Abstract
Exposure of plants to toxic concentrations of metals leads to disruption of the cellular redox status followed by an accumulation of reactive oxygen species (ROS). ROS, like hydrogen peroxide, can act as signaling molecules in the cell and induce signaling via mitogen-activated protein kinase (MAPK) cascades. MAPK cascades are evolutionary conserved signal transduction modules, able to convert extracellular signals to appropriate cellular responses. In this review, our current understanding about MAPK signaling in plant metal stress is discussed. However, this knowledge is scarce compared to research into the role of MAPK signaling in the case of other abiotic and biotic stresses. ROS production is a common response induced by different stresses and undiscovered analogies may exist with metal stress. Therefore, further attention is given to MAPK signaling in other biotic and abiotic stresses and its interplay with other signaling pathways to create a framework in which the involvement of MAPK signaling in metal stress may be studied.
Keywords: toxic metals, oxidative stress, reactive oxygen species (ROS), MAPK cascades, phosphatases, WRKY’s, ZAT12, antioxidative defense system, ethylene, jasmonic acid
1. Introduction
Pollution of soils with toxic metals is a worldwide problem of great concern [1]. The high amount of metals, locally present in the environment, is mainly caused by mining and industrial activities [2,3]. However, the metal content of soils can also be increased due to the agricultural use of phosphate fertilizers, metal-containing pesticides and fungicides and sewage sludge, or irrigation with wastewater [4]. Metals like copper (Cu), iron (Fe), zinc (Zn), cobalt (Co) and nickel (Ni) are essential micronutrients required for normal plant growth and development [5,6]. Although they are essential, these elements can become toxic for plants when they exceed the concentrations normally present in the environment. Other elements, like cadmium (Cd), lead (Pb), and aluminium (Al), are nonessential and adversely affect biochemical reactions and physiological processes in plants [7]. Uptake and accumulation of these metals by food and feed crops brings these potentially toxic metals into the food chain for consumption by animals (cattle) and humans [4]. Several in vitro as well as in vivo epidemiological studies demonstrated that enhanced metal exposure poses a tremendous risk to human health [8–12].
In plants, the main visible symptoms of metal toxicity are leaf chlorosis and growth inhibition, caused by its interference with photosynthesis, mineral nutrition and the water balance [7,13,14]. At the cellular level, metals can disturb the cellular redox status by the production of reactive oxygen species (ROS) or interference with the antioxidant defense system [15–18]. Although ROS can react with biomolecules, which can get irreversibly damaged leading to necrosis and cell death, they can also act as signals in diverse biological processes in plants. In this way, they can influence signal transduction pathways and gene expression, suggesting that cells have evolved strategies to utilize ROS as signals for controlling various biological programs. ROS are suited to act as signaling molecules since they are small and can diffuse over short distances. Among the different ROS, only hydrogen peroxide (H2O2) can cross plant membranes and therefore directly function in cell-to-cell signaling [19,20]. In plant cells, H2O2 acts as a signal molecule involved in acclimatory signaling triggering tolerance to various abiotic and biotic stresses, like metal stress, ultraviolet radiation, salt stress, drought stress, light stress, temperature stress and in plant-pathogen interactions [21–26]. H2O2 has also been shown to act as a key regulator in a broad range of physiological processes, such as senescence, stomatal movement, programmed cell death, and growth and development [27–30]. Downstream signaling events associated with ROS sensing involve Ca2+ and Ca2+-binding proteins, such as calmodulin; the activation of G-proteins; the activation of phospholipid signaling, which results in the accumulation of phosphatidic acid; and/or activation of mitogen-activated protein kinase (MAPK) pathways [19,31]. H2O2 can modulate the activity and regulation of different components of MAPK cascades, such as protein phosphatases, protein kinases and transcription factors [32,33].
After a general introduction to MAPK cascades and their regulation, this review will specifically focus on MAPKinase signaling under metal stress conditions and the possible outcomes of this signaling cascades. Because H2O2 production is involved in multiple abiotic stresses, comparison with other known ROS-induced signaling pathways is discussed.
