Abstract
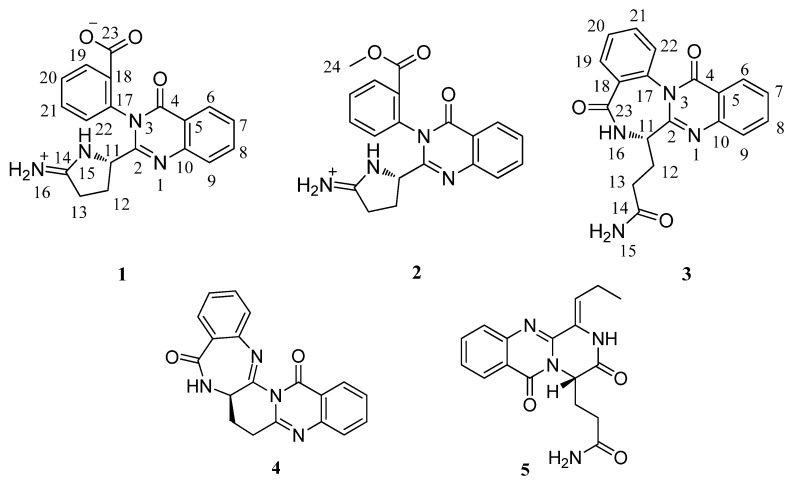
Three new alkaloids, including auranomides A and B (1 and 2), a new scaffold containing quinazolin-4-one substituted with a pyrrolidin-2-iminium moiety, and auranomide C (3), as well as two known metabolites auranthine (4) and aurantiomides C (5) were isolated from the marine-derived fungus Penicillium aurantiogriseum. The chemical structures of compounds 1–3 were elucidated by extensive spectroscopic methods, including IR, HRESIMS and 2D NMR spectroscopic analysis. The absolute configurations of compounds 1–3 were suggested from the perspective of a plausible biosynthesis pathway. Compounds 1–3 were subjected to antitumor and antimicrobial screening models. Auranomides A–C exhibited moderate cytotoxic activity against human tumor cells. Auranomides B was the most potent among them with an IC50 value of 0.097 μmol/mL against HEPG2 cells.
Keywords: marine-derived fungus, Penicillium aurantiogriseum, quinazolin-4-one, antitumor
1. Introduction
Quinazolin-4-one alkaloids are a class of natural scaffold which has been proved as a drug-like template in medicinal chemistry and considered a favored structure [1]. The quinazolin-4-one ring system has been consistently recognized as a promising pharmacophore because of its broad spectrum pharmacological activities such as antitumor [2], antitubercular [3], anti-HIV [4], anti-inflammatory [5], antiangiotensin [6], antibacterial [7], and antifungal [8]. Involved in centrosome separation and bipolar mitotic spindle formation, kinesin spindle protein (KSP) plays an important role in cell division [9]. Some quinazolin-4-one compounds are KSP inhibitors. They can arrest cells in mitosis and induce cell death [10], and have proved to be promising candidates for anticancer drugs [11]. The biological and pharmacological activities of quinazolin-4-one derived compounds have been documented not only from synthetic derivatives but also from several naturally occurring alkaloids isolated from families of the plant kingdom, and from microbes such as Streptomycetes and fungi [12,13,14,15,16,17].
During high throughput screening of novel compounds from marine derived microorganisms [18], our group have had identified two pyrone-type polyketides from the marine derived fungus Penicillium aurantiogriseum [19]. By varying the culture media according to the OSMAC (one strain-many compounds) approach [20,21], a drastically altered metabolite profile of the same strain was obtained. With the help of HPLC, we found the UV spectra of some compounds in the crude extract were similar to the specific absorbance of the quinazoline-4-one core. Using UV-guided fractionation, three new quinazolin-4-one derivatives (1−3), together with two known metabolites auranthine (4) [22] and aurantiomides C (5) [23] (Figure 1) were isolated from this marine-derived fungus. The novel quinazolinone derivatives were named as auranomides A, B and C (1−3). Herein, we report the isolation, structure elucidation and bioactivity evaluation of these alkaloids.
Figure 1.
Structures of compounds 1–5.
