Summary
Melanoma cell lines and normal human melanocytes were assayed for p53-dependent G1 checkpoint response to ionizing radiation-induced DNA damage. Sixty six percent of melanoma cell lines displayed a defective G1 checkpoint. Checkpoint function was correlated with sensitivity to ionizing radiation with checkpoint-defective lines being radio-resistant. Microarray analysis identified 316 probes whose expression was correlated with G1 checkpoint function in melanoma lines (P≤0.007) including p53 transactivation targets CDKN1A, DDB2 and RRM2B. The 316 probe list predicted G1 checkpoint function of the melanoma lines with 86% accuracy using a binary analysis and 91% accuracy using a continuous analysis. When applied to microarray data from primary melanomas, the 316 probe list was prognostic of four year distant metastases-free survival. Thus, p53 function, radio-sensitivity and metastatic spread may be estimated in melanomas from a signature of gene expression.
Keywords: gene, expression, signature, p53, function, checkpoint, melanoma
INTRODUCTION
The tumor suppressor gene TP53 encodes a homo-tetrameric transcription factor (Meek and Anderson, 2009) that regulates cell division, apoptosis and motility (Huarte et al., 2010; Muller et al., 2011). p53 transactivation of target genes is enhanced by phosphorylation by DNA damage checkpoint kinases (DNA-PK, ATM, ATR, CHK2), and by acetylation by p300 (Meek and Anderson, 2009). DNA damage also stimulates p53 protein translation (Ofir-Rosenfeld et al., 2008). p53 function is inhibited by ubiquitin-mediated proteosomal degradation (Dornan et al., 2004; Lu et al., 2008; Wang et al., 2010; Zhang et al., 2009). p53 transactivates many genes to arrest cell division, including the cyclin-dependent kinase inhibitor p21Waf1 as a primary effector (Abbas and Dutta, 2009; Berns et al., 2004). Inactivation of p53 signaling by any of several mechanisms including mutation or deletion of TP53, inactivation of ATM, amplification of MDM2, expression of viral oncoproteins, or alteration in cofactors or down-stream effectors is associated with enhanced growth and genomic instability (Chen et al., 2009; Lane and Levine, 2010; Levine, 2011; Simpson et al., 2005). Mutation/inactivation of TP53/p53 is a poor prognostic sign in cancer as it is associated with reduced survival and increased tumor relapse (Bild et al., 2009; Murakami et al., 2000). A recent analysis of a genetically engineered mouse model of melanoma concluded that wildtype p53 suppresses the progression of nevi to invasive melanoma (Terzian et al., 2010). This paper is concerned with developing a method to predict p53 protein function in melanoma by using a signature of gene expression.
Although large frequencies of melanoma cell lines and primary and metastatic melanomas (80–90%) displayed high levels of p53 protein (Stretch et al., 1991; Weiss et al., 1993), mutations in TP53 are uncommon in melanomas (10–25%), suggesting that stabilization of p53 protein may occur in melanoma without gene mutation (Daniotti et al., 2004). Consequently, genetic analysis of TP53 mutations may not provide an accurate assessment of p53 function in melanoma cells. p53 plays a key role in the G1 checkpoint response to ionizing radiation(IR)-induced DNA damage (Kastan et al., 1991; Kastan et al., 1992) transactivating numerous genes that participate as effectors of the cellular DNA damage response (Amundson et al., 2008; Zhou et al., 2007). IR is used to treat melanoma (Sloan et al., 2009) and alteration in p53 function has been associated with radioresistance in melanoma cell lines (Satyamoorthy et al., 2000). Although the induction of key p53-responsive transcripts provided an index of p53 signaling function, it was found that the basal level of expression of 175 transcripts provided a better predictor of clonogenic survival after IR than a different set of 25 IR-responsive transcripts (Amundson et al., 2008). In a prior analysis of melanoma cell lines, defective p53-dependent G1 checkpoint function was associated with an altered pattern of gene expression (Kaufmann et al., 2008). Under basal growth conditions checkpoint-defective lines displayed reduced expression of p53 transactivation targets CDKN1A, DDB2 and MDM2 and increased expression of cell cycle genes such as CDC7, cyclin B1, CDK1 and geminin. Here this analysis was extended to include a total of thirty five melanoma cell lines, applying new methods of bioinformatic analysis of gene expression to identify a signature of defective p53 function. The signature could distinguish radio-resistant melanoma cell lines and poor prognosis melanomas.
RESULTS
G1 checkpoint function in NHM and melanoma cell lines
NHMs responded to IR with an effective G1 checkpoint response, manifested as a significant reduction in the fraction of cells in the first half of S phase at 8 h after treatment with IR relative to the sham-treated control (Kaufmann et al., 2008). Thus, we report G1 checkpoint function as the fraction of G1 cells that evaded IR-induced arrest of entry to S. Inactivation of p53 by expression of a dominant-negative allele (TP53-H179Q) or HPV16 E6 oncoprotein ablated the IR-induced G1 arrest in the NHM2 melanocyte culture (Kaufmann et al., 2008).
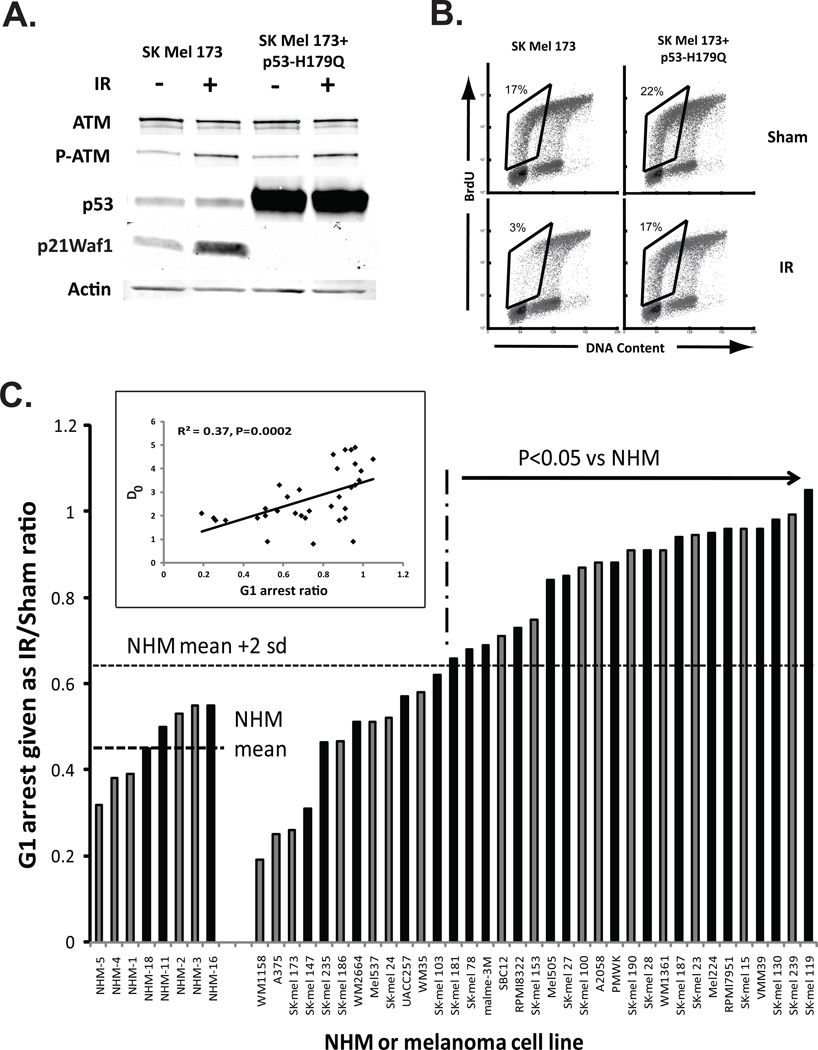
To determine whether IR-induced G1 arrest was p53-dependent in melanoma cell lines, the checkpoint-effective SK-Mel173 melanoma line was transduced with the dominant-negative TP53 allele (H179Q) to inactivate p53 signaling (Simpson et al., 2005) (Figure 1A). Expression of p53-H179Q in SK-Mel173 reduced the basal level of expression of p21Waf1, an effector of p53-dependent G1 checkpoint response (Dulic et al., 1994), and blocked the induction of p21Waf1 by IR. While the parental line responded to IR with 82% reduction in the fraction of cells in the first half of S, melanoma cells expressing the dominant-negative TP53 allele displayed only a 23% IR-induced reduction in early S cells (Figure 1B). A similar degree of reversal of IR-induced G1 arrest was observed in two other melanoma cell lines after expression of p53-H179Q (not shown). Our previous analysis demonstrated melanoma lines with defective G1 checkpoint function failed to induce p21Waf1 after treatment with IR while checkpoint-effective lines and NHM did induce p21Waf1 (Kaufmann et al., 2008). Thus, the radiation-induced G1 arrest was p53-dependent in melanomas as well as melanocytes.
