, 2006; Tirosh et al, 2011), predicting the effect of specific mutations on gene expression profiles presents a formidable problem. An even bigger systems biology challenge is to predict the effect of a mutation for different environmental conditions, thereby predicting genotype–environment interactions at the level of gene expression.
Figure 1.
A systematic examination of gene expression variation across genotypes and environments. (A) The measured gene expression levels across the five genotypes and five environments are shown for the scrm-4 gene. For each pairing, the colors in the periphery and center of the circle indicate the triplicate data and mean, respectively. Note the genotype–environment interaction. (B) Expression profiles for 50 other genes are shown in the same format. (C) Venn diagram indicating the number of genes with significant variation across genotypes (genotypic), environments (environmental), as well as genotype–environmental interactions (non-additive variation). These sets were delineated using two-way ANOVA with a threshold for significance established by randomization experiments (Supplementary Figure S5).
Genotype–environment interactions have been identified in a handful of cases for single- and multi-cellular organisms, and across both strains and species (Wittkopp et al, 2004; Li et al, 2006; Smith and Kruglyak, 2008; Tirosh et al, 2009; Gerke et al, 2010; Tirosh et al, 2011). In particular, evidence suggests that much of the observed gene expression variation within a species is due to changes at distant genomic positions (trans changes; Wittkopp et al, 2004; Li et al, 2006; Smith and Kruglyak, 2008; Wittkopp et al, 2008; Tirosh et al, 2009). Furthermore, in yeast, genes with high expression plasticity tend to have a TATA box in their promoter (Tirosh et al, 2006) and also a nucleosome occluded upstream region (Tirosh and Barkai, 2008). It is not well understood, however, how such trans effects targeting particular genes contribute to genotype–environment interactions. Here we investigate in detail the genomic properties of genes exhibiting genotype–environment interactions.
Results
To study genotype–environment interactions at a genomic level, mRNA was collected from C. elegans embryos extracted from animals of five distinct geographical isolates (genotypes) examined in five conditions (environments) and subjected to microarray analysis. Each of the 25 genotype–environment combinations was assayed by a pool of 50 embryos collected individually at the four-cell stage, in triplicates. The four-cell stage is easy to identify morphologically and allows query of the composition of the large maternal mRNA dowry deposited in the embryo with low variability, therefore providing high sensitivity to detecting differences (Baugh et al, 2003). The resulting data set exhibited expected distributions of expression levels, high reproducibility across replicates, linear expression values of spiked-in transcripts, and congruence with a previous data set (Supplementary Figures S1–4).
To systematically identify genes showing genotype–environment interactions, we invoked two-way ANOVA to compute the statistical significance of the variance across genotypes, environments, and their interaction. For example, the two-way ANOVA P-values for the scrm-4 gene were 10−300 (across genotypes), 10−6 (across environments), and 10−3 (genotype–environment interaction), indicating a high significance for the observed changes across all three factors (Figure 1A). Figure 1B shows the expression of other genes exhibiting different patterns of variation. We filtered the data set to score only those genes with a range of expression within the linear dynamic range of the microarray (2 to 5 log10 units, see Supplementary Figure S3) and a minimum level of variation (0.5 log10 units, see Supplementary Table S1 for robustness to this parameter). This filter reduced the set to 4083 genes, of which 787 and 767 show significant variation across genotypes (but not environments) and environments (but not genotypes), respectively (Figure 1C), and henceforth refer to these as genotypic and environmental genes. Consistent with previous work in yeast (Tirosh et al, 2006), we found that the set of genes that vary across genotypes and the set of genes that vary across environments significantly overlap (P<10−200, hypergeometric distribution). Similarly, we used two-way ANOVA to define 198 genes with genotype–environment interactions (Figure 1C and Supplementary Figure S6) and proceeded to query their defining properties.