2. Results and Discussion
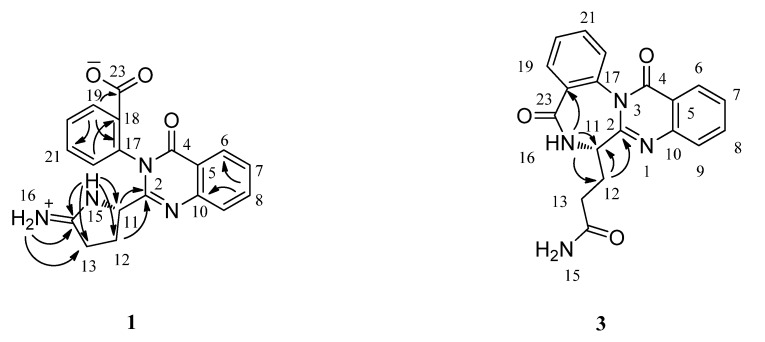
Compound 1 was obtained as a white amorphous powder with the specific absorbance for the quinazoline-4-one. The molecular formula of 1 was determined to be C19H16N4O3 (fourteen degrees of unsaturation) by analysis of its HRESIMS (m/z 349.1290 [M + H]+). The UV spectrum of 1 showed a specific absorbance for the quinazoline-4-one at 216.0, 259.0 and 296.0 nm. The 1H, 13C, and HSQC spectra of 1 (Table 1), showed 19 carbon signals for two methylene group, one sp3 hybrid methine group, eight aromatic methine carbons, five sp2 hybrid aromatic quaternary carbons, and three sp2 hybrid quaternary carbons at δC 161.3 (C-4), 165.7 (C-23) and 171.6 (C-14), suggesting the presence of an amide carbonyl, a carboxylic acid, and a C=N carbon, respectively. The 1H and 13C NMR spectra revealed the presence of an ortho-disubstituted benzene ring corresponding to the anthranilate moieties for 1, which is a fragment of the quinazoline-4-one moiety. Analysis of the 1H–1H COSY NMR data led to the identification of the fragment from C-11 through C-12 to C-13. In the HMBC spectrum (Figure 2), correlations from H-15 to C-11, C-12, C-13 and C-14, together with correlations from H2-N-16 to C-13 and C-14 revealed the pyrrolidin-2-iminium moiety. The HMBC correlations from H-11 and H2-12 to C-2 indicated that the pyrrolidin-2-iminium moiety was attached to C-2 through C-11. Protons and carbons signals for another ortho-disubstituted benzene ring were observed from the NMR data. Considering the molecular formula, as well as the unsaturation requirements for 1, this ortho-disubstituted benzene ring was attached to N-3 through C-17. In the 1H spectrum, three exchangeable protons were observed. The proton at δH 10.29 was assigned to N-15 by its HMBC correlations to C-11, C-12, C-13 and C-14. The HMBC correlations from the other two exchangeable protons at δH 9.25 and 8.75 to C-13 and C-14 indicated the carbon signal (δC 171.6) should be an iminium positive ion. On the basis of these data, the structure of compound 1 was established.
Table 1.
NMR spectroscopic data forauranomides A and B (1 and 2) in DMSO-d6.
