Abstract
Cells use both deterministic and stochastic mechanisms to generate cell–to-cell heterogeneity, which enables the population to better withstand environmental stress. Here we show that, within a clonal population of mycobacteria, there is significant deterministic heterogeneity in elongation rate that arises because mycobacteria grow in an unusual, unipolar fashion. Division of the asymmetrically growing mother cell gives rise to daughter cells that differ in elongation rate and size. Because the mycobacterial cell division cycle is governed by time not cell size, rapidly elongating cells do not divide more frequently than slowly elongating cells. Importantly, the physiologically distinct subpopulations of cells that arise through asymmetric growth and division are differentially susceptible to clinically important classes of antibiotics.
Tuberculosis, caused by infection with Mycobacterium tuberculosis, remains a major global health problem, killing more than 1.5 million people annually. While antibiotic therapy rapidly reduces the bacterial burden, eliminating the infection requires long courses of multiple antibiotics (1, 2). It has been suggested that this lengthy course of treatment is required because the mycobacterial population is functionally heterogeneous and contains cells that are differentially susceptible to antibiotics, non-replicating, or sequestered in the body (3, 4). Because almost all antibiotics target bacterial processes involved in cell growth, we hypothesized that there is heterogeneity in growth states of mycobacterial cells that underlies differential antibiotic susceptibility.
To measure the growth and antibiotic susceptibility of mycobacteria at a single cell level, we designed a microfluidic chamber to culture mycobacteria for live-cell imaging. This device allows cell movement in two-dimensions but constrains bacteria to a single focal plane (Fig. 1A). Cells are grown in shallow channels that are connected to a perpendicular media channel, which provides a homogeneous mixture of nutrients by diffusion. This device enables us to image mycobacterial growth for up to five generations at which point images become too crowded to score (Fig. 1B). In this device, we can expose cells to defined stressors such as antibiotics, and follow the responses of individual cells. As the growth and division machinery is highly conserved between pathogenic and nonpathogenic mycobacteria, we focused our live-cell imaging experiments on the experimentally tractable, model mycobacterium, Mycobacterium smegmatis.
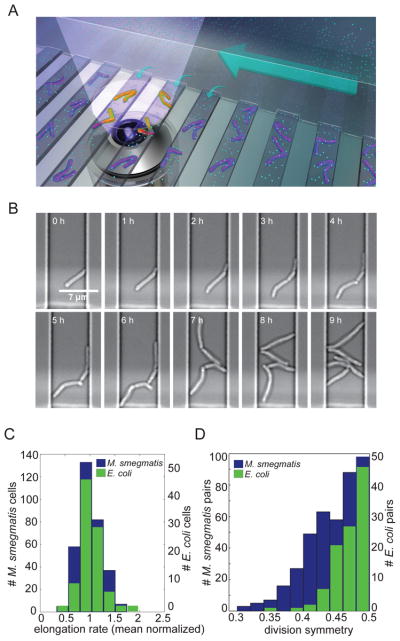
Figure 1. M. smegmatis exhibits heterogeneous growth characteristics.
(A) Schematic diagram of the microfluidic device used for long-term imaging of mycobacteria. Media flows through the main channel (large arrow) and provides nutrients (cyan circles) by diffusion (small arrows) to the cells. (B) Bright-field, time-lapse imaging of M. smegmatis in the microfluidic device.. (C) Distribution of average (mean centered) elongation rates of 322 individual M. smegmatis (blue; left axis) and 102 E. coli (green; right axis) cells averaged over the course of one cell cycle. The mean elongation rates were 1.15 μm/h for M. smegmatis and 3.72 μm/h for E. coli. (D) Distribution of division symmetry for 166 M. smegmatis (blue; left axis) and 105 E. coli (green; right axis) pairs of sister cells. Division symmetry is calculated as the ratio of the length of the smaller sister to the sum of the lengths of both sisters at division.
To assess population variation in growth states, we measured the elongation rates of individual M. smegmatis cells grown in rich medium. As a point of comparison, we also measured the growth parameters of individual Escherichia coli cells grown in rich medium in our microfluidic device. We found significantly more variability in the elongation rates of mycobacterial cells compared to E. coli cells (Fig. 1C; F<0.05 (5)). Mycobacteria lack the molecular rulers that ensure symmetric cell division, which place the division septum in the center of the cell in other rod shaped organisms such as E. coli and Bacillus subtilis (6). Thus, we wondered whether the variability in mycobacterial elongation rates was related to asymmetry in cell division (7, 8). We therefore assessed the symmetry of mycobacterial cell division and found that cell division is significantly less symmetric in M. smegmatis than in E. coli (Fig. 1D; F<.001 (5)). We observed similar asymmetry in cell division in M. tuberculosis (fig. S1).