Figure 1.
G1 checkpoint function in NHM strains and melanoma cell lines. Actively proliferating NHM and melanoma cell lines were treated with 1.5 Gy IR or sham-treated, then 6 h later incubated with BrdU for 2 h. Two-channel flow cytometry was used to determine DNA content and incorporation of BrdU in cell nuclei (Kaufmann et al., 2008). The fraction of cells with 2–3 N DNA content that incorporated BrdU (first half of S phase) was quantified. The p53-dependent G1 checkpoint response to IR-induced DNA damage arrested progression of G1 cells into S phase, producing an emptying of the S compartment. A. Western immunoblot analysis of phospho-ATM, p53 and p21Waf1 in SK-Mel173 cells before and after transduction of p53-H179Q (Simpson et al., 2005) was done as previously described (Kaufmann et al., 2008). Samples were harvested 6 h after sham-treatment or treatment with IR. Actin was included as a protein- loading control. B. Flow cytometric analysis of G1 checkpoint function in SK-Mel173 melanoma cells. The indicated fraction of cells in the first half of S is enclosed by the rhombus. Expression of the dominant-negative p53 allele (p53H179Q) reversed the IR-induced emptying of S. C. The ratio of the fractions of cells in the first half of S phase in IR-treated and sham-treated cultures (IR/Sham ratio) was determined as an index of G1 checkpoint function. Results shown as black columns are from the current analysis (mean + sd, N=2–5). Results shown as gray columns were reported previously (Kaufmann et al., 2008). The mean IR/Sham ratio in the eight NHM strains was 0.46 (sd = 0.9). Melanoma cell lines with ratio’s > 0.64 were defective for G1 checkpoint function (P < 0.05). Melanoma cell line annotations are given in the Supplement. The insert shows the correlation between the G1 arrest ratio (IR/sham) and radiosensitivity (D0).
In NHMs an average of 46% of G1 cells evaded the checkpoint and entered S phase in the first 8 h after irradiation with 1.5 Gy IR (n=8, sd=9%) (Figure 1C). Melanoma cell lines as a class displayed a significant defect in G1 checkpoint function. When the entire set of thirty five lines was analyzed and compared to the set of eight NHMs, the inhibition of G1 to S progression after IR was significantly attenuated in melanoma cell lines (P<0.01). Melanoma cell lines that were newly analyzed displayed the same range of variation in G1 checkpoint function as melanoma lines that were analyzed previously. When compared on a line-by-line basis, 23 of 35 lines (66%) were determined to express a significant defect in G1 checkpoint function (P<0.05, one sided Student’s t-test with correction for multiple comparisons). Some melanoma lines (e.g., WM1158, A375, SK-Mel173) displayed effective G1 checkpoint function with less than 30% of cells evading G1 arrest. Other lines (e.g., SK-Mel119, SK-Mel239) displayed full ablation of G1 checkpoint response with over 95% of G1 cells evading G1 arrest. The cutoff for effective versus defective G1 checkpoint function by empirical determination was 64% evasion of the checkpoint. Melanoma cell lines with less than 64% evasion of G1 arrest were not significantly different from the NHMs, while melanoma lines with greater than 64% evasion of G1 arrest were significantly different. The empirically determined cutoff for identification of G1 checkpoint-defective melanoma cell lines was equivalent to two standard deviations above the NHM mean (Figure 1C).
The p53-dependent DNA damage response can not only induce G1 arrest but also apoptosis. Under the conditions of the flow cytometric assay for G1 arrest, apoptotic cells with sub-diploid DNA content were not evident so a colony formation assay was used to quantify radiation-induced cell death. Radio-sensitivity was determined as the D0 value for IR-induced inactivation of clonal expansion (Sambade et al., 2011). The Spearman Correlation test detected a significant correlation between G1 checkpoint function and radio-sensitivity (R2=0.37, P=0.0002) (Figure 1C, insert). Melanoma cell lines with effective p53-dependent G1 checkpoint response to IR were radio-sensitive while lines with defective G1 checkpoint function were radio-resistant (see Supplement for listing of D0 values).
Signatures of defective G1 checkpoint function
Each of the melanoma lines that were evaluated for DNA damage G1 checkpoint function was also analyzed for gene expression using the 44,000 element Agilent printed oligonucleotide microarray. RNA was isolated from undamaged cells in logarithmic phase growth and mRNA expression levels were determined relative to a pooled universal reference (log2 ratio). Quantitative trait analysis (QTA) was performed using the Spearman Correlation test to identify probes whose expression was correlated with G1 checkpoint function. Because some genes are represented by more than one probe on the array, the number of genes that was correlated with G1 checkpoint function was less than the number of probes. Probe lists were derived with P-value cutoffs varying between 0.01 and 0.001 (Table 1). A P-value cutoff of 0.001 or less identified 31 probes while a P-value cutoff of 0.01 identified 441 probes. The probe lists that were obtained at each P-value cutoff were then utilized in a Least Absolute Shrinkage and Selection Operator (LASSO) array tool (Witten and Tibshirani, 2009; Zhao and Simon, 2010) using leave-one-out cross-validation (Table 1). This method returned a predicted value for G1 checkpoint function for each of the melanoma cell lines. The greater the correlation between the observed and predicted values for the G1 checkpoint, the greater the accuracy of the method for identifying G1 checkpoint-effective and -defective melanoma cell lines. The optimal P- value cutoff for predictive accuracy of G1 checkpoint function was 0.007. At this P-value cutoff 316 probes were identified that passed the test (Table 1, the probe list with gene annotations is given in the Supplement). When the observed values for G1 checkpoint function were compared to the values that were predicted by LASSO a high correlation was observed (R2=0.63). Using 64% evasion of G1 arrest as the cutoff for effective versus defective G1 checkpoint function, the LASSO tool predicted G1 checkpoint function in the melanoma lines with 91% accuracy (32 of 35 with correct classification) (Table 1, Figure 2A). Unsupervised hierarchical cluster analysis of the melanoma cell lines using the 316 probe list also separated the G1 checkpoint-effective and -defective lines, with 86% accuracy (Figure 2B). Of the nineteen melanoma lines that were deemed to have high radio-resistance (Sambade et al., 2011), sixteen (84%) were clustered in the G1 checkpoint-defective clade (Chi2, P<0.025).
Table 1.