2. MAPKinases: Nomenclature and Classification
MAPK cascades are important signaling modules that convert signals generated from the receptors/sensors to cellular responses. They are composed of three protein kinase modules: MAPKK kinases (MAPKKK), MAPK kinases (MAPKK or MKK) and MAP kinases (MAPK or MPK). When MAPKKKs, serine/threonine kinases, are activated, they can phosphorylate MAPKKs via serine/threonine residues in the S/T-X5-S/T motif. MAPKKs are dual-specificity kinases that activate MAPKs through phosphorylation of both tyrosine and serine/threonine residues in the TXY motif. MAPKs are kinases that phosphorylate a variety of substrates, including transcription factors, transcription regulators, splicing factors and other protein kinases [34].
In Arabidopsis, 60 MAPKKKs, 10 MAPKKs and 20 MAPKs have been identified (Table 1). The MAPKKKs form the largest and most heterogeneous group of MAPK components. They can be divided in two large subgroups: the MEKK-type, for which MAPKKK function is provided, and the Raf-like kinases, for which MAPKKK function is not yet known. In Arabidopsis, examples of MEKK-like kinases are ANP1/2/3 and MEKK1. CTR1 and EDR1 belong to the Raf-like kinases [35].
Table 1.
Classification of mitogen-activated protein (MAP) Kinases. In the first and second column, the different MAPKinase modules and the amino acid sequences phosphorylated by them are presented. Then MAPKKKs, MAPKKs and MAPKs are classified according to their phosphorylated amino acid sequence and placed in groups A to D based on sequence alignment. In the final two columns, examples of MAPKinases belonging to each group as well as the plant responses, in which they are involved, are provided. MAPkinases indicated in bold are discussed in this review.
| Classification of MAPKinases | |||||
|---|---|---|---|---|---|
|
| |||||
| MAPKinase | Amino acids phosphorylated by the kinase | Phosphorylated amino acid motif | Group | Examples | Response to/in |
| MAPKKK | Serine/threonine | MEKK1-type | A | MEKK1, MEKK2, MEKK3, MEKK4 | Redox control–oxidative stress; Abiotic stresses: drought, salt, mechanical; Biotic stress: bacterial pathogens; Hormones: salicylic acid. |
| ANP1, ANP2, ANP3 | Redox control–oxidative stress; Biotic stress: bacterial pathogens; Hormones: auxin; Cytokinesis. | ||||
| Raf-like | B | EDR1, CTR1 | Hormones: ethylene; Disease resistance signling. | ||
| C | ATN1, MRK1 | Unknown. | |||
|
| |||||
| MAPKK | Threonine/tyrosine | S/T-XXXXX-S/T | A | MKK1, MKK2, MKK6 | Redox control–oxidative stress; Abiotic stresses: cold, salt, low humidity, mechanical; Biotic stresses: bacterial pathogens; Hormones: salicylic acid; Cell division. |
| B | MKK3 | Oxidative stress; Abiotic stresses: mechanical; Biotic stresses: bacterial pathogens; Hormones: jasmonic acid. | |||
|
| |||||
| MAPKKK | Threonine/tyrosine | S/T-XXXXX-S/T | C | MKK4, MKK5 | Redox control–oxidative stress; Abiotic stresses: salt; Biotic stress: bacterial pathogens; Hormones. |
| D | MKK7, MKK8, MKK9, MKK10 | Oxidative stress; Biotic stress: bacterial pathogens. | |||
|
| |||||
| MAPK | Serine/threonine/tyrosine | TEY | A | MPK3, MPK6, MPK10 | Redox control–oxidative stress; Abiotic stresses: salt, cold; Biotic stress: bacterial pathogens; Hormones: jasmonic acid. |
| B | MPK4, MPK5, MPK11, MPK12, MPK13 | Redox control–oxidative stresses; Abiotic stresses: salt, cold, low humidity, mechanical; Hormones: salicylic acid; Cell division. | |||
| C | MPK1, MPK2, MPK7, MPK14 | Oxidative stress; Abiotic stresses: mechanical; Biotic stresses: bacterial pathogens; Hormones: jasmonic acid, abscisic acid; Circadian-rhythm-regulated. | |||
| TDY | D | MPK8, MPK9, MPK15/16/17/18/19/20 | Oxidative stress; Abiotic stresses: mechanical; Biotic: blast fungus; Hormones: jasmonic acid. | ||
MAPKKs (or MKKs) are the smallest group, only half as many as there are MAPKs. MAPKKs are probably able to activate multiple MAPKs, which suggests interplay between different signal transduction pathways occurring at this level. Plant MAPKKs are subdivided in four groups (A–D) based on sequence alignment. Group A MAPKKs seem to be involved in multiple abiotic stresses and cell division and are responsive to pathogens. MAPKKs belonging to group C are abiotic stress-responsive and function upstream of group A MAPKs, which play a role in environmental and hormonal responses [35].