Asymmetry in cell elongation could cause apparent asymmetry in cell division and subsequent variability in the elongation rates of daughter cells. To assess this possibility, we took advantage of the fact that mycobacteria elongate at their poles rather than along the lateral cell body as in E. coli (6, 9). This allowed us to quantify cell elongation by pulse labeling the cell wall with a fluorescent amine reactive dye and measuring the extension of the unlabeled poles (Fig. 2A; (10)). Strikingly, we found that mycobacterial cells elongate preferentially at the old pole (Fig. 2B and C). In static images, unipolar growth produces a “cigar-band” of cell wall labeling with the amine reactive dye where one pole has elongated significantly more than the other, which we also observe in M. tuberculosis (Fig. 2D).
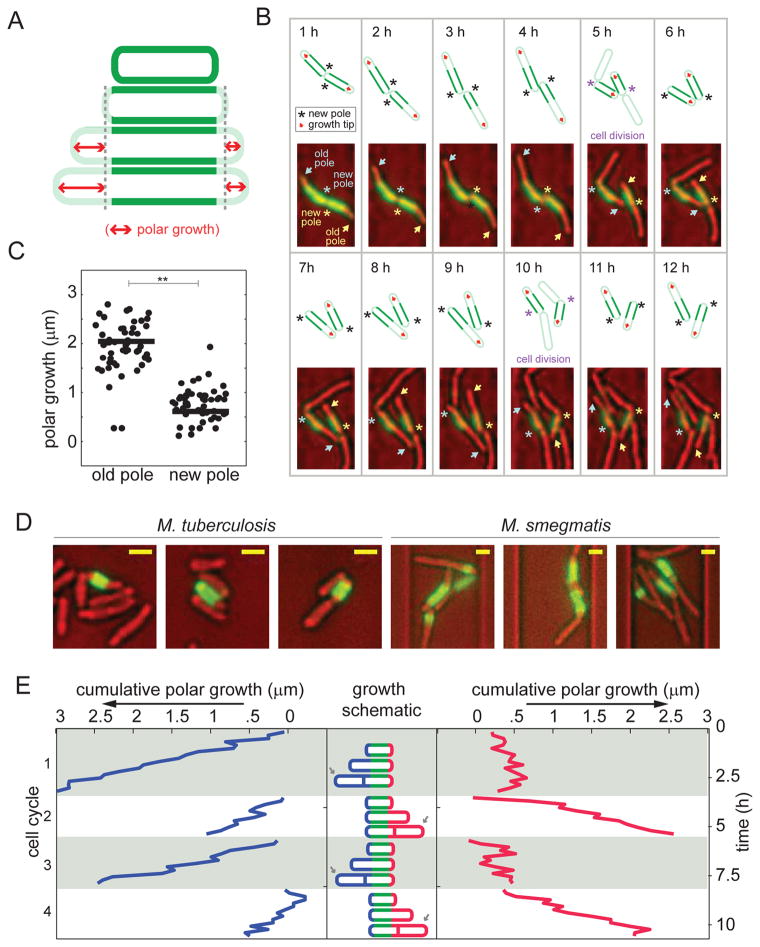
Figure 2. M. smegmatis growth is asymmetric and elongation occurs from the old pole.