Spearman correlation test to identify probes whose expression was correlated with G1 checkpoint function.
| Sig. levela | 0.001 | 0.002 | 0.003 | 0.004 | 0.005 | 0.006 | 0.007 | 0.008 | 0.009 | 0.010 |
|---|---|---|---|---|---|---|---|---|---|---|
| Probe listb | 31 | 80 | 135 | 184 | 236 | 285 | 316 | 358 | 400 | 441 |
| LASSO-R2c | 0.48 | 0.54 | 0.42 | 0.59 | 0.53 | 0.47 | 0.63 | 0.60 | 0.57 | 0.55 |
| Accuracyd | 29/35 | 31/35 | 27/35 | 30/35 | 31/35 | 29/35 | 32/35 | 30/35 | 31/35 | 30/35 |
Significance level of the Spearman correlation test.
Number of significant probes. The number of probes passing primary filtration was 23,354.
LASSO prediction of G1 checkpoint function using leave-one-out cross validation. R2 is the proportion of variation in G1 checkpoint function that is explained by the LASSO model.
Accuracy is the number of samples for which G1 checkpoint function (effective vs defective) was predicted correctly by LASSO using leave-one-out cross validation, divided by the total analyzed.
Figure 2.
G1 checkpoint probe list predicts G1 checkpoint function. A. The 316 probe list that was generated by QTA with P = 0.007 was subjected to LASSO with leave-one-out cross validation. LASSO returned an equation for prediction of G1 checkpoint function as: ∑icixi + 0.884 where ci and xi are the coefficient and expression for the i-th transcript, respectively. Values of G1 checkpoint function that were predicted by LASSO were plotted against the observed values. The cutoff value for effective versus defective G1 checkpoint function was 0.64. LASSO correctly classified G1 checkpoint function for 91% of the melanoma cell lines. B. Unsupervised hierarchical cluster of 35 melanoma cell lines using the 316 probes that were correlated with G1 checkpoint function (P = 0.007). Gene expression in melanoma cell lines was visualized using the method of Eisen et al. (Eisen et al., 1998), which organizes lines and transcripts according to similarity. Transcripts that were expressed at greater levels than the group median are shown in red and transcripts that were expressed at lesser levels than the group median are shown in green. The intensity of color is proportional to the separation from the median. The two major clades (branches) in the melanoma cell line dendogram largely represented checkpoint-defective and checkpoint-effective classes. There were five lines that were misclassified for a correct classification rate of 86%. Melanoma lines with mutations in B-Raf and N-Ras were represented in both classes. Melanoma lines with wildtype B-Raf and N-Ras were restricted to the checkpoint-defective class.
The predictive 316 probe list that was obtained by QTA was compared to a probe list of the same size that was obtained by using a Bayesian statistical test. The Bayesian tool generated probe lists based not on the P-value of the correlation of gene expression with G1 checkpoint function, but rather the false discovery rate (FDR). The top 316 probes on the Bayesian list had FDR’s of 0.14 or less. When subjected to LASSO, the Bayesian 316 probe list predicted G1 checkpoint function with 86% accuracy. Hierarchical clustering using the Bayesian list predicted defective G1 checkpoint function with 80% accuracy (results not shown). Thus, two different statistical tools, when applied to the melanoma cell line microarray dataset, returned similar sets of genes with comparable accuracies for prediction of G1 checkpoint function.
A total of 139 probes were common to the QTA and Bayesian 316 probe lists; these probes were correlated with G1 checkpoint function with low P-value (<0.007) and low FDR (<0.14). The common gene set was found to predict G1 checkpoint function with 83% accuracy using LASSO and 86% accuracy using hierarchical clustering (results not shown, the common probe list is given in the Supplement).
G1 checkpoint function is correlated with expression of CDKN1A and p21Waf1 but not CDKN2A and p53
Several p53-regulated transcripts (CDKN1A, DDB2, RRM2B, TRIAP1 and DRAM) were present on the QTA G1 checkpoint probe list and all of these transcripts were negatively correlated with evasion of the G1 checkpoint. Expression of these transcripts diminished as the percent of cells evading the G1 checkpoint increased. CDKN1A expression in the melanoma lines relative to G1 checkpoint function is shown in Figure 3A. The coefficient of correlation between expression of CDKN1A and the fraction of cells evading the G1 checkpoint was −0.54 (P = 0.001, Spearman Correlation Test). All melanoma lines with effective G1 checkpoint function displayed comparatively high expression of CDKN1A (>0.2 log2 ratio). Of the 23 lines with a significant defect in G1 checkpoint function, 9 displayed expression of CDKN1A at or above the level seen in the checkpoint-effective lines. Thus, when used as a marker of G1 checkpoint function, expression of CDKN1A alone misclassified 9/35 melanoma lines for an accuracy of 74%.
Figure 3.
Correlation of G1 checkpoint function with expression of CDKN1A and p21Waf1. A. Expression of CDKN1A mRNA in melanoma cell lines was determined by microarray analysis. The CDKN1A probe was represented ten times on the printed arrays and the average log2 ratio (mRNA in sample divided by mRNA in reference) was determined. The coefficient of correlation between expression of CDKN1A and the fraction of cells evading the G1 checkpoint was −0.54 (P = 0.001, Spearman Correlation Test). B. Expression of p21Waf1 in NHMs and melanoma cell lines was determined by western immunoblot analysis (Kaufmann et al., 2008). Equal amounts of cell lysate protein were separated by gel electrophoresis and after transfer to nylon membranes incubated with anti-p21Waf1 antibody. Immunoblots were scanned and pixel intensities of expression of p21Waf1 were normalized to expression of gamma-tubulin which served as an internal loading control. p21Waf1 expression in melanoma cell lines was expressed relative to the NHM16 culture. The immunoblot image was cropped to remove non-essential lanes. C. The coefficient of correlation between p21Waf1 expression and CDNK1A expression was 0.75 (P<0.0001). D. G1 checkpoint function in melanoma cell lines is shown as a function of expression of p21Waf1 protein (Spearman correlation coefficient = −0.63, P=0.002).
Western immunoblot was done on twenty one melanoma lines to determine the correlation between expression of CDKN1A mRNA and the protein expressed from this gene, p21Waf1. A representative portion of the analysis is shown in Figure 3B. The NHM16 strain expressed p21Waf1 during basal growth conditions and also expressed a smaller immuno-reactive species that may correspond to a C-terminal truncation of p21Waf1 that can be induced in normal skin fibroblasts by a high fluence of UVC (30 J/m2) (Poon and Hunter, 1998). Melanoma cell lines displayed variable expression of full length p21Waf1 (Figure 3B) that was well-correlated with mRNA expression (Spearman correlation coefficient = 0.75, P < 0.0001) (Figure 3C). Two of the twenty one melanoma lines that were examined displayed only the increased mobility protein (e.g., SK-Mel28, Figure 3B). This protein species is commonly observed in transformed cell lines (Poon and Hunter, 1998). The melanoma lines with minimal native protein, including the two with the faster mobility protein, had low CDKN1A mRNA expression (Figure 3C). Some melanoma lines with severe defects in G1 checkpoint function expressed p21Waf1 at or above levels seen in checkpoint-effective melanoma lines and NHMs (Figure 3D). For example, SK-Mel23 with a defective G1 checkpoint (Figure 1) expressed p21Waf1 at the same level as A375 with an effective G1 checkpoint. In spite of the examples in which expression of p21Waf1 was not correlated with G1 checkpoint function, the overall coefficient of correlation between G1 checkpoint function and expression of full length p21Waf1 was −0.63 (P=0.002). Expression of p21Waf1 appeared to have about the same accuracy for prediction of G1 checkpoint function (~80%) as expression of CDKN1A.
p53 is regulated by MDM2 and MDM2 is regulated by p14ARF, one of the protein products of the CDKN2A melanoma suppressor gene locus. Melanomas and melanoma cell lines often display deletion of the CDKN2A gene locus and reduced expression of p16INK4A (Kaufmann et al., 2008), the other protein product of the locus. Seventeen of the melanoma lines in this analysis (49%) displayed severely reduced expression of transcript from the locus (Supplement). Of the 23 melanoma lines with defective G1 checkpoint function, 9 (39%) displayed severely reduced expression of CDKN2A. Of the 12 melanoma lines with effective G1 checkpoint function, 8 (67%) displayed severely reduced CDKN2A mRNA. These percentages were not significantly different (P > 0.1, Chi-square). Thus, reduced expression of CDKN2A was not associated with defects in G1 checkpoint function.