We first asked whether intergenic lengths might vary across sets of genes with particular expression patterns, as the intergenic distance upstream of a gene’s coding region is a proxy for the length of the promoter (Davidson, 2006). Thus, longer intergenic regions generally reflect a higher complexity in regulation (Shen-Orr et al, 2010). Constitutively expressed genes—defined as those with high expression without significant genotypic or environmental variation (Figure 1B)—have significantly shorter intergenic regions (Figure 2A), consistent with their potentially simple requirements for regulation (Grishkevich et al, 2011; P<10−122, Kolmogorov–Smirnov test, henceforth KS test). Genes showing environmental changes do not have a different intergenic lengths distribution than the background, while genotypic genes have slightly longer intergenic regions (P<10−11, KS test). This result indicates that an extensive promoter region may be a liability in terms of an inherent bias for producing aberrant expression patterns. Strikingly, interaction genes have intergenic regions that are significantly longer, suggesting complex regulation upon these genes (P<10−7, KS test). Consistently, we found a higher motif concentration in the 1-kb promoter region immediately 5′ of the coding region of interaction genes relative to that of all genes (Figure 2B, P<0.039, KS test). The properties of intergenic length and motif concentration are significantly correlated (P<10−16, correlation coefficient, Supplementary Table S2) providing evidence for the notion that longer intergenic lengths indeed reflect increased regulation. C. elegans chromosomal ends are known to be gene poor (Barnes et al, 1995), but interaction genes showed no bias toward chromosome ends, so genomic location cannot explain the longer intergenic lengths (Supplementary Table S3). These results implicate the interaction genes as a class of highly regulated genes in which the promoter sequence is longer and includes more motifs. Examining other genomic properties, we further found that interaction genes are also enriched in their nucleosome occupancy at the promoter region consistent with our observation of their high expression variability (Supplemenatry Figure S8; Tirosh and Barkai, 2008).
Figure 2.
Genes with genotype–environment interactions show the hallmarks of highly regulated genes. Distribution of (A) intergenic lengths, (B) motif concentration, and (C) expression levels for the indicated gene categories. Expression levels were defined according to the median across genotypes and environments. The plots indicate the normalized frequencies of the measurements across each gene set. (D) SNP analysis. For each gene set, the fraction of genes with at least one independent SNP across the strains is indicated for each 100 nucleotide promoter bin.
To further query the properties of the interaction genes, we examined expression levels. Constitutively expressed genes were highly expressed (by their definition as highly and steadily expressed), while the genotypic and environmental genes had generally low expression (Figure 2C). By contrast, interaction genes occupied an intermediate position along this scale, expressed significantly higher than the environmental and genotypic genes (P<10−19, KS test). This predisposition toward higher expression provides additional support for the notion that interaction genes are under distinct regulation relative to the other gene classes. As intergenic distance and basal expression levels may be thought of as proxies for highly regulated genes, we asked whether such a class of genes is enriched for genes with genotype–environment interactions. We defined a set of presumably highly regulated genes as those with long intergenic distance (>5 kb) and a mid-range of expression (>2.5 and <3.5 log10 units); these two properties are only weakly correlated (Supplementary Table S2). This set of 477 genes is enriched for genotype–environment interactions (P<0.007, hypergeometric distribution CDF), while lowly expressed genes (<2.5 log10 units) are depleted in interactions (P<0.02, hypergeometric distribution CDF). These trends are supported by the complete pattern of enrichments for interactions along the dimensions of intergenic distance and expression level as shown in Supplementary Figure S9 (see also Supplementary Figure S7).
Changes in expression in the interaction genes may be due to local changes to the promoter (cis) or to changes to either the regulators or remote regulatory regions (trans). To distinguish between these we attempted to map the genomic changes that correlate with expression differences. We sequenced the four non-Bristol (N2) strains (see Supplementary information) and mapped single-nucleotide polymorphisms (SNPs) across the strains to the motif-rich 1-kb promoter region upstream of the start of translation of all genes. We first examined the number of promoter SNPs found in the constitutively expressed genes. These show a paucity of SNPs relative to all genes suggesting strong selection on maintaining the coherence of the promoter region (P<10−11, KS test relative to background, Figure 2D). Interestingly, the genotypic genes showed a higher SNP density, suggesting that a significant fraction of the changes in these genes are caused by local (cis) changes as opposed to changes to other factors that impinge upon its expression (P<10−13, KS test). However, the genes showing genotype–environment interactions (interaction genes) were not significantly distinguished in their SNP content (P=0.93, KS test relative to all genes), suggesting that their expression changes are predominantly caused by trans effects.