(A) Schematic diagram of the pulse-chase experiment used to measure polar growth. Cells were labeled with amine reactive dye (green) and growth was assessed by measuring the extension of the unlabeled region (red arrows). Cell wall-labeling did not alter cell elongation rate (fig. S5A), other labeling chemistries such as hydroxylamine labeling via periodate oxidation led to similar staining patterns (data not shown), and similar labeling patterns were seen in cells grown in broth and in microfluidic channels (fig. S5B). (B) Time-lapse imaging of two sister cells following the pulse-labeling (green) of the cell wall. The bright field images were pseudo-colored red. New poles are annotated with asterisks and the old poles with arrows. Each cell’s poles are annotated with the same color asterisk and arrow. A schematic is drawn above each image, marking the new poles and growth poles with an asterisk and red arrow, respectively. We also used a fluorescein-vancomycin conjugate (Van-FL) to stain nascent peptidoglycan (30) and found preferential labeling of the old pole over the new pole (fig. S5C). (C) Growth over one cell cycle at new versus old poles in 50 cells (*p<0.001 by a Mann-Whitney rank sum test). There was no cell in which the new pole elongated more than the old pole. (D) Three representative images of M. tuberculosis (left) and M. smegmatis (right) following the pulse-labeling (green) of the cell wall (after 48 hours in broth culture for M. tuberculosis and six hours in the microfluidic device for M. smegmatis). The bright field images are pseudo-colored red. Size bars represent 1.3 μm. Comparison images of E. coli labeled under similar conditions may be found in fig. S5D. (E) Cumulative polar growth for one labeled cell plotted for each pole through four cell cycles. The cell inheriting the new pole alternates the direction of growth after division events.
Unipolar growth per se does not explain cell-to-cell variability in elongation rates or cell sizes but it does create different types of cells at division. One daughter cell inherits the growing pole while the other daughter cell must create a new growth pole after every division (schematic in Fig. 2E). The new growth pole is generated at the older pole (opposite the division septum), and therefore the direction of growth changes with every cell cycle. We have quantified this for a single, representative cell over four generations in Fig. 2E. By contrast, in the daughter cell that inherits the growing pole (indicated by an arrow in Fig. 2E), elongation continues from the inherited growth pole (fig. S2).
We hypothesized that the daughter cell inheriting the growth pole would elongate at a different rate than its sister cell, which must assemble a new growth pole. We tested this hypothesis by computing the differences in elongation rate between pairs of sister cells. We found that on average, the sister cell inheriting the growth pole elongates faster than the sister cell that establishes a new growth pole (Fig. 3A; p<0.05). The cell inheriting the growth pole is also longer at birth than its sister cell, consistent with a model in which elongation remains asymmetric during septation (Fig. 3B; p<0.05). Thus, each division results in two distinctive sister cells. We term these cells accelerators, which inherit the mother’s growth pole and tend to elongate faster, and alternators, which must regenerate a new growth pole and tend to elongate more slowly (Fig. 3C).
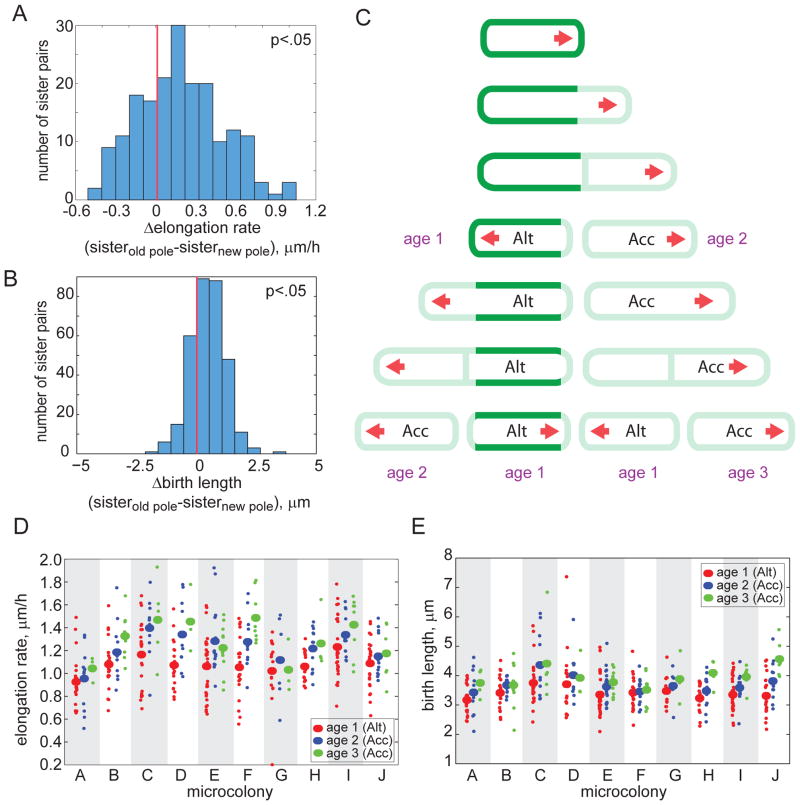
Figure 3. Division creates sister cells with different growth properties.