Melanomas have been tested for expression of p53 protein with as many as 85% reported to express high levels of p53 relative to that seen in quiescent nevi (Stretch et al., 1991). The primary melanocyte culture and melanoma cell lines that were tested for expression of p21Waf1 also were tested for expression of p53 (Figure 4). Melanoma cell lines with defective G1 checkpoint function had highly variable levels of p53 protein ranging from 2-times the NHM16 value (SK-Mel119) to 209-times the NHM16 value (Mel505). Similarly, melanoma lines with effective G1 checkpoint function expressed p53 at levels ranging between 1.5- and 55-fold the levels seen in NHM16. Consequently, p53 protein expression was not correlated with G1 checkpoint function in the melanoma cell lines (correlation coefficient = 0.04, P=0.9) (Figure 4B).
Figure 4.
Expression of p53 is not correlated with G1 checkpoint function. A. p53 protein expression was determined by western immunoblot. Pixel intensities of p53 expression were normalized to expression of gamma-tubulin which served as a loading control. Normalized p53 expression in melanoma cell lines was expressed relative to the NHM16 culture. The immunoblot image was cropped to remove non-essential lanes. B. G1 checkpoint function in melanoma cell lines (fraction evading G1 arrest) was expressed relative to levels of expression of p53 protein (normalized pixel intensities). The Spearman correlation coefficient was 0.04, P=0.9).
Transactivation of CDKN1A requires numerous post-translational modifications of p53. One of these modifications, methylation of lysine 372, is produced by SETD7 in response to DNA damage (Ivanov et al., 2007). SETD7 mRNA expression was identified by QTA to correlate with G1 checkpoint function (P=0.002, Supplement); reduced expression of SETD7 was associated with reduced G1 checkpoint function.
Pathway analysis of the 316 probe list identified two major categories of over-represented genes, p53 signaling and protein translation. Fourteen ribosomal protein genes were on the list and all were negatively correlated with checkpoint evasion. By comparing melanoma lines to normal melanocytes it was evident that these ribosomal genes were over-expressed in G1 checkpoint-effective melanoma cell lines (results not shown, the full microarray dataset is deposited in Gene Expression Omnibus). In contrast, ribosomal genes were expressed at levels seen in normal melanocytes in G1 checkpoint-defective melanoma cell lines.
The predictive 316 probe list and cancer prognosis
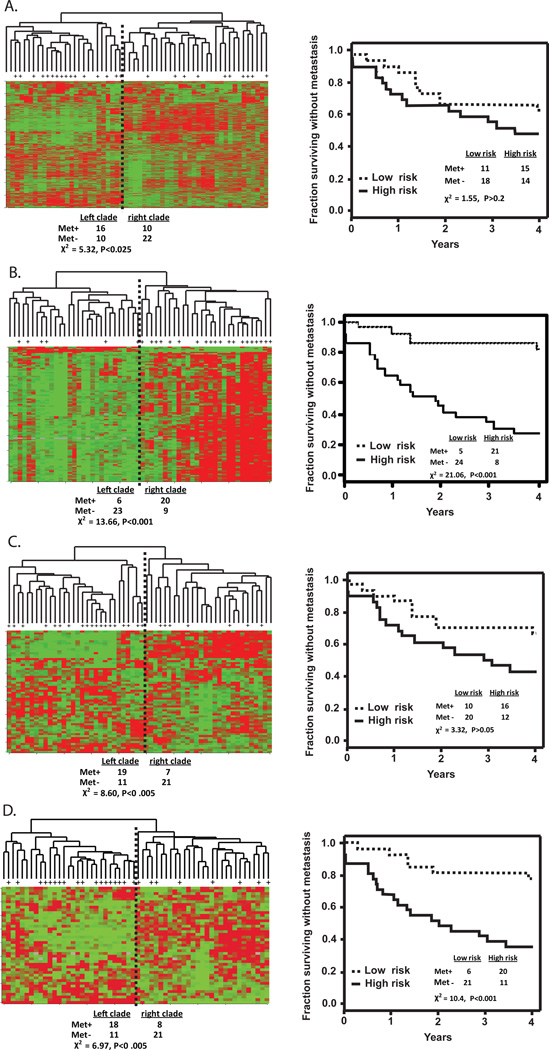
To validate the biological significance of the 316 probe G1 checkpoint signature, an independent sample of melanomas was analyzed. Gene expression was determined for fifty eight primary melanomas for which four year distant metastasis-free survival rates were known (Winnepenninckx et al., 2006). Cy3 and Cy5 pixel intensity values were obtained from an online source (http://www.ebi.ac.uk/arrayexpress/experiments/E-TABM-1) and loaded into BRB array tools to compute Cy3/Cy5 ratios. A total of 6307 probes passed filtration. Unsupervised hierarchical analysis of the 6307 probe set reproduced the original published result showing that there was significant (P=0.02) separation of the incidence of distant metastasis between the two clades (Figure 6A). A survival risk prediction (SRP) tool was applied which used the first two principal components in the microarray data set to stratify samples into high-risk and low-risk groups. SRP with the entire 6307 probe set did not distinguish primary melanomas with or without metastatic spread (Figure 6A). Significance analysis of microarray (SAM) was applied to identify genes whose expression was correlated significantly (FDR<5%) with metastasis-free survival. SAM returned 101 probes that were correlated with development of distant metastases. The SAM probe list segregated the melanoma samples into two clades with significant differences in metastasis (Figure 6B, P=0.0002)) and when the SRP tool was applied to this 101 probe list, a statistically significant separation of low and high risk groups was obtained (P<0.0001). A 254 probe set was originally reported in Winnepenninckx et al. to distinguish primary melanomas with and without metastatic spread. A total of 62 probes from this set were present in the 6307 probe set. These 62 probes also segregated the melanoma samples into clades with significant (P=0.003) differences in development of distant metastases (Figure 6C) but with these 62 probes the separation of low risk and high risk groups by the SRP tool failed to achieve statistical significance (P=0.07).
The 6307 probe set contained 43 probes that overlapped with the 316 probe G1 checkpoint signature. These 43 probes were used with LASSO to predict G1 checkpoint function in the melanoma cell lines, yielding a high correlation with values that were predicted using the 316 probe set (R2=0.61, P<0.0001, Supplement). Thus, the 43 probe set could serve as a surrogate for the predictive 316 probe signature. Hierarchical clustering of the primary melanoma samples using these 43 probes also produced two clades with significant (P=0.008) differences in distant metastasis (Figure 6D). Using the 43 probe list the SRP tool generated two groups with significantly different frequencies of metastatic spread (P=0.001). Thus a signature of G1 checkpoint function appeared to be prognostic for development of distant metastases in melanoma patients.
DISCUSSION
DNA damage checkpoints suppress carcinogenesis by slowing clonal expansion and enhancing genetic stability (Kaufmann and Kaufman, 1993). Thus, defects in checkpoint function enhance growth and genetic instability to accelerate malignant progression (Negrini et al., 2010). DNA damage checkpoints also influence the responses of cancer cells to radio- and chemo-therapies (Schwartz et al., 2000; Shao et al., 1997). The quantitative assay of IR-induced G1 arrest indicated that p53 signaling was defective in a large fraction (66%) of melanoma cell lines. Although mutational inactivation of p53 is seen in a minority of melanomas (Daniotti et al., 2004; Weiss et al., 1993), inactivation of the p53 signaling network occurred in a majority of the melanoma cell lines.