If trans effects dominate genotype–environment interactions, our set of interaction genes are expected to be enriched for particular functions reflecting a coordinated change due to a common source. To test for this, we screened through sets of functionally related genes using Gene Ontology, Pfam, and Wormbase Expression Clusters, and queried for enrichment in similarity among the gene expression in our data set. We found 16 gene sets with an enrichment for genotype–environment interactions (P<0.01, Supplementary Table S4, hypergeometric distribution). One such gene set comprises the potential targets of the deps-1 gene, initially defined by the upregulation after deps-1 loss of function (Spike et al, 2008). Of these potential targets, scrm-4 was shown in Figure 1A with elevated expression in heat and Hawaiian, and 10 other genes from this set are shown in Figure 1B. These show striking interactions as also evidenced by the significant ANOVA interaction P-values associated with this gene set (Supplementary Figure S10). Interestingly, the deps-1 gene itself does not show expression variation across the strains in our data set, suggesting that the difference in expression across strains may be post-transcriptional, or in a different co-regulator of these targets. The causal changes may also have occurred specifically in each of the targets, but this is unlikely as the promoters of deps-1 targets do not show enrichment in SNPs relative to background (P<0.96, KS test).
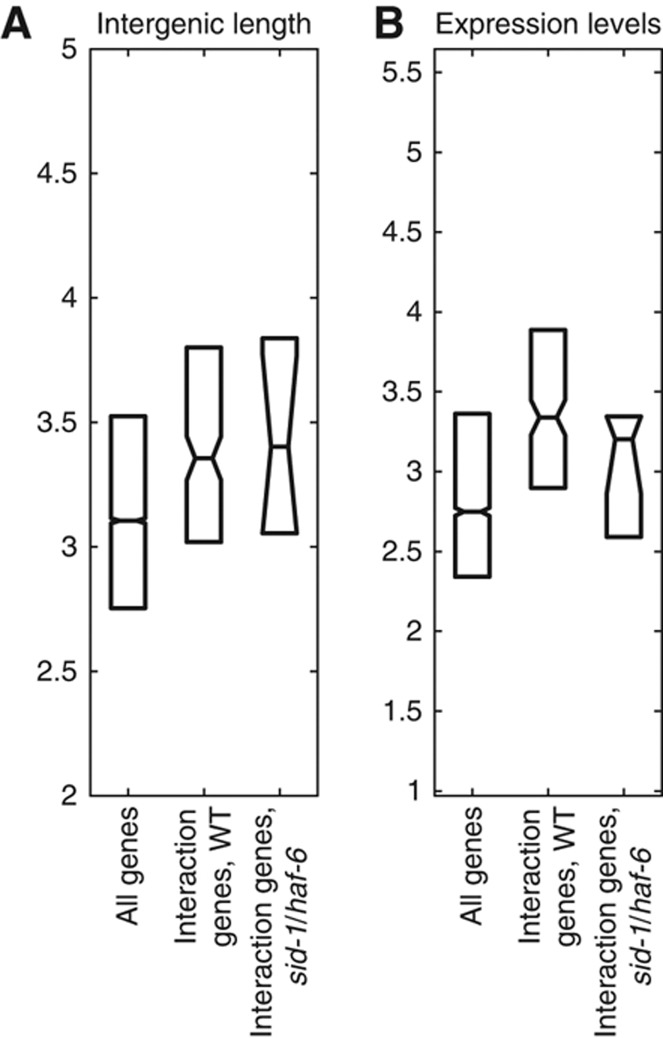
Our results indicate that genes with long promoters and a mid-range level of expression have a disproportionately higher likelihood to develop genotype–environment interactions following trans changes. We next asked if a transgenic strain with introduced mutations will produce genotype–environment interactions with this same pattern. Therefore, we compared expression levels across the five conditions on the same microarray platform in triplicate for the N2 strain and a nematode strain deficient for sid-1 and haf-6 function in the N2 background (HC445). As expected, sid-1 and haf-6 transcripts were significantly reduced (P<10−200 and 10−70, respectively). Querying the data for genotype–environment interactions, we detected 12 genes with significant genotype (N2 versus sid-1/haf-6)–environment interactions (P<0.005, two-way ANOVA, Supplementary Tables S5 and S8). Consistent with the above results, these 12 interaction genes also showed increased intergenic distances and higher expression on average (Figure 3). Although the P-value for the intergenic genes was >0.1, when examining the 100 genes with the best P-values, we found a P<0.001. This independent analysis provides strong support for our findings from the geographically distributed strains that interaction genes are highly regulated and that the genotype–environment interaction is due to trans effects.
