Abstract
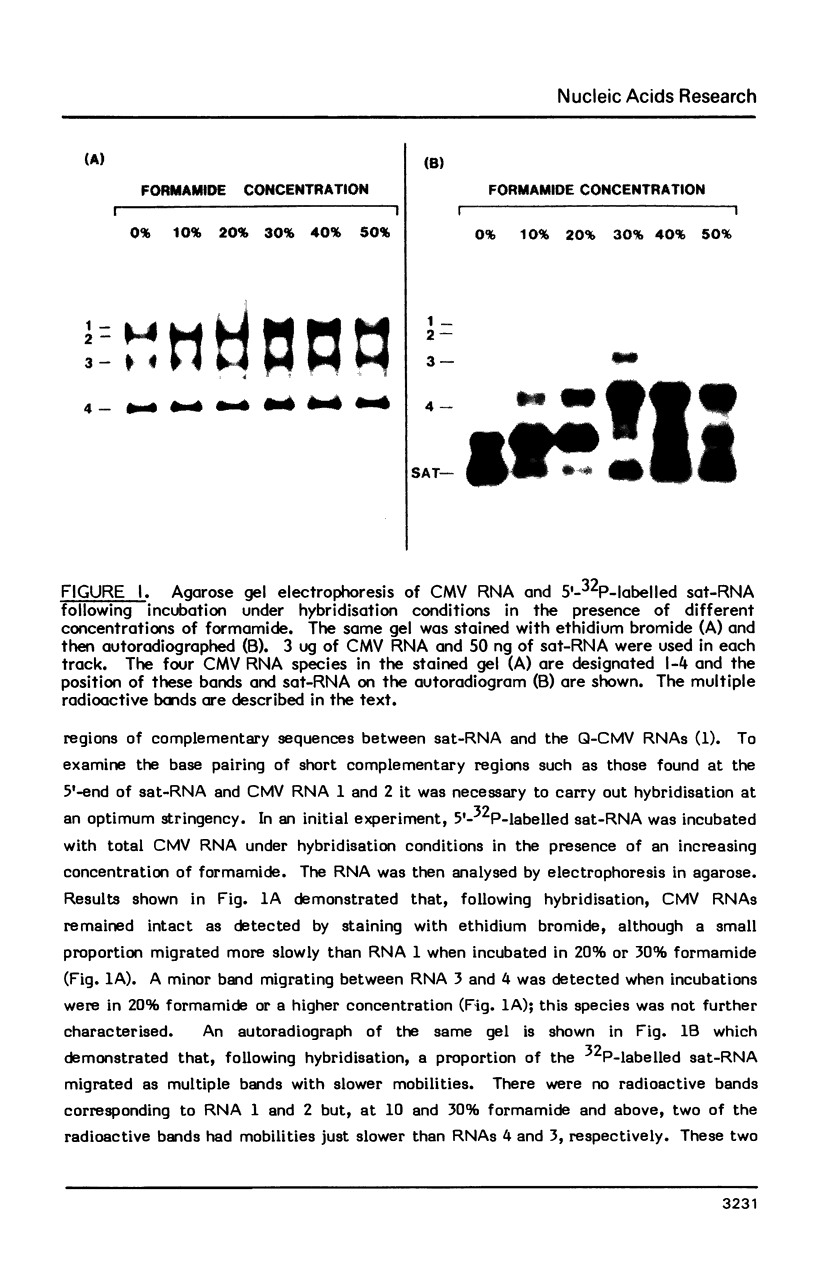
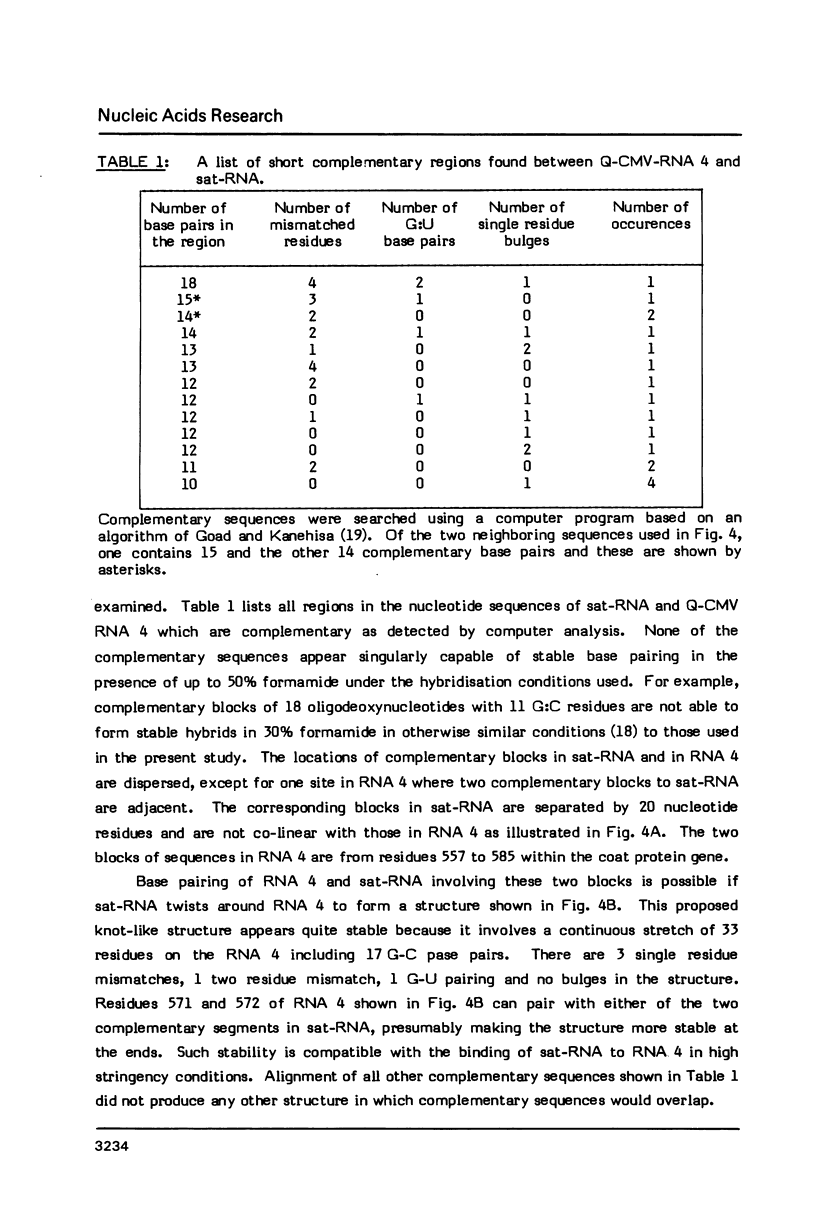
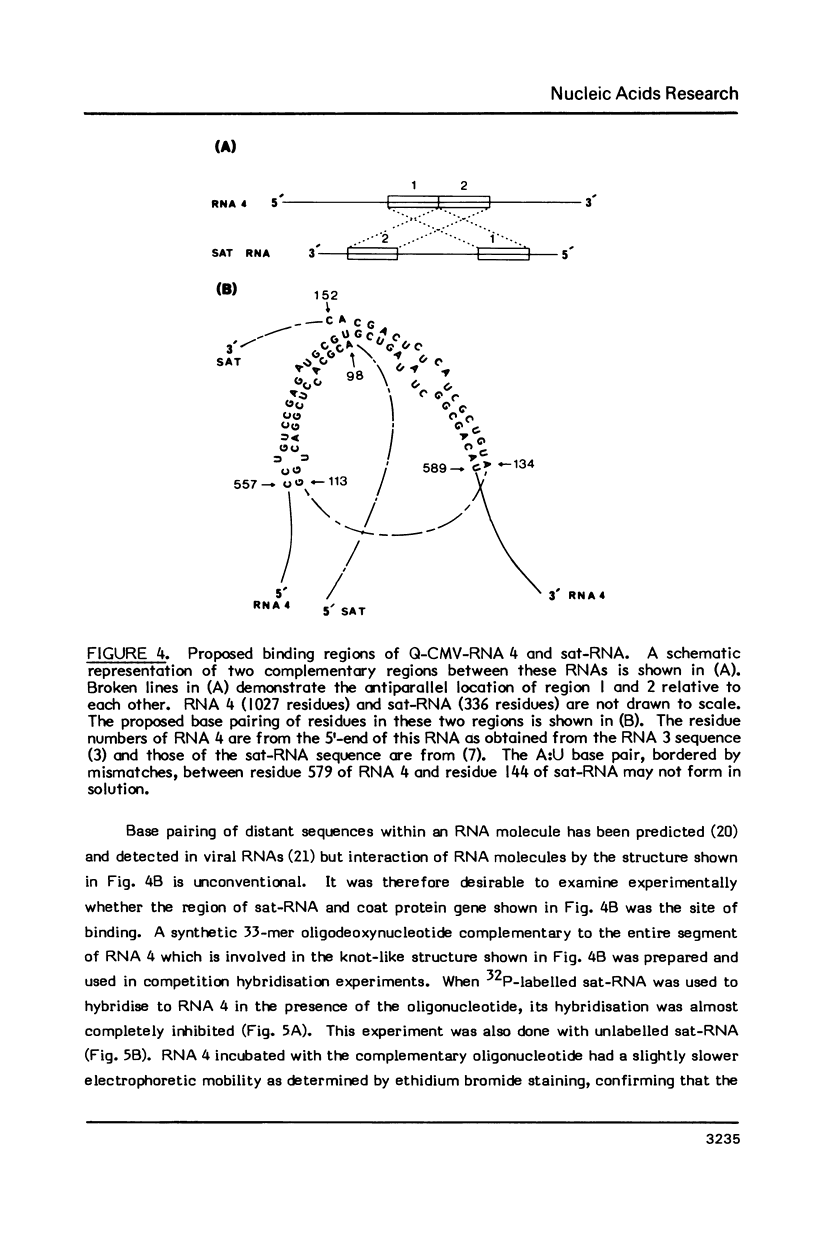
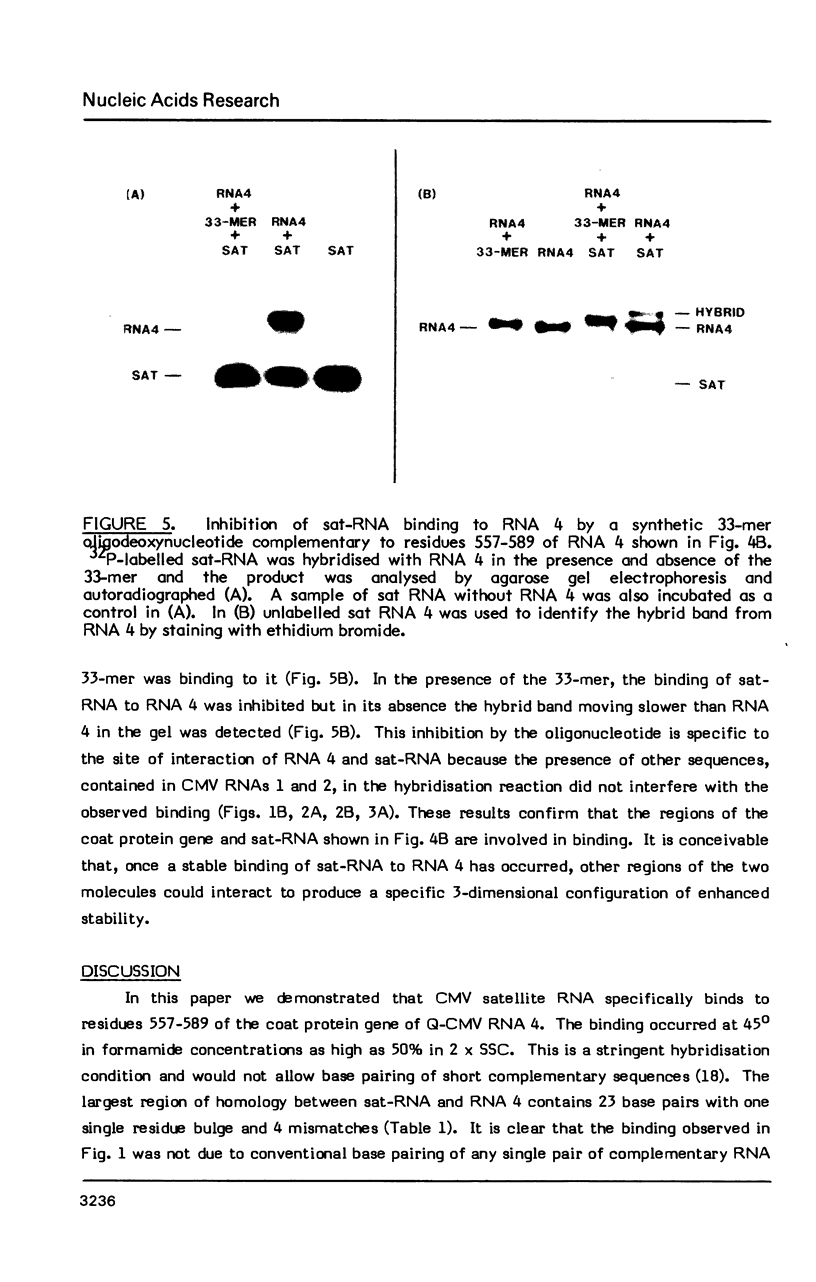
The interaction in vitro of the RNA of the Q-strain of cucumber mosaic virus (CMV) with its satellite RNA (sat-RNA) has been studied. In hybridisation reactions containing 30% formamide at 45 degrees, sat-RNA binds to CMV RNA 3 and 4 but not to CMV RNA 1 and 2 or RNA from tobacco mosaic virus and alfalfa mosaic virus. The viral coat protein gene present in RNA 3 and 4 contains the site of binding but this region does not contain complementary sequences of any significant length to the sat-RNA sequence. However, the optimum alignment of short complementary sequences present in these regions revealed a stable structure in which it is proposed that sat-RNA twists around the coat protein gene so that two separate blocks of nucleotides in sat-RNA base pair in opposite directions with two adjacent blocks in the coat protein gene to form a knot-like structure. The binding site is a region of 33 nucleotides within the coding region of the coat protein gene which base pairs with residues 98-113 and 134-152 of sat-RNA. The possibility of the binding region of sat-RNA functioning as an "anti-sense" sequence in regulation of the viral coat protein synthesis is discussed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Coleman J., Hirashima A., Inokuchi Y., Green P. J., Inouye M. A novel immune system against bacteriophage infection using complementary RNA (micRNA). Nature. 