Arabidopsis MAPKs (or MPKs) can be classified into two subtypes: those containing a TEY amino acid motif and those containing a TDY amino acid motif. The TEY subtype consists of three groups: A, B and C. MAPKs of group A are particularly involved in environmental and hormonal responses and examples of this group are MPK3 and MPK6. Group B MAPKs, to which MPK4 belongs, also play a role in environmental stress responses and seem to be involved in cell division. Little is known about group C MAPKs, but they appear to be circadian-rhythm-regulated. The TDY subtype forms group D. These MAPKs have a more extended C-terminal region in relation to groups A, B and C. On the other hand, they lack the C-terminal CD DOMAIN found in groups A, B and C, which serves as a docking site for MAPKKs, phosphatases and protein substrates. Group D MAPKs are shown to be induced by blast fungus and wounding [35].
The MAPKKKs are the most divergent group of kinases in the MAPK cascade. Therefore, different MAPKKKs can initiate similar MAPK cascades, which finally activate the same downstream MAPK. This is one of the mechanisms by which different stimuli converge onto one MAPK. The sharing of a single component by different cascades also leads to interaction between different pathways [35,36].
3. Regulation of MAPKinases
The outcome of a MAPK activation depends on the duration of its activation. The length of time that a MAPK remains active depends on the upstream specific regulation mechanisms, of which scaffolding (co-localization) and attenuation through phosphatases are best known. In addition, attention is given to lipid signaling which can initiate MAPK cascades.
3.1. Scaffolding
Specificity of different MAPK cascades functioning within the same cell is conferred by docking domains of scaffold proteins, which anchor different MAPK modules in one complex [37]. MAPK components themselves can also function as scaffolds. An example is the MAPKKK “Oxidative stress-activated MAP triple-kinase 1” (OMTK1), discovered in Medicago sativa. OMTK1 is able to activate MMK3 in response to H2O2 but not in case of cell treatment with yeast elicitor or 1-aminocyclopropane-1-carboxylic acid (ACC). Pull-down analysis between recombinant proteins showed that OMTK1 directly interacts with MKK3 and that both kinases are found together in a protein complex in vivo, suggesting that OMTK1 can determine specificity through its scaffolding function [38]. In Arabidopsis, no scaffold proteins of MAPK components with scaffold function are known to date.
3.2. Phosphatases
Negative regulation of MAPK cascades is performed by MAPK phosphatases, which dephosphorylate threonine and tyrosine residues within the activation motif of MAPKs. Three different forms of phosphatases were identified to date: protein tyrosine phosphatases (PTPs), protein serine-threonine phosphatases (PSTPs) or dual-specificity (Ser/Thr and Tyr) phosphatases (DSPs) [39]. In A. thaliana, members of all three classes are linked with MAPK inactivation, but dual-specificity MAPK phosphatases are the most important group because full inactivation of MAPKs requires dephosphorylation of both residues. The Arabidopsis genome encodes five possible candidate dual-specificity MAPK phosphatases (MKP1, MKP2, DsPTP1, PHS1 and IBR5) [39].