Figure 3.
Genes with genotype–environment interactions following functional disruption of sid-1/haf-6 also show the hallmarks of highly regulated genes. (A) Distributions of intergenic distances (log10), shown as boxplots, comparing the 12 genes with a genotype–environment interaction in the sid-1/haf-6 analysis (mutant and N2 strain across the five environments, P<0.005) with the background set and the 198 interaction genes in the geographical isolates analysis (Figures 1 and 2). (B) The data for expression levels (log10) are in the same format.
Discussion
The idea that trans effects have a dominant role in genotype–environment appears to be supported by several works. For example, studies in yeast observed that changes leading to constitutive expression across environments tended to be cis, while trans changes were typically condition-specific and the consequence of genetic changes to sensory genes (Tirosh et al, 2009). Moreover, in Drosophila and C. elegans, cis changes were implicated as the dominant mechanism of expression divergence among species, while trans changes appeared to account for most of the expression diversity within a species (Li et al, 2006; Wittkopp et al, 2008). Finally, a recent comparison of gene expression across four yeast species and four environmental conditions found that genes specifically induced in a particular environment in some species often showed high and constitutive expression across all conditions in the other species, suggesting that interactions may occur by transitions between alternative expression programs (Tirosh et al, 2011).
We observed that genes with complex promoters and mid-range expression are more highly regulated. This supports the idea that genes under the regulation of multiple transcription factors require longer promoters to accommodate more binding sites (Davidson, 2006). The relationship of highly regulated genes and mid-range expression levels is less clear, but could result from the complex regulation itself, which could increase the likelihood of expression at any particular instance or create higher basal expression levels due to leaky activation from multiple regulating TFs.
The ease by which new expression can arise in an environment-specific manner may itself be under selection. Such facilitated variation (Gerhart and Kirschner, 2007) for gene expression diversity may also explain in part the large amounts of divergent expression observed between species (Khaitovich et al, 2004; Yanai et al, 2004; Yanai and Hunter, 2009). We expect that future work will be directed toward generalizing the approach to developmental time points, cell types, and conditions. These can be expected to provide an understanding of how genotype–environment interactions arise in the transcriptome; a readily assessable and quantifiable phenotype of the genetic material. However, gene expression in the fullest sense must include protein activity and contributions to fitness (Feder and Walser, 2005) and these provide a challenging goal for the greater understanding of the influence of the genotype and the environment on the organism.
Materials and methods
Strains and conditions
The five C. elegans strains used in this study are previously collected geographical isolates. N2 was originally collected by LN Staniland from a mushroom compost near Bristol, England, (Nicholas et al, 1959) and is the standard lab strain used in C. elegans research (Brenner, 1974). CB4857 was collected from mushrooms in Claremont, California by EM Hedgecock (Hodgkin, 1993). RC301 was collected in 1983 by R Cassada from a compost heap in the Botanical Garden of the University of Freiburg in Germany (Hodgkin, 1993). CB4856 was isolated from a pineapple field in Hawaii in 1972 by L Hollen (Hodgkin, 1993). AB2 was collected from soil in Adelaide, Australia by D Riddle and A Bird (Hodgkin, 1993). The strains were propagated under control conditions: nematode growth medium (NG) with B. subtilis as a non-pathogenic food source. Embryos were collected by bleaching and ∼2000 were placed into each of five conditions: (1) Control: 20°C with B. subtilis on NG plates; (2) Heat: 25°C with B. subtilis on NG plates, (3) pH/salt/E. coli: 20°C with E. coli on high salt (4 × regular NG) and high pH (8.5 relative to pH of 6 for NG) plates; (4) Liquid culture: 20°C with B. subtilis in S-medium in a shaker incubator; and (5) Pathogen: 20°C with M. nematophilum on NG plates. B. subtilis was used here as the standard food source in all but one of the conditions as it is preferred by C. elegans relative to the E. coli OP50 strain (Garsin et al, 2003).