| Auranomide A | Auranomide B | |||||
|---|---|---|---|---|---|---|
| Position | δC mult. | δH (J in Hz) | HMBC a | δC mult. | δH (J in Hz) | |
| 2 | 155.4, C | 155.3, C | ||||
| 4 | 161.3, C | 161.3, C | ||||
| 5 | 120.9, C | 120.8, C | ||||
| 6 | 126.5, CH | 8.12, dd (8.0, 1.0) | 4, 8, 10 | 126.5, CH | 8.12, dd (8.0, 1.0) | |
| 7 | 127.2, CH | 7.58, t (8.0) | 5, 9 | 127.3, CH | 7.59, t (8.0) | |
| 8 | 134.9, CH | 7.89, td (8.0, 1.0) | 6, 10 | 135.0, CH | 7.90, td (8.0, 1.0) | |
| 9 | 127.1, CH | 7.72, d (8.0) | 5, 7 | 127.2, CH | 7.73, d (8.0) | |
| 10 | 146.7, C | 146.6, C | ||||
| 11 | 60.3, CH | 4.52, dd (8.5, 5.0) | 2, 14 | 60.2, CH | 4.51, dd (8.5, 5.0) | |
| 12 | 27.0, CH2 | 1.98, m | 2, 11, 13, 14 | 26.9, CH2 | 2.00, m | |
| 2.30, m | 2.31, m | |||||
| 13 | 29.3, CH2 | 2.76, m | 11, 12, 14 | 29.3, CH2 | 2.81, 2.75, m | |
| 14 | 171.6, C | 171.7, C | ||||
| 15 | N-H, br s, 10.29 | 11, 12, 13, 14 | N-H, br s, 10.32 | |||
| 16 | N-H, br s, 8.75 | 13, 14 | N-H, br s, 8.90 | |||
| N-H, br s, 9.25 | 13, 14 | N-H, br s, 9.29 | ||||
| 17 | 135.7, C | 135.9, C | ||||
| 18 | 128.8, C | 127.4, C | ||||
| 19 | 131.9, CH | 8.16, dd (7.5, 1.0) | 17, 21, 23 | 131.8, CH | 8.19, dd (7.5, 1.0) | |
| 20 | 130.3, CH | 7.72, t (7.5) | 18, 22 | 130.4, CH | 7.76, td (7.5, 1.0) | |
| 21 | 133.7, CH | 7.84, td (7.5, 1.0) | 17, 19 | 134.3, CH | 7.89, td (7.5, 1.0) | |
| 22 | 131.3, CH | 7.65, d (7.5) | 18, 20 | 131.5, CH | 7.71, d (7.5) | |
| 23 | 165.7, C | 164.5, C | ||||
| 24 | 52.5, CH3 | 3.66, s | ||||
a HMBC correlations, optimized for 8 Hz, are from proton(s) stated to the indicated carbon.
Figure 2.
Key HMBC correlations for compounds 1 and 3.
High resolution ESIMS(+) analysis of 2 revealed a pseudomolecular ion at m/z 363.1451 [M]+, consistent with the molecular formula C20H18N4O3, and corresponding to fourteen degrees of unsaturation. Compound 2 had a very similar UV spectrum to that of 1. The 1H, 13C, and HSQC spectra of 2 showed 20 carbon signals which were very similar to those of 1 except for the presence of methoxyl group signals (δH 3.36, δC 52.5). In the HMBC spectrum, correlation from H3-24 to C-23 indicated that the methoxyl group was attached to the carboxyl and generated a carboxylic acid methyl ester. The exchangeable proton at δH 10.32 was assigned to H-N-15 by the HMBC correlations to C-11, C-12, C-13 and C-14. The other exchangeable protons at δH 8.89 and 9.24 were assigned to H-N-15 by the HMBC correlations of H-N-16 to C-13 and C-14. On the basis of these data, the structure of compound 2 was established as the methyl ester of 1.
The molecular formula of 3 was determined to be C19H16N4O3 (fourteen degrees of unsaturation) by analysis of its HRESIMS (m/z 371.1132 [M + Na]+) and NMR data (Table 2), the same as that of auranomide A (1). It also showed the specific UV spectrum (λmax 219, 259 and 301 nm) for quinazolin-4-one. The 1H, 13C, and HSQC spectra of 3 showed 19 carbon signals for two methylene groups, one sp3 methine group, eight aromatic methine carbons, five sp2 aromatic quaternary carbons, and three sp2 quaternary carbons at δC 161.0 (C-4), 166.9 (C-23) and 173.7 (C-14), suggesting the presence of three carbonyl carbons. The 1H and 13C NMR spectra revealed the presence of an ortho-disubstituted benzene ring corresponding to two anthranilate moieties in compound 3. Analysis of the 1H–1H COSY NMR data led to the identification of the fragment from C-11 through C-12 to C-13. In the HMBC spectrum (Figure 2), correlations from H-N-16 to C-11, C-12 and C-18, as well as from H2-12 to C-2 and C-11 revealed the connection from C-2 through C-11, N-16 to C-23. On the basis of these data, the structure of compound 3 was established.
Table 2.