MKP1 was first identified by its involvement in genotoxic stress resistance. When 5-day-old seedlings were subjected to UV-C radiation (0.5–1 kJ/m2) or 50 mg L−1 methyl methane sulphonate, MKP1 was required for maintaining proper MAPK activity levels. Yeast-two-hybrid assays showed that MKP1 interacts with the stress-activated MAPKs MPK3, MPK4 and MPK6. Interaction of MKP1 with MPK6 was the most pronounced and MKP1 was reported to regulate MPK6 activity in vivo [40,41]. In addition, a role for MKP1 as a negative regulator of MPK3 and MPK6 activities was suggested in resistance against the bacterial pathogen Pseudomonas syringae and tolerance against UV-B and salt stresses [41–43]. Lee et al. [44] demonstrated that MKP1 activity increased after binding to calmodulin, indicating that Ca2+- and MAPK signaling are connected via the regulation of MKP1. MKPs can act together with other protein phosphatases to control MAPK activity. The protein tyrosine phosphatase 1 (PTP1), which was also shown to interact with MPK6, acts together with MKP1 to repress the biosynthesis of salicylic acid (SA) and camalexin, and pathogenesis-related gene expression, which makes plants more vulnerable to infection with Pseudomonas syringae [45].
Another MKP, MKP2, is also reported to bind and dephosphorylate MPK3 and MPK6 in vitro as well as in vivo [46,47]. After acute exposure to 500 ppb ozone, MKP2 acts as a positive regulator of the cellular redox status by repressing the activity of MPK3/6 [46]. In case of plant-pathogen interactions, MKP2 exerts differential and specific functions depending on the invading pathogen and is required for maintaining adequate levels of MPK3/6 activation. The appearance of bacterial wilting symptoms was delayed with one day in mkp2 homozygous knockout plants infected with Ralstonia solanacearum. In contrast, mkp2 knockout plants infected with Botrytis cinerea showed a systemic spread of the fungus throughout the whole plant after 15 days of inoculation, whereas in wild type plants lesions were local and restricted to the inoculated leaves [47].
3.3. Lipid Signaling
Besides the regulation of MAPKs by protein phosphatases, MAPK cascades can also be regulated via lipid signaling. Plasma membrane-associated phospholipase D (PLD) enzymes release phosphatidic acid (PA) from phosphatidylcholine, phosphatidylethanolamine and phosphatidylglycerol. PA has been implicated as a secondary messenger in many different stress responses, such as the production of ROS [48].
In Arabidopsis, Yu et al. [49] showed that PLD-derived PA binds to MPK6, leading to its activation during salt stress. Activated MPK6 is responsible for the phosphorylation of the Na+/H+ antiporter SOS1, which reduces Na+ accumulation in Arabidopsis leaves. In Arabidopsis, Anthony et al. [50] reported that binding of PA to “3-phosphinositide-dependent protein kinase 1” (PDK1) stimulates phosphorylation and activation of the serine/threonine protein kinase “Oxidative-signal inducible 1” (OXI1). Activity of OXI1 was induced within 30 min after treatment of Arabidopsis plants with plant growth factors, like auxin and cytokinin, suggesting a role for OXI1 in plant growth and cell division. Rentel et al. [51] revealed that OXI1 is involved in H2O2-dependent activation of MPK3/6 in ROS-dependent processes such as root-hair elongation and basal resistance to the fungal pathogen Peronospora parasitica. In addition, PA-stimulated activation of the PDK1/OXI1/MPK6 pathway was shown to promote plant growth in Arabidopsis seedlings after co-cultivation with the endophytic fungus Piriformospora indica [52]. Activation of MAPKs by OXI1 is mediated by serine/threonine protein kinases of the Pto-interacting 1 (PTI1) like family or NDP kinase 2 (NDPK2) [53–55]. However, further research is required to unravel the OXI1–MAPK cascade.
4. Role of MAPK Cascades in Stress Response Signaling
MAPK cascades are involved in normal cell metabolism like physiological, developmental and hormonal responses [34,56,57]. However multiple studies have shown that MAPK cascades play important roles in plant responses to biotic and abiotic stresses, such as pathogen infection, wounding, low temperature, drought, hyper- and hypo-osmolarity, high salininty, mechanical stress, metals and ROS [57–59].