Embryo collection and RNA processing
Four-cell stage embryos were isolated by mouth pipette (Baugh et al, 2003). Each sample comprised 50 pooled embryos. For each genotype/environment combination there were triplicates, thus the data set comprises 25 × 50 × 3=3750 individually isolated embryos. RNA was isolated using Trizol as previously described. RNA was amplified using the Ambion MessageAmpII for two rounds in order to produce sufficient quantities for microarray analysis. mRNA was isolated, amplified, and hybridized along with Agilent Spike-ins onto one color microarrays as previously described (Yanai and Hunter, 2009).
Gene expression
We designed a custom 15-K C. elegans microarray which was then manufactured by Agilent. The 60-mer probes were determined using OligoWiz2 (Wernersson et al, 2007) to target the coding region based upon the following factors: melting temperature, position along the transcript, folding potential, low complexity in the sequence, and cross-hybridization to other coding sequences. The probes were also restricted against spanning splice junctions to avoid missing transcripts due to errors in gene structure predictions. For each gene, the best scoring probe with no significant match in other coding sequences (E-value<0.001) was selected. This procedure yielded 16 831 gene-specific probes out of the total 20 074 genes searched. We then selected the 15 208 best scoring probes for the microarray. Data were extracted using Feature Extraction (Agilent). The raw data were normalized using quantile normalization. Analysis was done on log10 of the normalized data. The complete data set and array platforms have been deposited in the Gene Expression Omnibus with accession codes GSE34650 and GPL15046. The data are also available in Supplementary Table S6.
Genome sequencing of C. elegans strains
The strains RC301, CB4856, CB4857, and AB2 were sequenced so that together with the previously published strain, the N2 strain (Consortium, 1998), the genomes of all examined strains were known. Genomic DNA was extracted by proteinase K digestion followed by two rounds of phenol–chloroform extraction, with an intermediate step of RNase A digestion in TE. Genomic DNA libraries were built using Illumina’s standard paired-end protocol and 100 × 2 bp were sequenced on the Illumina HiSeq 2000 following the manufacturer’s recommendations. The number of reads mapped to the N2 genome (Wormbase release 220) were: 119 071 331 (CB4857), 109 807 250 (RC301), 58 309 757 (CB4856), and 113 429 439 (AB2) with a coverage of 116X, 107X, 58X, and 111X, respectively. SNP calling was performed using samtools utilities with the N2 genome as reference. SNPs with a variant quality score of at least 30 were selected. Overall 100 919 (CB4857), 85 776 (RC301), 184 912 (CB4856), and 98 415 (AB2) SNPs relative to N2 strain were detected. Probes on the microarray that were found to include SNPs in one or more of the strains were excluded from analysis. For this exclusion we used SNPs with all range of variant quality scores, i.e., even those with a quality score <30. 600 genes were excluded from analysis based upon this criterion. The complete sequencing data have been submitted to the NCBI SRA database with accession ID SRP011413.1 for the study. The accessions for the particular strains are SRS299995.1 (CB4857), SRS299996.1 (RC301), SRS299997.1 (CB4856), and SRS299999.1 (AB2). The SNPs in mpileup format are included as Supplementary Table S7.
Gene properties
Intergenic distances and expression clusters were retrieved from Wormbase (Yook et al, 2011). Constitutively expressed genes were defined as those genes with a mean expression greater than 4 log10 units in all strains/conditions and an absolute expression range <0.2. Gene regulatory information in terms of the number of regulatory motifs per 1-kb region of a genes’ promoter was identified using the CISRED server ( http://www.cisred.org). Motifs were required to have a P-value <0.05 and be conserved between C. elegans and C. briggsae.
Supplementary Material
Supplementary Figures S1-10, Supplementary Tables S1-5,8
Acknowledgments
We thank Merck for a small grant, the CGC for the strains, and E Troemel for M. nematophilum. We also thank D Silver for helpful assistance with the analyses.