NMR spectroscopic data forauranomide C (3) in DMSO-d6.
| Position | δH (J in Hz) | HMBC a | |
|---|---|---|---|
| 2 | 155.8, C | ||
| 4 | 161.0, C | ||
| 5 | 121.0, C | ||
| 6 | 126.8, CH | 8.19, dd (8.0, 1.0) | 4, 8, 10 |
| 7 | 128.8, CH | 7.60, t (8.0) | 5, 9 |
| 8 | 135.2, CH | 7.91, td (8.0, 1.0) | 6, 10 |
| 9 | 127.4, CH | 7.76, d (8.0) | 5, 7 |
| 10 | 145.9, C | ||
| 11 | 53.2, CH | 4.16, m | 2, 12, 13 |
| 12 | 24.1, CH2 | 2.15, 2.34, m | 2, 11, 13, 14 |
| 13 | 30.8, CH2 | 2.29, m | 11, 12, 14 |
| 14 | 173.7, C | ||
| 15 | N-H, 6.76, brs | 13, 14 | |
| N-H, 7.26, brs | 14 | ||
| 16 | N-H, 8.82, d, (7.0) | 11, 12, 18 | |
| 17 | 130.7, C | ||
| 18 | 131.2, C | ||
| 19 | 128.9, CH | 7.78, dd (8.5, 1.5) | 17, 21, 23 |
| 20 | 127.6, CH | 7.59, td (8.5, 1.5) | 18, 22 |
| 21 | 128.6, CH | 7.67, td (8.5, 1.5) | 17, 19 |
| 22 | 133.0, CH | 7.64, dd (8.5, 1.5) | 18, 20 |
| 23 | 166.9, C |
a HMBC correlations, optimized for 8 Hz, are from proton(s) stated to the indicated carbon.
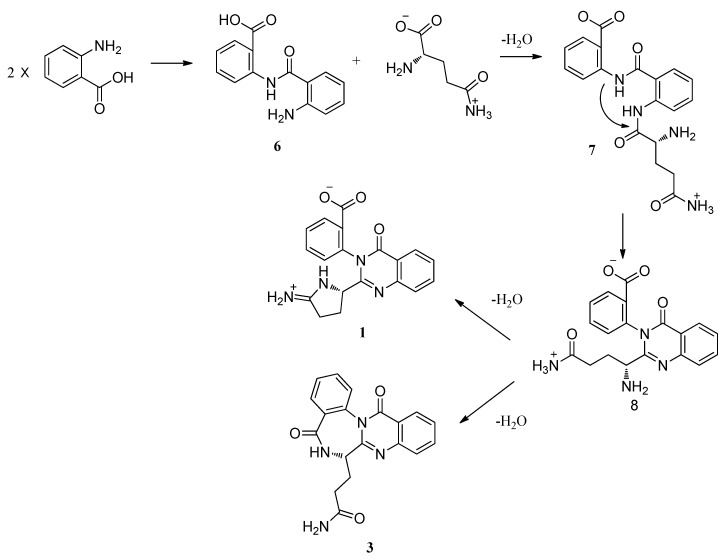
Auranomides A and B are a new class of alkaloids which contain the moiety of 2-pyrrolidin-2-iminium quinozoline-4-one. Quinozolin-4-one derivatives have been isolated from several fungi and originated via similar biosynthesis pathways [17,22,23]. Auranthine (4) was biosynthesized by Penicillium aurantiogriseum [24,25]. Scheme 1 shows a plausible biosynthesis pathway for quinozolin-4-ones analogues (1–3). Two molecules of anthranilic acid were incorporated into 2-(2-aminobenzamido)-benzoic acid (6). A subsequent incorporation of glutamine yielded 7. The amino group of anthranilic acid reacted with the carbonyl carbons of glutamine to yield 8. The primary amino group of glutamine could then react with the terminal amide and the carbonyl carbons of anthranilic acid to form 1 and 3, respectively. From the perspective of biosynthesis, the absolute configurations of C-11 for 1–3 were assigned as 11S.
Antitumor activity of auranomides A–C (1–3) was evaluated in several cells lines by the CCK-8 method. As shown in Table 3, auranomide B exhibited the most potent inhibitory effect against human myelogenous leukemia HEPG2 cells, with an IC50 value of 0.097 μmol/mL. These compounds were also assessed for activities against methicillin-resistant Staphylococcus aureus (MRSA, Clinical isolates, Beijing Chao-yang Hospital, Beijing, China), Candida albicans and synergistic antifungal activity with ketoconazole. None of them showed activities at low concentration (MICs > 100 μg/mL).
Scheme 1.
Plausible biosynthesis pathway for quinozolin-4-ones analogues.