1985 Jun 13;315(6020):601–603. doi: 10.1038/315601a0. [DOI] [PubMed] [Google Scholar]
- Dreher T. W., Bujarski J. J., Hall T. C. Mutant viral RNAs synthesized in vitro show altered aminoacylation and replicase template activities. Nature. 1984 Sep 13;311(5982):171–175. doi: 10.1038/311171a0. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Vournakis J. N., Donis-Keller H., Chaconas G., Dougall D. K., Kafatos F. C. End labeling of enzymatically decapped mRNA. Nucleic Acids Res. 1977 Dec;4(12):4165–4174. doi: 10.1093/nar/4.12.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francki R. I. Plant virus satellites. Annu Rev Microbiol. 1985;39:151–174. doi: 10.1146/annurev.mi.39.100185.001055. [DOI] [PubMed] [Google Scholar]
- Francki R. I., Randles J. W., Chambers T. C., Wilson S. B. Some properties of purified cucumber mosaic virus (Q strain). Virology. 1966 Apr;28(4):729–741. doi: 10.1016/0042-6822(66)90257-1. [DOI] [PubMed] [Google Scholar]
- Goad W. B., Kanehisa M. I. Pattern recognition in nucleic acid sequences. I. A general method for finding local homologies and symmetries. Nucleic Acids Res. 1982 Jan 11;10(1):247–263. doi: 10.1093/nar/10.