(A–B) Distribution of the differences in elongation rate (A) and birth length (B) between sister cells inheriting the old pole and the new pole. The distributions are skewed (p<0.05; red lines denote zero), indicating that the sister inheriting the older pole elongates faster (in 71% of the 161 sister pairs) and is larger at division (in 74% of the 161 sister pairs). In 7.5% of the 161 sister pairs, the sister that inherited the new pole elongated faster and was longer at birth than its sister. (C) Schematic model for mycobacterial growth. A labeled cell (green) is shown to elongate from one pole (red arrow). Two sister cells are created at division: an accelerator (Acc) cell inheriting the old (growing) pole and an alternator (Alt) cell inheriting the new pole. Growth pole age (in generations) is labeled in purple. (D–E) Elongation rate (D) and birth lengths (E) are plotted for ten microcolonies, with cell subpopulations grouped by growth pole age. Growth pole age was scored by mapping the pedigrees of unlabeled cells through several generations via live cell imaging. Subpopulation means (large ovals) are plotted along with data from individual cells (small circles). Elongation rate and birth length increase within a colony as the growth poles age (p<0.05 for alternator vs. accelerator elongation rates; and p<0.05 for alternator vs. accelerator and age two vs. age three birth lengths).
By definition, all alternator cells have new growth poles, while accelerator cells inherit growth poles of varying ages. Some accelerator cells inherit growth poles created in the previous generation while others inherit growth poles created several generations earlier. To understand whether growth pole age impacts the elongation rate of accelerator cells, we mapped the pedigrees of single cells. We assigned an “age” to a cell based on the number of generations its growth pole had experienced; alternator cells have an age of one and accelerator cells have an age of two or greater (Fig. 3C). We then compared the elongation rate of cells of different ages in populations arising from a single cell, which we term a microcolony. Cells with older growth poles elongate faster than cells with younger growth poles (Fig. 3D, p<0.05 for accelerator vs. alternator cells). In addition, the birth length of cells increases as the growth pole matures (Fig. 3E, p<0.05 for accelerator vs. alternator cells and age three vs. age two cells). Taken together, these data suggest that as the growth pole matures, cells elongate faster and are larger. Of note, we occasionally observed cells with older growth poles that elongated more slowly than cells with younger growth poles in the same microcolony (e.g. Fig. 3D, colony G), suggesting there may be a mechanism to “reset” the elongation rate.
These data show that the mycobacterial growth pattern generates a population of cells that is heterogeneous in size and elongation rates. We assessed whether rapidly elongating cells also divide more frequently than slowly elongating cells, as would be expected if mycobacteria control cell cycle entry using sized based regulation like E. coli (11, 12). Alternatively, mycobacteria might regulate entry into the cell cycle using a time-based mechanism. In yeast, investigators have differentiated between size- and time-based cell cycle regulation using the relationship between birth length and elongation length (13, 14). In cells that employ size-based cell cycle regulation, small cells must grow more before dividing causing the birth length to be negatively correlated with elongation length, while in time-based regulation, these lengths are uncorrelated. We therefore measured the association between the birth and elongation lengths of M. smegmatis cells. We found no correlation between the two lengths in M. smegmatis (regression line slope of 0.00; Fig. 4A), while in E. coli we found these lengths to be negatively correlated, as expected (regression line slope of −0.75; fig. S3A). These data suggest that the mycobacterial division cycle, which we use as a measure of the cell cycle more broadly, is regulated by time rather than size. Consistent with time-based regulation of cell cycle progression, cell division is synchronized in a microcolony, with closely related cells dividing at similar times (Fig. 4B and fig. S3B). We calculated the microcolony division cycle length by characterizing the distribution of division events in time and in the frequency domain using a Fourier transform (Fig. 4B and fig. S3B). The major frequency corresponds to the division cycle length (and is unbiased by the increasing number of cells as the colony grows) and the amplitude of the peak is a metric for synchronization in the colony. The colony cycle times calculated from the mean and the frequency domain are very similar (within 0.2 h for each microcolony). However, there were significant differences in cycle times between some microcolonies (fig. S3C). These data suggest that as yet unidentified factors may modulate the cell cycle timer, compounding the diversity of elongation states within the population. It is also important to note that the cell cycle length is much longer in slow-growing mycobacteria (~22 hours in M. tuberculosis as compared to ~3 hours in M. smegmatis). It is also therefore possible that growth and division cycle timing in M. tuberculosis is subject to additional layers of regulation that are not reflected in these studies.