Table 3.
Inhibitory effect of auranomides A–C on the proliferation of tumor cell lines assayed by the CCK-8 method.
| Compound | Antitumor (Inhibition Rate at 100 μg/mL) | |||
|---|---|---|---|---|
| K562 | ACHN | HEPG2 | A549 | |
| Auranomide A | 20.48 | 16.45 | 16.68 | 1.04 |
| Auranomide B | 76.36 | 75.31 | 73.28 | 30.46 |
| Auranomide C | 5.78 | 8.74 | 10.72 | 16.90 |
3. Experimental Section
3.1. General Experimental Procedures
UV data were recorded on a Mariner System 5304 instrument. IR spectra were recorded on a Nicolet 5700 FT-IR Microscope spectrometer (FT-IR Microscope Transmission). NMR spectra were recorded on a Varian Inova 500 MHz spectrometer at 500.103 MHz for 1H and 125.762 MHz for 13C in DMSO-d6 using solvent signals (DMSO; δH 2.50/δC 39.5) as reference; the coupling constants were in Hz. ESIMS spectra were recorded with a ABI Mariner ESI-TOF. Column chromatography was performed with silica gel (200–300 mesh, Qingdao Haiyang Chemical Factory) and Sephadex LH-20 (Pharmacia Co.) columns. HPLC was performed using an Agilent Chromatorex C18 (5 μm) semipreparative column (9.4 × 250 mm). ODs were read by Envision 2103 multilabel reader (PerkinElmer, Waltham, Massachusetts, USA).
3.2. Fungal Material and Cultivation
The fungus Penicillium aurantiogriseum was obtained from marine mud of the Bohai Sea and identified by analysis of internal transcribed spacer (ITS) regions including ITS1, 5.8S rRNA and ITS2 (GenBank Accession Number: HM587449) and morphology. The strain was deposited at the China General Microbiological Culture Collection Center (CGMCC) in the Institute of Microbiology, Chinese Academy of Sciences, Beijing. The fermentation medium of the strain consisted of 200 g potato infusion, 20 g glucose, 0.25 g (NH4)2HPO4 and 20 g agar powder in 1 L artificial sea water. Altogether, thirty 1 L Erlenmeyer flasks containing 200 mL of the fermentation medium were incubated without rotation at 25 °C for 14 days.
3.3. Extraction and Isolation
The fermentation product was exhaustively extracted with EtOAc:MeOH:AcOH (80:15:5) to yield an extract (3.4 g). The residue was suspended in H2O and then partitioned with EtOAc. The EtOAc fraction was chromatographed on a reduced pressure silica gel column using a gradient of CH2Cl2 in MeOH to afford 10 fractions. The third fraction was subjected to a Sephadex LH-20 column [petroleum ether-CH2Cl2-MeOH (5:5:1)] to give compound 4 (6.5 mg) and four sub-fractions. The third sub-fraction was further purified by reversed-phase HPLC to afford auranomides A (1, 1.2 mg), B (2, 1.3 mg) and C (3, 2.1 mg). The eighth fraction was subjected to a Sephadex LH-20 column [petroleum ether-CH2Cl2-MeOH (5:5:1)] to give five sub-fractions. The second sub-fraction was further purified by reversed-phase HPLC to afford compound 5 (7.2 mg).
Auranomide A (1): White amorphous powder; [α]20D +14.9 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 296 (3.22), 259 (3.40), 216 (3.93), 213 (3.95) nm; IR νmax 3394, 3189, 1681, 1609, 1473, 1431, 1401, 1275, 1205, 1136, 1049, 1027, 1007 cm−1; 1H and 13C NMR data, see Table 1; ESIMS m/z 349 [M + H]+; HRESIMS m/z 349.1290 [M + H]+ (calcd for C19H17N4O3, 349.1295).
Auranomide B (2): Yellow oil; [α]20D +10.8 (c 0.10, MeOH); UV (MeOH) λmax 296 (3.28), 259 (3.61), 217 (4.26), 213 (4.26) nm; IR νmax 3194, 2851, 1682, 1609, 1574, 1473, 1436, 1296, 1277, 1203, 1136, 1049, 1027, 1007, 964 cm−1; 1H and 13C NMR data, see Table 1; ESIMS m/z 363 [M]+; HRESIMS m/z 363.1451 [M]+ (calcd for C20H19N4O3, 363.1452).