The p53-dependent G1 checkpoint response to IR-induced DNA damage was predicted in melanoma cell lines by analysis of basal levels of gene expression. A large number of probes were correlated with G1 checkpoint function and the 316 probe list was found to correctly classify 86–91% of the melanoma lines using two different tools (hierarchical cluster and LASSO). The probe list also appeared to provide prognostic information for primary melanomas. These results suggest that it may be possible to predict p53-dependent G1 checkpoint function, radiosensitivity and propensity for metastatic spread with reasonable accuracy by analysis of gene expression in primary melanomas. Given the size of primary melanomas, methods that are suited for small samples such as quantitative reverse transcription polymerase chain reaction will be needed to quantify mRNAs in the predictive probe set. p53 signaling function may be useful to clinicians for decisions about post-surgery adjuvant radio- and chemo-therapy. It is notable that in addition to its roles to arrest cell division, induce apoptosis and suppress genomic instability, p53 also retards cell migration and suppresses invasion (Muller et al., 2011). Knowledge of the functionality of the p53 signaling pathway may provide an indication to use drugs that target p53-defective cancers. For example, p53-defective cancer cells are sensitive to inhibitors of PLK-1 (Degenhardt et al., 2010) and Chk1 (Ma et al., 2011). Alternatively, p53 function may be restored by using nutlin-3 to inhibit MDM2-p53 interaction ((Ji et al., 2012)
Defective G1 checkpoint function in melanoma lines was in part attributed to reduced expression of CDKN1A mRNA and p21Waf1 protein. p21Waf1 is the p53-regulated cyclin-dependent kinase inhibitor that enforces IR-induced G1 arrest (Dulic et al., 1994). p21Waf1 has been tested as a biomarker of malignant progression in melanoma. In two studies high expression of p21Waf1 was associated with poor prognosis (Alonso et al., 2004; Poyraz et al., 2004); high expression of p21 Waf1 was associated with good prognosis in a third recent study (Gould Rothberg et al., 2009). However, when analyzed in quartiles, survival in this study varied with p21Waf1 expression in a “U”-shaped curve. The hazard ratio was higher for the lowest and highest quartiles than for the two middle quartiles (Gould Rothberg et al., 2009). Given the results presented here indicating that expression of p21Waf1 is correlated with effective p53-dependent G1 checkpoint function, the results indicate either that effective p53 signaling enhances malignant progression or over-expression of p21Waf1 in a subset of melanomas reflects a further loss of growth control. We favor the latter interpretation. It is conceivable that primary melanomas with over-expression of p21Waf1 have acquired additional genetic alterations that render the cancer cells resistant to growth inhibition by the cyclin-dependent kinase inhibitor. The stability of p21Waf1 is regulated by ubiquitin-mediated proteolysis (Abbas and Dutta, 2009) and AKT can phosphorylate p21Waf1 to force cytoplasmic localization (Abukhdeir and Park, 2008) so mechanisms for enhanced expression of p21Waf1 in poor-prognosis melanoma are apparent.
Our previous analysis of gene expression and G1 checkpoint function in melanoma cell lines (Kaufmann et al., 2008) included NHMs in the G1 checkpoint-effective class. NHMs and melanomas display substantial differences in gene expression, especially in known p53-responsive transcripts (Yu et al., 2009). The G1 checkpoint-effective samples were reported to have lower expression of cell proliferation-associated genes such as CDC7, cyclin B1 and CDK1 than G1 checkpoint-defective samples. In the current analysis in which only melanoma cell lines were used to identify genes that were correlated with G1 checkpoint function, proliferation genes were not evident. Thus, melanoma lines with effective and defective p53-dependent G1 checkpoint function expressed proliferation genes at similar levels.
In addition to the reduced expression of five p53 target genes in G1 checkpoint-defective melanoma lines (CDKN1A, DDB2, RRM2, TRIAP1, DRAM), checkpoint-defective lines displayed reduced expression of fourteen ribosomal protein genes relative to checkpoint-effective melanoma lines. Pathway analysis indicated that ribosomal genes were significantly over-represented in the 316 G1 checkpoint-correlated probe list. There is growing evidence that p53 function is responsive to changes in ribosomal proteins (Deisenroth and Zhang, 2010). This is attributed to interactions between certain ribosomal proteins and MDM2, the ubiquitin ligase that regulates p53 protein expression. Depletion of individual ribosomal protein subunits can activate p53 and induce G1 arrest. Of the fourteen ribosomal genes that were correlated with G1 checkpoint, only one, RPS3, has been reported to influence p53 function (Yadavilli et al., 2009). Inspection of mRNA expression revealed that the correlation with G1 checkpoint function was due to the over-expression of the ribosomal genes in the checkpoint-effective lines. The checkpoint-defective lines expressed these mRNA’s at levels similar to those seen in NHMs. Thus, transformation of melanocytes with effective p53 signaling was associated with enhanced expression of a large set of ribosomal protein genes.
G1 checkpoint function was not associated with the major subtypes of melanoma as determined by oncogene mutation status. B-Raf mutations tend to predominate in melanomas in sunlight-damaged skin. N-Ras mutations, while evident on sunlight-damaged skin, predominate in congenital nevi and melanomas (Blokx et al., 2010; Carr and Mackie, 1994; Papp et al., 1999). Mutations in N-Ras and B-Raf were equally distributed between G1 checkpoint-effective and -defective lines indicating that the G1 checkpoint-defective phenotype was not related to the Ras or Raf oncogene mutation that initiates melanomagenesis. However, melanomas with wildtype B-Raf and N-Ras, which we have designated the “epithelial” subtype (Shields et al., 2007), did appear to be more common among the G1 checkpoint-defective lines. While 56% of N-Ras mutant melanoma lines and 65% of B-Raf mutant lines displayed a significant defect in G1 checkpoint function, five of five (100%) of the wildtype lines were defective for the G1 checkpoint. More epithelial subtype melanomas must be examined to determine if this apparent predilection to inactivation of p53 signaling is an element of the subtype.
We previously reported that the epithelial subtype melanoma cell lines with defective G1 checkpoint function all displayed reduced expression of MITF and MITF-regulated transcripts (Shields et al., 2007). Based on this characteristic these lines are in the “invasive” melanoma subtype (Hoek et al., 2006). The “invasive” and “proliferative” subtypes of melanoma (Hoek et al., 2006) that may represent reversible elements of the epithelial-mesenchymal transition (Hanahan and Weinberg, 2011; Hoek and Goding, 2010) were evident in the melanoma cell line hierarchical cluster (results not shown). However, among melanoma cell lines with mutations in N-Ras and B-Raf, expression of MITF was not correlated with G1 checkpoint function, with 11 of 16 MITF-high lines (69%) and 7 of 14 MITF-low lines (50%) being checkpoint-defective (P>0.2).
A limitation of this analysis is the reliance upon established melanoma cell lines for quantitative assessment of p53-dependent G1 checkpoint function. Melanoma cell lines differ from primary melanomas in the frequency of homozygous deletion of the CDKN2A tumor suppressor gene (Walker et al., 1998), but they resemble primary melanomas for expression of proliferative and invasive subtypes (Hoek et al., 2006) and for the distributions and frequencies of DNA copy number variants (Lin et al., 2008; Stark and Hayward, 2007). The inactivation of G1 checkpoint function seen in the melanoma cell lines was not an inevitable consequence of in vitro cell culture as melanoma lines with effective G1 checkpoint function were identified and all NHMs displayed effective G1 checkpoint function. The melanoma cell lines had not acquired mutations in TP53 as the frequency of mutations in TP53 exons was 18% (William Jeck and Norman Sharpless, personal communication), a value similar to that observed in primary melanoma. Ectopic expression of telomerase can extend NHM proliferative lifespan in vitro and high passage telomerase-expressing melanocytes retain effective G1 checkpoint function (unpublished). Although DNA damage response capacity as measured in the melanoma cell lines is presumed to resemble what was present in melanoma cells when harvested from patient tumors, this presumption requires empirical confirmation using primary and secondary explant cultures.