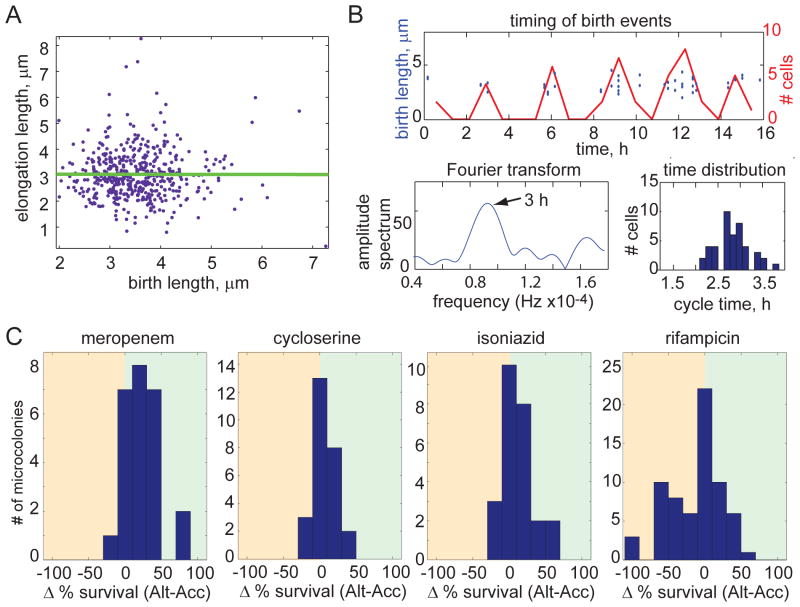
Figure 4. Population heterogeneity of growth characteristics is maintained by time-based cell division cycle regulation and contributes to differential susceptibility to antibiotic stress.
(A) Birth length and elongation length, which is defined as the length that the cell elongates between birth and division, are uncorrelated for 322 M. smegmatis cells (regression slope of 0.00), suggesting that mycobacteria use time to regulate their cell cycle. (B) Cell cycle timing is characterized for one microcolony (a second representative microcolony is shown in fig. S3B). Division events are plotted in the time domain as discrete events (blue circles; left axis) and as a histogram (red line; right axis). The histogram of birth events was used to generate an amplitude spectrum with a Fourier transform, shown in the lower left plot. The peak represents the cycle time of the microcolony (arrow). The spread of cycle times for individual cells is shown here as histogram in the lower right. (C) Distributions of the difference in bacterial survival between accelerator and alternator cells for each microcolony following treatment with meropenem, cycloserine, isoniazid, and rifampicin at minimal inhibitory concentrations (2.3 mM, 0.04 mg/ml, 25 μM, and 50 μM, respectively; p<0.05 for each distribution). Survival was scored by determining the percentage of cells that were able to regrow after antibiotic was removed. The analysis includes 317 cells in 25 microcolonies for meropenem, 374 cells in 26 microcolonies for cycloserine, 334 cells in 25 microcolonies for isoniazid, and 453 cells in 66 microcolonies for rifampicin.
Thus, asymmetric elongation and cell division and a timed cell cycle quickly create a population of mycobacterial cells that vary in their elongation rates, sizes and perhaps other physiologic properties. Since antibiotics target processes essential for growth and division, we hypothesized that these cells might be differentially sensitive to antibiotics. We therefore sought to determine the susceptibility of alternator and accelerator cells to treatment with different classes of antibiotics. To do this, we used live cell imaging to establish the pedigrees of growing cells and challenged them with the indicated antibiotics at the minimum inhibitory concentrations. We identified bacterial survival by scoring for post-treatment regrowth in 25–66 independent microcolonies. Because we observed variability between microcolonies in the efficacy of some antibiotics, especially isoniazid and rifampicin, we calculated the difference in bacterial survival between accelerator and alternator cells for each microcolony and assessed the distribution of this differential across all microcolonies.