Auranomide C (3): Yellow oil; [α]20D −63.0 (c 0.10, MeOH); UV (MeOH); λmax (log ε) 301 (2.72), 259 (3.10), 219 (3.69), 213 (3.68) nm; IR νmax 3404, 2958, 1679, 1619, 1456, 1439, 1392, 1335, 1302, 1257, 1207, 1137, 1028, 1006 cm−1; 1H and 13C NMR data, see Table 2; ESIMS m/z 371 [M + Na]+; HRESIMS m/z 371.1132 [M + Na]+ (calcd for C19H16N4O3Na, 371.1120).
3.4. Antitumor Activity
Human myelogenous leukemia cell line K562, human renal cell carcinoma cell line ACHN, human hepatocellular liver carcinoma cell line HEPG2 and human lung adenocarcinoma cell line A549 were routinely cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum at 37 °C for 4 h, in an incubator with a humidified atmosphere of 5% CO2. The adherent cells at their logarithmic growth stage were digested and were inoculated onto 96-well culture plates at a density of 1.0 × 104 cell/well for the determination of proliferation. Test samples were added to the medium, and incubation was continued for 72 h. Coloration substrate, cell counting kit-8 (CCK-8), was added to the medium followed by further incubation for 3 h. Absorbance at 450 nm with a 600 nm reference was measured thereafter. Medium and DMSO control wells, in which the compound was absent, were included in all of the experiments in order to eliminate the influence of DMSO. The inhibitory rate of cell proliferation was calculated by the following formula (Equation 1):
Inhibition Rate (%) = (ODcontrol − ODtreat)/ODcontrol (1)
The IC50 values (the concentration of a compound that is required for 50% inhibition) were calculated from the corresponding log-dose inhibition curve by the LOGIT method.
3.5. Antibacterial Assay
The clinical methicillin-resistant Staphylococcus aureus (MRSA) strain was used as the test strain for antibacterial bioassay. Fresh Mueller-Hinton Broth medium (40 μL) was added to each well of a sterilized 96-well microtiter plate (Greiner, Germany), 2 μL of the samples to be tested were added to the test wells, then 40 μL of the test strain solutions were added to each well. The plate was incubated at 37 °C overnight. The anti-MRSA positive control was vancomycin and the minimal inhibition concentrations (MICs) were checked by measuring and comparing the optical densities of the blank control and tested wells. All the experiments were tested in triplicate.
3.6. Antifungal and Synergistic Antifungal Assay
Candida albicans SC5314 was used as a test strain for the antifungal and synergistic antifungal bioassay according to a previous paper [18]. All the experiments were carried out in flat bottom, 96-well microtiter plates (Greiner, Germany), using a broth microdilution protocol modified from the Clinical and Laboratory Standards Institute M-27A methods [26]. Overnight cultures were picked to prepare the strain solution with medium RPMI 1640 at a concentration of 1 × 104 cfu/mL. Two μL of the samples to be tested was added to the test wells in 96 well-plates followed by 80 μL strain solution. The test plates were incubated at 35 °C for 16 h. The antifungal positive control was ketoconazole and MICs were determined by measuring and comparing the optical densities of the blank control and tested wells. For the synergistic antifungal assay, ¼ of the concentration of ketoconazole needed for inhibition in the antifungal assay was supplemented into the strain solution. All other procedures were the same as the antifungal assay. All the experiments were tested in triplicate.
4. Conclusions
By changing the fermentation conditions, such as changing the carbon and nitrogen source as well as the marine derived fungus Penicillium aurantiogriseum, completely different natural products were produced including three novel quinazolin-4-ones auranomides A–C. Among them, auranomide B showed moderate antitumor activity. The possible biosynthetic pathway for these novel alkaloids was proposed which begins with two molecules of anthranilic acid and involves one molecule of glutamine. This study proved that the OSMAC approach seems effective in searching for natural products with new structures.
Acknowledgments
This work was supported in part by grants from National Natural Science Foundation of China (30911120483, 81102356, 30901849, 30911120484), the CAS Pillar Program (XDA04074000) and the Ministry of Science and Technology of China (2011ZX11102-011-11, 2012CB721006, 2007DFB31620). L. Z. is an Awardee for National Distinguished Young Scholar Program in China.