4.1. MAPK Cascades Are Involved in Metal Stress
Several authors reported the involvement of MAPK signaling in metal stress for different plant species (Table 2). In general, mRNA as well as activity levels are increased quickly after metal exposure, ranging from 5 min to 1 h, and activation of MAPKs is transient. In Arabidopsis, it is proven that MPK3 and MPK6 are activated in response to short-term exposure (less than 1 h) to CdCl2 concentrations as low as 1 μM, via the accumulation of ROS [60]. However, so far, evidence for the involvement of a complete MAPK cascade pathway in metal stress responses is rather scarce in plants.
Table 2.
Induction of MAPKinases under metal stress. MAPK cascade modules affected by exposure to metals are categorized based upon plant species and type of kinase. Exposure to metals influences MAPKinase mRNA levels as well as the activity at the protein level.
| Metal-Induced MAPKinases | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Plant | Component of MAPK cascade | Metal | Concentration | Exposure Time | Observations | Reference | |
| Arabidopsis thaliana | MAPKKK | MEKK1 | Cd | 500 μM CdCl2 | 1–3 h | ↑ mRNA levels | [66] |
| MAPK | MPK3/MPK6 | Cd | 1 μM CdCl2 | 10 min | ↑ activity | [60] | |
| Cu/Cd | 2 μM CuSO4/5 μM CdSO4 | 2–24 h | ↑ mRNA levels | [21] | |||
|
| |||||||
| Brassica juncea | MAPK | 46 kDa MAPK | As(III) | 50 μM As(III) | 15 min–1 h | ↑ activity | [67] |
|
| |||||||
| Medicago sativa | MAPKK | SIMKK | Cu | 100 μM CuCl2 | 30 min | induces SAMK and SIMK | [62] |
| MAPK | SAMK | Cu/Cd | 100 μM CuCl2/CdCl2 | 10 min–1 h (Cu)/30 min–3 h (Cd) | ↑ activity | [62] | |
| SIMK | Cu/Cd | 100 μM CuCl2 /CdCl2 | 5 min–6 h (Cu)/10 min–6 h (Cd) | ↑ activity | [62] | ||
| MMK2 | Cu/Cd | 100 μM CuCl2/CdCl2 | 10 min–1 h (Cu)/10 min–3 h (Cd) | ↑ activity | [62] | ||
| MMK3 | Cu/Cd | 100 μM CuCl2/CdCl2 | 10 min–1 h (Cu)/10 min–1 h (Cd) | ↑ activity | [62] | ||
|
| |||||||
| Oryza sativa | MAPKK | OsMKK4 | As | 50 μM As(III) | 3–9 h | ↑ mRNA levels | [61] |
| MAPK | OsMSRMK2 | Cu/Cd/Hg | 100 μM CuSO4/CdCl2/HgClO3 | 30 min | ↑ mRNA levels | [68] | |
| OsMSRMK3 | Cu/Cd/Hg | 100 μM CuSO4/CdCl2/HgClO3 | 15 min–2 h | ↑ mRNA levels | [69] | ||
| OsWJUMK | Cu/Cd/Hg | 100 μM CuSO4/CdCl2/HgClO3 | 15 min–2 h | ↑ mRNA levels | [69] | ||
| OsMPK2 | Cd | 400 μM CdCl2 | 3–12 h | ↑ mRNA levels | [70] | ||
| Cu | 100 μM CuCl2 | 3–12 h | ↑ mRNA levels | [71,72] | |||
| OsMPK3 | Cu/Cd | 50 μM CuCl2/100 μM CdCl2 | 1 h | ↑ activity | [73] | ||
| As | 50 μM As(III) | 30 min–9 h | ↑ mRNA and activity levels | [61] | |||
| OsMPK4 | As | 50 μM As(III) | 30 min–9 h | ↑ mRNA and activity levels | [61] | ||
| OsMPK6 | Cu/Cd | 50 μM CuCl2/100 μM CdCl2 | 1 h | ↑ activity | [73] | ||
| 40 kDa MAPK | Zn | 1 mM ZnCl2 | 15 min–8 h | ↑ activity | [74] | ||
| Pb | 10 mM Pb(NO3)2 | 30 min–8 h | ↑ activity | [75] | |||
| 42 kDa MAPK | Zn | 1 mM ZnCl2 | 15 min–8 h | ↑ activity | [74] | ||
| I | 500 μM FeSO4 | 15–30 min | ↑ activity | [76] | |||
| Pb | 10 mM Pb(NO3)2 | 15 min–8 h | ↑ activity | [75] | |||
|
| |||||||
| Zea mays | MAPK | ZmMPK3 | Cd | 500 CdCl2 | 30 min–1 h | ↑ mRNA levels | [77] |