Given their different rates of elongation and potential differences in cell wall composition, we predicted that accelerator cells would be more susceptible to cell wall synthesis inhibitors than alternator cells. Indeed, accelerator cells were significantly more sensitive than alternator cells to the peptidoglycan synthesis inhibitors, cycloserine and meropenem (Fig. 4C and fig. S4; p<0.05). Accelerator cells were also more sensitive than alternator cells to isoniazid, which blocks synthesis of cell wall mycolic acids, although there was more variability between microcolonies in the effectiveness of isoniazid (Fig. 4C and fig. S4; p<0.05). However, accelerator cells are not universally more susceptible to antibiotic treatment. Strikingly, when microcolonies are treated with rifampicin, which inhibits RNA polymerase, alternator cells were more susceptible than accelerator cells (Fig. 4C and fig. S4; p<0.05). As in the case of isoniazid, there was variability among microcolonies in response to rifampicin, suggesting that other potentially heritable factors may also contribute to a cell’s susceptibility to these drugs. Thus, we find that alternator and accelerator cells vary in their susceptibility to different classes of antibiotics, consistent with the model that asymmetric growth and division creates physiologically distinct subpopulations of cells.
In this study, we show that growth and division in mycobacteria is distinct from better characterized model bacteria. Although unusual, the mycobacterial growth pattern contains spatial and temporal elements that are similar to those used by other bacteria to achieve rapid functional diversification of closely related cells (15–21). The most obvious example of this is Caulobacter crescentis, which also exploits polar asymmetry, asymmetry at cell division, and age-dependent changes in pole function to rapidly create a dimorphic population (15, 22–25). In these organisms and in mycobacteria, variable growth and division patterns create deterministic population diversity at a very high frequency. Mycobacteria may also use other lower frequency mechanisms, both deterministic and stochastic, to generate a tapestry of cell types (26–29). We anticipate that if these mechanisms for bacterial diversification operate in M. tuberculosis as they do in M. smegmatis, they may contribute to the highly variable outcomes of tuberculosis infection and treatment.
Supplementary Material
Acknowledgments
We thank Eric Rubin, Barry Bloom, Chris Sassetti, Douglas Lauffenburger, Elson Liu and Joshua Model for helpful discussions, Peter Sorger for access to his live cell imaging systems, Alexander J. Aranyosi for assistance with microfabrication, Jacqueline Goldstein for help with image annotation, and Janet Sinn-Hanlon for the device illustration. We thank Robert Husson and Mushtaq Mir for the kind gift of Van-Alexa. B.B.A. and S.M.F. designed the experiments and wrote the initial manuscript. B.B.A. performed the experiments, annotated images, and developed the analysis programs. M.F.S., V.A., D.I., and M.T. designed the microfluidic devices that were built by M.F.S. D.H. performed the initial pulse-chase experiment and annotated images. This work was partially supported by a National Institute of Health Director’s New Innovator Award 1DP20D001378, Doris Duke Charitable Foundation under Grant 2010054, and United States Army Medical Research Acquisition Activity under Grant W81XWH-10-2-0161 to S.M.F., the National Institute of Biomedical Imaging and Bioengineering under Grant P41 EB002503 to M.T. (BioMEMS Resource Center), and by the Massachusetts General Hospital Executive Committee on Research through a post-doctoral fellowship to M.F.S.
Footnotes
REFERENCES AND NOTES
- 1.Cohn DL, Catlin BJ, Peterson KL, Judson FN, Sbarbaro JA. A 62-dose, 6-month therapy for pulmonary and extrapulmonary tuberculosis. A twice-weekly, directly observed, and cost-effective regimen. Ann Intern Med. 1990;112:407–415. doi: 10.7326/0003-4819-76-3-112-6-407. [DOI] [PubMed] [Google Scholar]
- 2.Elzinga G, Raviglione MC, Maher D. Scale up: meeting targets in global tuberculosis control. Lancet. 2004;363:814–819. doi: 10.1016/S0140-6736(04)15698-5. [DOI] [PubMed] [Google Scholar]
- 3.McCune RM, Feldmann FM, Lambert HP, McDermott W. Microbial persistence. I. The capacity of tubercle bacilli to survive sterilization in mouse tissues. J Exp Med. 1966;123:445–468. doi: 10.1084/jem.123.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connolly LE, Edelstein PH, Ramakrishnan L. Why Is Long-Term Therapy Required to Cure Tuberculosis? PLoS Med. 2007;4:e120. doi: 10.1371/journal.pmed.0040120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Materials and methods are available as supporting material on Science online.