Analysis of melanocyte transformation by mutant B-Raf and N-Ras oncogenes suggests that both pRB and p53 may suppress transformation. The mutant oncogenes produce a phenotype of oncogene-induced senescence (OIS) when expressed in NHM (Bandyopadhyay et al., 2007; Cheung et al., 2008; Gray-Schopfer et al., 2006; Haferkamp et al., 2009a; Haferkamp et al., 2009b; Leikam et al., 2008; Michaloglou et al., 2005; Wajapeyee et al., 2008; Yu et al., 2009; Zhuang et al., 2008). In NHM that were transduced with mutant N-Ras, inactivation of pRB delayed expression of senescence and inactivation of both RB and p53 prevented OIS (Haferkamp et al., 2009b). Expression of mutant B-Raf with a dominant-negative p53, a p16INK4A-resistant CDK4 and telomerase generated growth-factor-independent melanocytes (Garraway et al., 2005). Expression of mutant N-Ras with telomerase, mutant p53 and mutant CDK4 produced transformed melanocytes that grew as invasive melanoma in human skin xenografts (Chudnovsky et al., 2005). CDK4 inactivates pRB and pRB may be required during OIS to induce a closed chromatin state that represses expression of genes that are required for cell proliferation (Cheung et al., 2008). High level expression of mutant B-Raf arrests the growth of melanocytes but lower level expression of mutant B-Raf enhances melanocyte proliferation (Cheung et al., 2008; Yu et al., 2009). Growth of mutant B-Raf-expressing melanocytes was further enhanced by depletion of p53 using shRNA thereby generating a transformed melanocyte line that grew slowly in immuno-compromised mice and displayed deletion of the pRB gene locus (Yu et al., 2009). Bypass of the pRB-dependent OIS barrier may permit melanocytes to proliferate until a p53-dependent replicative senescence checkpoint is activated. Normal melanocytes express very low levels of telomerase, as do neval melanocytes (Miracco et al., 2000), and so proliferative clones of oncogene-transformed melanocytes may arrest growth behind a p53-dependent DNA damage response that is triggered when telomeres have eroded to a deprotected state (Simpson et al., 2005; Soo et al., 2011). Thus, p53 serves to arrest the division of oncogene-transformed melanocytes that were not collected behind the pRB barrier. A recent analysis also demonstrated that the p53 signaling pathway is dysfunctional in a majority of melanomas (Yu et al., 2009).
In summary, analysis of gene expression was used to predict p53-dependent G1 checkpoint function in melanoma cell lines with 86–91% accuracy. This method may provide a means to assess p53 signaling capacity in primary cancer specimens prior to selection of post-surgery adjuvant radio- or chemo-therapy.
METHODS
Cell culture
NHMs and melanoma cell lines were grown and assayed for G1 checkpoint function as previously described (Kaufmann et al., 2008). NHM cultures that were not tested previously were obtained from Clonetech, Inc. A list of the melanoma cell lines, their sources, and growth media are given in the Supplement. Cells were routinely assayed for mycoplasma contamination using commercial kits (Geneprobe, Gene-Probe and PlasmoTest™, Invivogen). Contaminated cultures were cleansed with ciprofloxacin and/or a commercial antibiotic mixture (Plasmocin, Invivogen). Results are reported only for mycoplasma-negative cell cultures. G1 checkpoint function was quantified by flow cytometric determination of incorporation of 5’-bromo-deoxyuridine (BrdU) into DNA 6–8 h after irradiation with 1.5 Gy IR using in the current analysis a RS2000 Biological Irradiator (Rad-Source) or in the previous analysis a Gammacell 40 (Atomic Energy of Canada) cesium irradiator. The two sources were calibrated to produce equivalent G1 and G2 checkpoint responses at equivalent IR doses. The fraction of cells in the first half of S phase in IR-treated cultures was expressed as a percentage of the same fraction scored in sham-treated controls. Western immunoblot analysis to quantify expression of p21Waf1 and p53 was done as previously described (Kaufmann et al., 2008). The melanoma cell lines were previously characterized for sensitivity to IR (Sambade et al., 2011). D0 values (D0 = dose required to reduce the initial surviving fraction to 0.37) were determined by nonlinear regression analysis using GraphPad Prism 5.0.
Biostatistics
G1 checkpoint function was compared among individual melanoma cell lines and a group of seven NHMs, based on a linear mixed-effects model to account for the potential correlation of multiple measurements (median of 3 and with inter-quartile range of 1 to 6) per cell line. A one-sided t-test used the predicted empirical Bayesian estimate for each cell line from the output of the linear mixed-effects model to identify melanoma cell lines with significantly reduced IR-induced G1 arrest in comparison to the group of NHMs. P values were adjusted for multiple comparisons using the method of Benjamini and Hochberg to control for the false discovery rate (Benjamini and Hochberg, 1995).
Bioinformatics
Analysis of gene expression using 4X44K printed oligonucleotide microarrays (Agilent catalog# G4112F) was performed as previously described (Kaufmann et al., 2008). mRNA expression in NHM and melanoma cell lines was expressed relative to a universal human mRNA standard (Stratagene catalog#74000). Analysis of microarray data was done using BRB array tools software (Simon et al., 2007). The quantitative trait analysis (QTA) tool used the Spearman Correlation Coefficient to identify genes whose expression was correlated with G1 checkpoint function in melanoma cell lines. The Least Absolute Shrinkage and Selection Operator (LASSO) tool was used with leave-one-out cross validation to predict G1 checkpoint function from gene expression signatures. The survival risk prediction (SRP) tool used the first two principal components to stratify melanoma samples (Winnepenninckx et al., 2006) into low risk and high risk groups for prediction of outcome. Hierarchical clustering was also used to predict G1 checkpoint-effective and -defective melanoma cell lines. A Bayesian method for selection of significant genes was applied as previously described (Kaufmann et al., 2008).
Supplementary Material
Figure 5.
Analysis of melanoma using G1 checkpoint probe list. Global gene expression profiles of 58 primary melanomas (Winnepenninckx et al., 2006) were analyzed for prediction of four year distant metastasis-free survival. Samples with distant metastasis are indicated by a plus sign. A. Hierarchical cluster analysis and survival risk prediction of 58 primary melanomas using 6307 probe list. B. Cluster analysis and risk prediction with the 101 probe SAM list. C. Cluster analysis and risk prediction with the 62 probes from a 254 probe list that was prognostic for development of distant metastasis (Winnepenninckx et al., 2006). D. Cluster analysis and risk prediction with the 43 probes from the 316 probe G1 checkpoint signature.
Significance.
Successful therapy for metastatic cancer often depends upon p53-dependent apoptosis so it is important to determine p53 function in primary lesions. While mutations in p53 are uncommon in melanoma, abnormalities in p53 function are common. A method is described for estimating p53 signaling function in melanomas from a signature of gene expression. The signature was prognostic for development of distant metastases in melanoma patients. Expression profiling of primary melanomas should be clinically useful.
Acknowledgements
We are grateful to Dr. Alan Houghton, Memorial Sloan-Kettering Cancer Center, for providing melanoma cell lines and Alexandra Balaban for assistance with analysis of melanoma microarray datasets. Supported by PHS grants ES10126, CA16086, CA106991, ES07126 and ES014635.