References
- 1.Liu J.-F., Kaselj M., Isome Y., Ye P., Sargent K., Sprague K., Cherrak D., Wilson C.J., Si Y., Yohannes D., et al. Design and synthesis of a quinazolinone natural product-templated library with cytotoxic activity. J. Comb. Chem. 2005;8:7–10. doi: 10.1021/cc050108g. [DOI] [PubMed] [Google Scholar]
- 2.Birch H.L., Buckley G.M., Davies N., Dyke H.J., Frost E.J., Gilbert P.J., Hannah D.R., Haughan A.F., Madigan M.J., Morgan T., et al. Novel 7-methoxy-6-oxazol-5-yl-2,3-dihydro-1H-quinazolin-4-ones as IMPDH inhibitors. Bioorg. Med. Chem. Lett. 2005;15:5335–5339. doi: 10.1016/j.bmcl.2005.06.108. [DOI] [PubMed] [Google Scholar]
- 3.Al-Deeb A.O., Alafeefy A.M. Sythesis of Some new 3H-quinazolin-4-one derivatives as potential antitubercular agents. World Appl. Sci. J. 2008;5:94–99. [Google Scholar]
- 4.Alagarsamy V., Giridhar R., Yadav M.R., Revathi R., Ruckmani K., Clercq E.D. AntiHIV, antibacterial and antifungal activities of some novel 1,4-disubstituted-1,2,4-triazolo[4,3-a] quinazolin-5(4H)-ones. Indian J. Pharm. Sci. 2006;68:532–535. doi: 10.4103/0250-474X.27840. [DOI] [Google Scholar]
- 5.Kumar A., Rajput C.S. Synthesis and anti-inflammatory activity of newer quinazolin-4-one derivatives. Eur. J. Med. Chem. 2009;44:83–90. doi: 10.1016/j.ejmech.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Ismail M.A.H., Barker S., Abou El Ella D.A., Abouzid K.A.M., Toubar R.A., Todd M.H. Design and synthesis of new tetrazolyl- and carboxy-biphenylylmethyl-quinazolin-4-one derivatives as angiotensin II AT1 Receptor Antagonists. J. Med. Chem. 2006;49:1526–1535. doi: 10.1021/jm050232e. [DOI] [PubMed] [Google Scholar]
- 7.Mohamed M.S., Kamel M.M., Kassem E.M.M., Abotaleb N., Abd El-moez S.I., Ahmed M.F. Novel 6,8-dibromo-4(3H)quinazolinone derivatives of anti-bacterial and anti-fungal activities. Eur. J. Med. Chem. 2010;45:3311–3319. doi: 10.1016/j.ejmech.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Pandey S.K., Singh A., Singh A. Nizamuddin antimicrobial studies of some novel quinazolinones fused with [1,2,4]-triazole, [1,2,4]-triazine and [1,2,4,5]-tetrazine rings. Eur. J. Med. Chem. 2009;44:1188–1197. doi: 10.1016/j.ejmech.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 9.Blangy A., Lane H.A., d’Herin P., Harper M., Kress M., Nigg E.A. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 10.Theoclitou M.-E., Aquila B., Block M.H., Brassil P.J., Castriotta L., Code E., Collins M.P., Davies A.M., Deegan T., Ezhuthachan J., et al. Discovery of (+)-N-(3-Aminopropyl)-N-[1-(5-benzyl-3-methyl-4-oxo-[1,2]thiazolo[5,4-d]pyrimidin-6-yl)-2-methylpropyl]-4-methylbenzamide (AZD4877), a kinesin spindle protein inhibitor and potential anticancer agent. J. Med. Chem. 2011;54:6734–6750. doi: 10.1021/jm200629m. [DOI] [PubMed] [Google Scholar]
- 11.Kantarjian H., Padmanabhan S., Stock W., Tallman M., Curt G., Li J., Osmukhina A., Wu K., Huszar D., Borthukar G., et al. Phase I/II multicenter study to assess the safety, tolerability, pharmacokinetics and pharmacodynamics of AZD4877 in patients with refractory acute myeloid leukemia. Invest. New Drugs. 2012;30:1107–1115. doi: 10.1007/s10637-011-9660-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wattanapiromsakul C., Forster P.I., Waterman P.G. Alkaloids and limonoids from bouchardatia neurococca: systematic significance. Phytochemistry. 2003;64:609–615. doi: 10.1016/s0031-9422(03)00205-x. [DOI] [PubMed] [Google Scholar]