Author contributions: IY, CPH, and DHS designed the experiments. IY designed the custom microarray, and DHS and IY performed the microarray experiments from culturing worms to scanning the hybridized microarrays. TH sequenced the strains. VG, SB, and IY analyzed the data. VG and IY wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barnes TM, Kohara Y, Coulson A, Hekimi S (1995) Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141: 159–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP (2003) Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130: 889–900 [DOI] [PubMed] [Google Scholar]
- Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium. (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018 [DOI] [PubMed] [Google Scholar]
- Davidson EH (2006) The Regulatory Genome: Gene Regulatory Networks in Development and Evolution Amsterdam, Boston: Elsevier/Academic Press, [Google Scholar]
- Feder ME, Walser JC (2005) The biological limitations of transcriptomics in elucidating stress and stress responses. J Evol Biol 18: 901–910 [DOI] [PubMed] [Google Scholar]
- Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, Ausubel FM (2003) Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300: 1921. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart J, Kirschner M (2007) The theory of facilitated variation. Proc Natl Acad Sci USA 104 Suppl 1: 8582–8589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke J, Lorenz K, Ramnarine S, Cohen B (2010) Gene-environment interactions at nucleotide resolution. PLoS Genet 6: e1001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishkevich V, Hashimshony T, Yanai I (2011) Core promoter T-blocks correlate with gene expression levels in C. elegans. Genome Res 21: 707–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J (1993) Molecular cloning and duplication of the nematode sex-determining gene tra-1. Genetics 133: 543–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Weiss G, Lachmann M, Hellmann I, Enard W, Muetzel B, Wirkner U, Ansorge W, Paabo S (2004) A neutral model of transcriptome evolution. PLoS Biol 2: E132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Alvarez OA, Gutteling EW, Tijsterman M, Fu J, Riksen JA, Hazendonk E, Prins P, Plasterk RH, Jansen RC, Breitling R, Kammenga JE (2006) Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet 2: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Stone EA, Ayroles JF (2009) The genetics of quantitative traits: challenges and prospects. Nat Rev Genet 10: 565–577 [DOI] [PubMed] [Google Scholar]
- Nicholas WL, Dougherty EC, Hansen EL (1959) Axenic cultivation of Caenorhabditis briggsae with chemically undefined supplements; comparative studies with related nematodes. Ann NY Acad Sci 77: 218–236 [Google Scholar]
- Rifkin SA, Kim J, White KP (2003) Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet 33: 138–144 [DOI] [PubMed] [Google Scholar]
- Shen-Orr SS, Pilpel Y, Hunter CP (2010) Composition and regulation of maternal and zygotic transcriptomes reflects species-specific reproductive mode. Genome Biol 11: R58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EN, Kruglyak L (2008) Gene-environment interaction in yeast gene expression. PLoS Biol 6: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike CA, Bader J, Reinke V, Strome S (2008) DEPS-1 promotes P-granule assembly and RNA interference in C. elegans germ cells. Development 135: 983–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N (2008) Two strategies for gene regulation by promoter nucleosomes. Genome Res 18: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N (2009) A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324: 659–662 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, Carmi M, Barkai N (2006) A genetic signature of interspecies variations in gene expression. Nat Genet 38: 830–834 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Wong KH, Barkai N, Struhl K (2011) Extensive divergence of yeast stress responses through transitions between induced and constitutive activation. Proc Natl Acad Sci USA 108: 16693–16698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernersson R, Juncker AS, Nielsen HB (2007) Probe selection for DNA microarrays using OligoWiz. Nat Protoc 2: 2677–2691 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG (2004) Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG (2008) Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet 40: 346–350 [DOI] [PubMed] [Google Scholar]
- Yanai I, Graur D, Ophir R (2004) Incongruent expression profiles between human and mouse orthologous genes suggest widespread neutral evolution of transcription control. Omics 8: 15–24 [DOI] [PubMed] [Google Scholar]
- Yanai I, Hunter CP (2009) Comparison of diverse developmental transcriptomes reveals that coexpression of gene neighbors is not evolutionarily conserved. Genome Res 19: 2214–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook K, Harris TW, Bieri T, Cabunoc A, Chan J, Chen WJ, Davis P, de la Cruz N, Duong A, Fang R, Ganesan U, Grove C, Howe K, Kadam S, Kishore R, Lee R, Li Y, Muller HM, Nakamura C, Nash B et al. (2011) WormBase 2012: more genomes, more data, new website. Nucleic Acids Res 40: D735–D741 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-10, Supplementary Tables S1-5,8