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon K. H., Symons R. H. Satellite RNA of cucumber mosaic virus forms a secondary structure with partial 3'-terminal homology to genomal RNAs. Nucleic Acids Res. 1983 Feb 25;11(4):947–960. doi: 10.1093/nar/11.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould A. R., Palukaitis P., Symons R. H., Mossop D. W. Characterization of a satellite RNA associated with cucumber mosaic virus. Virology. 1978 Feb;84(2):443–455. doi: 10.1016/0042-6822(78)90261-1. [DOI] [PubMed] [Google Scholar]
- Gould A. R., Symons R. H. Cucumber mosaic virus RNA 3. Determination of the nucleotide sequence provides the amino acid sequences of protein 3A and viral coat protein. Eur J Biochem. 1982 Aug;126(2):217–226. [PubMed] [Google Scholar]
- Hobden A. N., Read M. J., Dykes C. W., Harford S. M13 clones carrying point mutations: identification by solution hybridization. Anal Biochem. 1985 Jan;144(1):75–78. doi: 10.1016/0003-2697(85)90085-5. [DOI] [PubMed] [Google Scholar]
- Izant J. G., Weintraub H. Inhibition of thymidine kinase gene expression by anti-sense RNA: a molecular approach to genetic analysis. Cell. 1984 Apr;36(4):1007–1015. doi: 10.1016/0092-8674(84)90050-3. [DOI] [PubMed] [Google Scholar]
- Kaper J. M., Tousignant M. E., Lot H. A low molecular weight replicating RNA associated with a divided genome plant virus: defective or satellite RNA? Biochem Biophys Res Commun. 1976 Oct 18;72(4):1237–1243. [PubMed] [Google Scholar]