- 6.Hett EC, Rubin EJ. Bacterial growth and cell division: a mycobacterial perspective. Microbiol Mol Biol Rev. 2008;72:126–156. doi: 10.1128/MMBR.00028-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farnia P, et al. Growth and cell-division in extensive (XDR) and extremely drug resistant (XXDR) tuberculosis strains: transmission and atomic force observation. Int J Clin Exp Med. 2010;3:308–314. [PMC free article] [PubMed] [Google Scholar]
- 8.Singh B, Ghosh J, Islam NM, Dasgupta S, Kirsebom LA. Growth, cell division and sporulation in mycobacteria. Antonie Van Leeuwenhoek. 2010;98:165–177. doi: 10.1007/s10482-010-9446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thanky NR, Young DB, Robertson BD. Unusual features of the cell cycle in mycobacteria: polar-restricted growth and the snapping-model of cell division. Tuberculosis (Edinb) 2007;87:231–236. doi: 10.1016/j.tube.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Mentinova M, McLuckey SA. Covalent Modification of Gaseous Peptide Ions with N-Hydroxysuccinimide Ester Reagent Ions. Journal of the American Chemical Society. 2010;132:18248–18257. doi: 10.1021/ja107286p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Messer W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol Rev. 2002;26:355–374. doi: 10.1111/j.1574-6976.2002.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 12.Løbner-Olesen A, Skarstad K, Hansen FG, von Meyenburg K, Boye E. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell. 1989;57:881–889. doi: 10.1016/0092-8674(89)90802-7. [DOI] [PubMed] [Google Scholar]
- 13.Sveiczer A, Novak B, Mitchison JM. The size control of fission yeast revisited. J Cell Sci. 1996;109(Pt 12):2947–2957. doi: 10.1242/jcs.109.12.2947. [DOI] [PubMed] [Google Scholar]
- 14.Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 2007;448:947–951. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- 15.Shapiro L, McAdams HH, Losick R. Generating and Exploiting Polarity in Bacteria. Science. 2002;298:1942–1946. doi: 10.1126/science.1072163. [DOI] [PubMed] [Google Scholar]
- 16.Chen YE, et al. Spatial gradient of protein phosphorylation underlies replicative asymmetry in a bacterium. Proc Natl Acad Sci USA. 2011;108:1052–1057. doi: 10.1073/pnas.1015397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carniol K, Ben-Yehuda S, King N, Losick R. Genetic dissection of the sporulation protein SpoIIE and its role in asymmetric division in Bacillus subtilis. J Bacteriol. 2005;187:3511–3520. doi: 10.1128/JB.187.10.3511-3520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veening J-W, et al. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc Natl Acad Sci USA. 2008;105:4393–4398. doi: 10.1073/pnas.0700463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Yehuda S, Losick R. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell. 2002;109:257–266. doi: 10.1016/s0092-8674(02)00698-0. [DOI] [PubMed] [Google Scholar]
- 20.Stewart EJ, Madden R, Paul G, Taddei F. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 2005;3:e45. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huh D, Paulsson J. Non-genetic heterogeneity from stochastic partitioning at cell division. Nat Genet. 2011;43:95–100. doi: 10.1038/ng.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsokos CG, Perchuk BS, Laub MT. A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev Cell. 2011;20:329–341. doi: 10.1016/j.devcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci USA. 2002;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen RB, Wang SC, Shapiro L. Dynamic localization of proteins and DNA during a bacterial cell cycle. Nat Rev Mol Cell Biol. 2002;3:167–176. doi: 10.1038/nrm758. [DOI] [PubMed] [Google Scholar]
- 25.Viollier PH, Sternheim N, Shapiro L. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J. 2002;21:4420–4428. doi: 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chai Y, Norman T, Kolter R, Losick R. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes & Development. 2010;24:754–765. doi: 10.1101/gad.1915010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial Persistence as a Phenotypic Switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 28.Bordes P, et al. SecB-like chaperone controls a toxin-antitoxin stress-responsive system in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2011;108:8438–8443. doi: 10.1073/pnas.1101189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Micro. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 30.Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology (Reading, Engl) 2008;154:725–735. doi: 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.