- 13.Sun H.H., Barrow C.J., Cooper R. Benzomalvin D, A new 1,4-benzodiazepine atropisomer. J. Nat. Prod. 1995;58:1575–1580. doi: 10.1021/np50124a015. [DOI] [Google Scholar]
- 14.Witt A., Bergman J. Recent developments in the field of quinazoline chemistry. Curr. Org. Chem. 2003;7:659–677. doi: 10.2174/1385272033486738. [DOI] [Google Scholar]
- 15.Jiao R.H., Xu S., Liu J.Y., Ge H.M., Ding H., Xu C., Zhu H.L., Tan R.X. Chaetominine, a cytotoxic alkaloid produced by endophytic Chaetomium sp. IFB-E015. Org. Lett. 2006;8:5709–5712. doi: 10.1021/ol062257t. [DOI] [PubMed] [Google Scholar]
- 16.Maskey R.P., Shaaban M., Grün-Wollny I., Laatsch H. Quinazolin-4-one derivatives from Streptomyces isolates. J. Nat. Prod. 2004;67:1131–1134. doi: 10.1021/np0305425. [DOI] [PubMed] [Google Scholar]
- 17.Xin Z.H., Fang Y., Du L., Zhu T., Duan L., Chen J., Gu Q.-Q., Zhu W.-M. Aurantiomides A−C, quinazoline alkaloids from the sponge-derived fungus Penicillium aurantiogriseum SP0-19. J. Nat. Prod. 2007;70:853–855. doi: 10.1021/np060516h. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L.X., Yan K.Z., Zhang Y., Huang R., Bian J., Zheng C.S., Sun H.X., Chen Z.H., Sun N., An R., et al. High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections. Proc. Natl. Acad. Sci. USA. 2007;104:4606–4611. doi: 10.1073/pnas.0609370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu K., Ren B., Wei J., Chen C., Sun J., Song F., Dai H., Zhang L. Verrucisidinol and verrucosidinol acetate, two pyrone-type polyketides isolated from a marine derived fungus, Penicillium aurantiogriseu. Mar. Drugs. 2010;8:2744–2754. doi: 10.3390/md8112744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X., Bolla K., Ashforth E., Zhuo Y., Gao H., Huang P., Stanley S., Hung D., Zhang L. Systematics-guided bioprospecting for bioactive microbial natural products. Antonie van Leeuwenhoek. 2012;101:55–66. doi: 10.1007/s10482-011-9671-1. [DOI] [PubMed] [Google Scholar]
- 21.Yang N., Ren B., Wang J., Zhang L. Isolation and novel natural products from un- or under explored marine actinomycetes. Bull. BISMiS. 2011;2:181–188. [Google Scholar]
- 22.Yeulet S.E., Mantle P.G., Bilton J.N., Rzepa H.S., Sheppard R.N. Auranthine, a new benzodiazepinone metabolite of Penicillium aurantiogriseum. J. Chem. Soc. Perkin Trans. 1. 1986:1891–1894. [Google Scholar]
- 23.Larsen T.O., Frydenvang K., Frisvad J.C. UV-guided screening of benzodiazepine producing species in Penicillium. Biochem. Syst. Ecol. 2000;28:881–886. doi: 10.1016/S0305-1978(99)00126-X. [DOI] [PubMed] [Google Scholar]
- 24.Yeulet S.E., Mantle P.G. Biosynthesis of auranthine by Penicillium aurantiogriseum. FEMS Microbiol. Lett. 1987;41:207–210. doi: 10.1111/j.1574-6968.1987.tb02198.x. [DOI] [Google Scholar]
- 25.Larsen T.O., Franzyk H., Jensen S.R. UV-guided isolation of verrucines A and B, novel quinazolines from Penicillium verrucosum structurally related to anacine from Penicillium aurantiogriseum. J. Nat. Prod. 1999;62:1578–1580. doi: 10.1021/np990251p. [DOI] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standard. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts: Approved Standard; Document M27-A2. National Committee for Clinical Laboratory Standards; Wayne, PA, USA: 2002. [Google Scholar]