- Kemper B. Inactivation of parathyroid hormone mRNA by treatment with periodate and aniline. Nature. 1976 Jul 22;262(5566):321–323. doi: 10.1038/262321a0. [DOI] [PubMed] [Google Scholar]
- Mathews M. B. Binding of adenovirus VA RNA to mRNA: a possible role in splicing? Nature. 1980 Jun 19;285(5766):575–577. doi: 10.1038/285575a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Melton D. A. Injected anti-sense RNAs specifically block messenger RNA translation in vivo. Proc Natl Acad Sci U S A. 1985 Jan;82(1):144–148. doi: 10.1073/pnas.82.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. A., Dreher T. W., Hall T. C. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (-)-sense genomic RNA. Nature. 1985 Jan 3;313(5997):68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- Peden K. W., Symons R. H. Cucumber mosaic virus contains a functionally divided genome. Virology. 1973 Jun;53(2):487–492. doi: 10.1016/0042-6822(73)90232-8. [DOI] [PubMed] [Google Scholar]
- Pestka S., Daugherty B. L., Jung V., Hotta K., Pestka R. K. Anti-mRNA: specific inhibition of translation of single mRNA molecules. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7525–7528. doi: 10.1073/pnas.81.23.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij C. W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985 Mar 11;13(5):1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaian M. A., Williams R. H., Gordon K. H., Gould A. R., Symons R. H. Nucleotide sequence of cucumber-mosaic-virus RNA 2 reveals a translation product significantly homologous to corresponding proteins of other viruses. Eur J Biochem. 