Abbreviations
- BrdU
5’-bromo-deoxyuridine
- IR
ionizing radiation
- NHM
normal human melanocyte
- QTA
quantitative trait analysis
- LASSO
least absolute shrinkage and selection operator
References
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abukhdeir AM, Park BH. P21 and p27: roles in carcinogenesis and drug resistance. Expert Rev Mol Med. 2008;10:e19. doi: 10.1017/S1462399408000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso SR, Ortiz P, Pollan M, Perez-Gomez B, Sanchez L, Acuna MJ, Pajares R, Martinez-Tello FJ, Hortelano CM, Piris MA, et al. Progression in cutaneous malignant melanoma is associated with distinct expression profiles: a tissue microarray-based study. Am J Pathol. 2004;164:193–203. doi: 10.1016/s0002-9440(10)63110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundson SA, Do KT, Vinikoor LC, Lee RA, Koch-Paiz CA, Ahn J, Reimers M, Chen Y, Scudiero DA, Weinstein JN, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res. 2008;68:415–424. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay D, Curry JL, Lin Q, Richards HW, Chen D, Hornsby PJ, Timchenko NA, Medrano EE. Dynamic assembly of chromatin complexes during cellular senescence: implications for the growth arrest of human melanocytic nevi. Aging Cell. 2007;6:577–591. doi: 10.1111/j.1474-9726.2007.00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statistical Society, Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Bild AH, Parker JS, Gustafson AM, Acharya CR, Hoadley KA, Anders C, Marcom PK, Carey LA, Potti A, Nevins JR, et al. An integration of complementary strategies for gene-expression analysis to reveal novel therapeutic opportunities for breast cancer. Breast Cancer Res. 2009;11:R55. doi: 10.1186/bcr2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokx WA, Van Dijk MC, Ruiter DJ. Molecular cytogenetics of cutaneous melanocytic lesions - diagnostic, prognostic and therapeutic aspects. Histopathology. 2010;56:121–132. doi: 10.1111/j.1365-2559.2009.03452.x. [DOI] [PubMed] [Google Scholar]
- Carr J, Mackie RM. Point mutations in the N-ras oncogene in malignant melanoma and congenital naevi. Br J Dermatol. 1994;131:72–77. doi: 10.1111/j.1365-2133.1994.tb08460.x. [DOI] [PubMed] [Google Scholar]
- Chen B, Simpson DA, Zhou Y, Mitra A, Mitchell DL, Cordeiro-Stone M, Kaufmann WK. Human papilloma virus type16 E6 deregulates CHK1 and sensitizes human fibroblasts to environmental carcinogens independently of its effect on p53. Cell Cycle. 2009;8:1775–1787. doi: 10.4161/cc.8.11.8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung M, Sharma A, Madhunapantula SV, Robertson GP. Akt3 and mutant V600E B-Raf cooperate to promote early melanoma development. Cancer Res. 2008;68:3429–3439. doi: 10.1158/0008-5472.CAN-07-5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudnovsky Y, Adams AE, Robbins PB, Lin Q, Khavari PA. Use of human tissue to assess the oncogenic activity of melanoma-associated mutations. Nat Genet. 2005;37:745–749. doi: 10.1038/ng1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniotti M, Oggionni M, Ranzani T, Vallacchi V, Campi V, Di Stasi D, Torre GD, Perrone F, Luoni C, Suardi S, et al. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene. 2004;23:5968–5977. doi: 10.1038/sj.onc.1207780. [DOI] [PubMed] [Google Scholar]
- Degenhardt Y, Greshock J, Laquerre S, Gilmartin AG, Jing J, Richter M, Zhang X, Bleam M, Halsey W, Hughes A, et al. Sensitivity of cancer cells to Plk1 inhibitor GSK461364A is associated with loss of p53 function and chromosome instability. Mol Cancer Ther. 2010;9:2079–2089. doi: 10.1158/1535-7163.MCT-10-0095. [DOI] [PubMed] [Google Scholar]
- Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010 doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O'rourke K, Koeppen H, Dixit VM. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- Dulic V, Kaufmann WK, Wilson SJ, Tlsty TD, Lees E, Harper JW, Elledge SJ, Reed SI. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Gould Rothberg BE, Berger AJ, Molinaro AM, Subtil A, Krauthammer MO, Camp RL, Bradley WR, Ariyan S, Kluger HM, Rimm DL. Melanoma prognostic model using tissue microarrays and genetic algorithms. J Clin Oncol. 2009;27:5772–5780. doi: 10.1200/JCO.2009.22.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-Schopfer VC, Cheong SC, Chong H, Chow J, Moss T, Abdel-Malek ZA, Marais R, Wynford-Thomas D, Bennett DC. Cellular senescence in naevi and immortalisation in melanoma: a role for p16? Br J Cancer. 2006;95:496–505. doi: 10.1038/sj.bjc.6603283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferkamp S, Scurr LL, Becker TM, Frausto M, Kefford RF, Rizos H. Oncogene-induced senescence does not require the p16(INK4a) or p14ARF melanoma tumor suppressors. J Invest Dermatol. 2009a;129:1983–1991. doi: 10.1038/jid.2009.5. [DOI] [PubMed] [Google Scholar]
- Haferkamp S, Tran SL, Becker TM, Scurr LL, Kefford RF, Rizos H. The relative contributions of the p53 and pRb pathways in oncogene-induced melanocyte senescence. Aging (Albany NY) 2009b;1:542–556. doi: 10.18632/aging.100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Goding CR. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res. 2010;23:746–759. doi: 10.1111/j.1755-148X.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov GS, Ivanova T, Kurash J, Ivanov A, Chuikov S, Gizatullin F, Herrera-Medina EM, Rauscher F, 3rd, Reinberg D, Barlev NA. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol. 2007;27:6756–6769. doi: 10.1128/MCB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Njauw CN, Taylor M, Neel V, Flaherty KT, Tsao H. p53 rescue through HDM2 antagonism suppresses melanoma growth and potentiates MEK inhibition. J Invest Dermatol. 2012;132:356–364. doi: 10.1038/jid.2011.313. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Onyekwere O, Sidransky D, Volgelstein B, Craig R. Participation of p53 protein in the cellular responses to DNA damage. Cancer Research. 1991;51:6304–6311. [PubMed] [Google Scholar]
- Kastan MB, Zhan Q, El-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Kaufmann WK, Kaufman DG. Cell cycle control, DNA repair and initiation of carcinogenesis. Faseb J. 1993;7:1188–1191. doi: 10.1096/fasebj.7.12.8375618. [DOI] [PubMed] [Google Scholar]
- Kaufmann WK, Nevis KR, Qu P, Ibrahim JG, Zhou T, Zhou Y, Simpson DA, Helms-Deaton J, Cordeiro-Stone M, Moore DT, et al. Defective cell cycle checkpoint functions in melanoma are associated with altered patterns of gene expression. J Invest Dermatol. 2008;128:175–187. doi: 10.1038/sj.jid.5700935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikam C, Hufnagel A, Schartl M, Meierjohann S. Oncogene activation in melanocytes links reactive oxygen to multinucleated phenotype and senescence. Oncogene. 2008;27:7070–7082. doi: 10.1038/onc.2008.323. [DOI] [PubMed] [Google Scholar]
- Levine AJ. Introduction: The Changing Directions of p53 Research. Genes Cancer. 2011;2:382–384. doi: 10.1177/1947601911413463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, Laine E, Greulich H, Tseng H, Gates C, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Nguyen TA, Zhang X, Donehower LA. The Wip1 phosphatase and Mdm2: cracking the "Wip" on p53 stability. Cell Cycle. 2008;7:164–168. doi: 10.4161/cc.7.2.5299. [DOI] [PubMed] [Google Scholar]
- Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med. 2011;17:88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek DW, Anderson CW. Posttranslational Modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol. 2009;1:a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, Van Der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Miracco C, Pacenti L, Santopietro R, Laurini L, Biagioli M, Luzi P. Evaluation of telomerase activity in cutaneous melanocytic proliferations. Hum Pathol. 2000;31:1018–1021. doi: 10.1053/hupa.2000.9779. [DOI] [PubMed] [Google Scholar]
- Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami I, Hiyama K, Ishioka S, Yamakido M, Kasagi F, Yokosaki Y. p53 gene mutations are associated with shortened survival in patients with advanced non-small cell lung cancer: an analysis of medically managed patients. Clin Cancer Res. 2000;6:526–530. [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp T, Pemsel H, Zimmermann R, Bastrop R, Weiss DG, Schiffmann D. Mutational analysis of the N-ras, p53, p16INK4a, CDK4, and MC1R genes in human congenital melanocytic naevi. J Med Genet. 1999;36:610–614. [PMC free article] [PubMed] [Google Scholar]
- Poon RY, Hunter T. Expression of a novel form of p21Cip1/Waf1 in UV-irradiated and transformed cells. Oncogene. 1998;16:1333–1343. doi: 10.1038/sj.onc.1201897. [DOI] [PubMed] [Google Scholar]
- Poyraz A, Akyurek N, Gonul Ii, Erdem O. P21 and Bax expression in cutaneous malignant melanomas: correlation with histologic prognostic parameters. J Exp Clin Cancer Res. 2004;23:625–631. [PubMed] [Google Scholar]
- Sambade MJ, Peters EC, Thomas NE, Kaufmann WK, Kimple RJ, Shields JM. Melanoma cells show a heterogeneous range of sensitivity to ionizing radiation and are radiosensitized by inhibition of B-RAF with PLX-4032. Radiother Oncol. 2011 doi: 10.1016/j.radonc.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyamoorthy K, Chehab NH, Waterman MJ, Lien MC, El-Deiry WS, Herlyn M, Halazonetis TD. Aberrant regulation and function of wild-type p53 in radioresistant melanoma cells. Cell Growth Differ. 2000;11:467–474. [PubMed] [Google Scholar]
- Schwartz JL, Jordan R, Kaufmann WK, Rasey J, Russell KJ, Weichselbaum RR. Evidence for the expression of radiation-induced potentially lethal damage being a p53-dependent process. Int J Radiat Biol. 2000;76:1037–1043. doi: 10.1080/09553000050111505. [DOI] [PubMed] [Google Scholar]
- Shao RG, Cao CX, Shimizu T, O'connor PM, Kohn KW, Pommier Y. Abrogation of an S-phase checkpoint and potentiation of camptothecin cytotoxicity by 7-hydroxystaurosporine (UCN-01) in human cancer cell lines, possibly influenced by p53 function. Cancer Res. 1997;57:4029–4035. [PubMed] [Google Scholar]
- Shields JM, Thomas NE, Cregger M, Berger AJ, Leslie M, Torrice C, Hao H, Penland S, Arbiser J, Scott G, et al. Lack of Extracellular Signal-Regulated Kinase Mitogen-Activated Protein Kinase Signaling Shows a New Type of Melanoma. Cancer Res. 2007;67:1502–1512. doi: 10.1158/0008-5472.CAN-06-3311. [DOI] [PubMed] [Google Scholar]
- Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of Gene Expression Data Using BRB-Array Tools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- Simpson DA, Livanos E, Heffernan TP, Kaufmann WK. Telomerase expression is sufficient for chromosomal integrity in cells lacking p53 dependent G1 checkpoint function. J Carcinog. 2005;4:18. doi: 10.1186/1477-3163-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan AE, Nock CJ, Einstein DB. Diagnosis and treatment of melanoma brain metastasis: a literature review. Cancer Control. 2009;16:248–255. doi: 10.1177/107327480901600307. [DOI] [PubMed] [Google Scholar]
- Soo JK, Mackenzie Ross AD, Kallenberg DM, Milagre C, Heung Chong W, Chow J, Hill L, Hoare S, Collinson RS, Hossain M, et al. Malignancy without immortality? Cellular immortalization as a possible late event in melanoma progression. Pigment Cell Melanoma Res. 2011;24:490–503. doi: 10.1111/j.1755-148X.2011.00850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–2642. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- Stretch JR, Gatter KC, Ralfkiaer E, Lane DP, Harris AL. Expression of mutant p53 in melanoma. Cancer Res. 1991;51:5976–5979. [PubMed] [Google Scholar]
- Terzian T, Torchia EC, Dai D, Robinson SE, Murao K, Stiegmann RA, Gonzalez V, Boyle GM, Powell MB, Pollock PM, et al. p53 prevents progression of nevi to melanoma predominantly through cell cycle regulation. Pigment Cell Melanoma Res. 2010;23:781–794. doi: 10.1111/j.1755-148X.2010.00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GJ, Flores JF, Glendening JM, Lin AH, Markl ID, Fountain JW. Virtually 100% of melanoma cell lines harbor alterations at the DNA level within CDKN2A, CDKN2B, or one of their downstream targets. Genes Chromosomes Cancer. 1998;22:157–163. doi: 10.1002/(sici)1098-2264(199806)22:2<157::aid-gcc11>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Wang L, He G, Zhang P, Wang X, Jiang M, Yu L. Interplay between MDM2, MDMX, Pirh2 and COP1: the negative regulators of p53. Mol Biol Rep. 2010 doi: 10.1007/s11033-010-0099-x. [DOI] [PubMed] [Google Scholar]
- Weiss J, Schwechheimer K, Cavenee WK, Herlyn M, Arden KC. Mutation and expression of the p53 gene in malignant melanoma cell lines. Int J Cancer. 1993;54:693–699. doi: 10.1002/ijc.2910540427. [DOI] [PubMed] [Google Scholar]
- Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, Avril MF, Ortiz Romero PL, Robert T, Balacescu O, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- Witten DM, Tibshirani R. Covariance-regularized regression and classification for high-dimensional problems. J R Stat Soc Series B Stat Methodol. 2009;71:615–636. doi: 10.1111/j.1467-9868.2009.00699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadavilli S, Mayo LD, Higgins M, Lain S, Hegde V, Deutsch WA. Ribosomal protein S3: A multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair (Amst) 2009;8:1215–1224. doi: 10.1016/j.dnarep.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Mcdaid R, Lee J, Possik P, Li L, Kumar SM, Elder DE, Van Belle P, Gimotty P, Guerra M, et al. The role of BRAF mutation and p53 inactivation during transformation of a subpopulation of primary human melanocytes. Am J Pathol. 2009;174:2367–2377. doi: 10.2353/ajpath.2009.081057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lin L, Guo H, Yang J, Jones SN, Jochemsen A, Lu X. Phosphorylation and degradation of MdmX is inhibited by Wip1 phosphatase in the DNA damage response. Cancer Res. 2009;69:7960–7968. doi: 10.1158/0008-5472.CAN-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Simon R. Development and validation of predictive indices for a continuous outcome using gene expression profiles. Cancer Inform. 2010;9:105–114. doi: 10.4137/cin.s3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Chou J, Zhou Y, Simpson DA, Cao F, Bushel PR, Paules RS, Kaufmann WK. Ataxia telangiectasia-mutated dependent DNA damage checkpoint functions regulate gene expression in human fibroblasts. Mol Cancer Res. 2007;5:813–822. doi: 10.1158/1541-7786.MCR-07-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang D, Mannava S, Grachtchouk V, Tang WH, Patil S, Wawrzyniak JA, Berman AE, Giordano TJ, Prochownik EV, Soengas MS, et al. C-MYC overexpression is required for continuous suppression of oncogene-induced senescence in melanoma cells. Oncogene. 2008;27:6623–6634. doi: 10.1038/onc.2008.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.