1984 Sep 3;143(2):277–284. doi: 10.1111/j.1432-1033.1984.tb08370.x. [DOI] [PubMed] [Google Scholar]
- Rezaian M. A., Williams R. H., Symons R. H. Nucleotide sequence of cucumber mosaic virus RNA. 1. Presence of a sequence complementary to part of the viral satellite RNA and homologies with other viral RNAs. Eur J Biochem. 1985 Jul 15;150(2):331–339. doi: 10.1111/j.1432-1033.1985.tb09025.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg U. B., Preiss A., Seifert E., Jäckle H., Knipple D. C. Production of phenocopies by Krüppel antisense RNA injection into Drosophila embryos. Nature. 1985 Feb 21;313(6004):703–706. doi: 10.1038/313703a0. [DOI] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- Studnicka G. M., Rahn G. M., Cummings I. W., Salser W. A. Computer method for predicting the secondary structure of single-stranded RNA. Nucleic Acids Res. 1978 Sep;5(9):3365–3387. doi: 10.1093/nar/5.9.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A., Abrahams J. P., Pleij C. W., Bosch L. Five pseudoknots are present at the 204 nucleotides long 3' noncoding region of tobacco mosaic virus RNA. Nucleic Acids Res. 1985 Nov 11;13(21):7673–7686. doi: 10.1093/nar/13.21.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]