| ZmMPK5 | Cr(VI) | 250 μM Cr(VI) | 30 min | ↑ activity | [78] | ||
Rao et al. [61] predicted a possible MAPK cascade in rice namely OsMKK4/OSMPK3. Two weeks old rice plants exposed to 50 μM arsenite showed increased transcript levels of OsMPK3 in leaves and roots already after 30 min of exposure. These results were confirmed at the protein level: activity of OsMPK3 was elevated within 3h. Gene expression levels of OsMKK4 were also elevated in leaves and roots after 3 h exposure to arsenite. In Medicago sativa roots, transient activation of MAPKs (SIMK, SAMK, MKK2 and MMK3) was rapidly induced (less than 10 min) after exposure to 100 μM CuCl2, whereas treatment with 100 μM CdCl2 delayed this profile. In addition, transient expression assays in Arabidopsis protoplasts with HA-tagged SIMK, SAMK, MMK2 and MKK3, and a myc-tagged MAPKK (SIMKK), showed that SIMKK specifically activated SIMK and SAMK after exposure to 100 μM CuCl2 [62]. Opdenakker et al. [21] showed that 24 h exposure of Arabidopsis thaliana seedlings to environmental realistic concentrations of Cu and Cd increased transcript levels of MAPKinases in a time-dependent manner. After 2 h of exposure to 2 μM Cu, gene expression of OXI1, the MAPKKK “Arabidopsis NPK1-like protein kinase 1” (ANP1) and the MAPKs MPK3 and MPK6 was already affected in the roots and leaves of Arabidopsis plants. After exposure to 5 μM Cd, no changes in gene expression of these enzymes were observed before 24 h. These changes in gene expression seemed to be related to the production of H2O2 by these metals, either directly and fast by Cu (Fenton-HaberWeiss reactions) or indirectly and delayed by Cd (e.g., via NADPH oxidases).
Activation of ANP1 and OXI1 by H2O2 and induction of a phosphorylation cascade involving MPK3 and MPK6 has been reported before in Arabidopsis leaf cells and whole plants [51,63]. Application of 200 μM H2O2 to Arabidopsis protoplasts, increased the activity of ANP1, MPK3 and MPK6 within 10 min. Co-transfection of protoplasts with ANP1 and MPK3 or MPK6 revealed that ANP1 could further enhance the activity of MPK3 and MPK6 after H2O2 treatment [63]. Rentel et al. [51] showed that gene expression of OXI1 was already enhanced after 30 min in 7-day-old seedlings treated with 10 mM H2O2 and oxi1 knockout mutants failed to activate MPK3 and MPK6 after treatment with H2O2. Additionally, a toxicity test based on primary root elongation showed that oxi1 and mpk6 knock-outs were more tolerant to excess Cu, but not Cd, suggesting that OXI1 and MPK6 play important roles in the observed stress response following Cu exposure [64].
However, knowledge about the downstream signaling targets of MAPKs is rather scarce under metal stress. Roelofs et al. [65] compared known signaling pathways induced by metals stress as well as by other abiotic stresses (cold, heat, salt, drought) between soil invertebrates and plants. They showed that all abiotic stresses switched on more than one stress-responsive pathway, seen in the overlap of transcription factors used by each stressor, and they speculated that bZIP, MYB and MYC transcription factors could be downstream targets of MAPK signaling in plant metal stress.
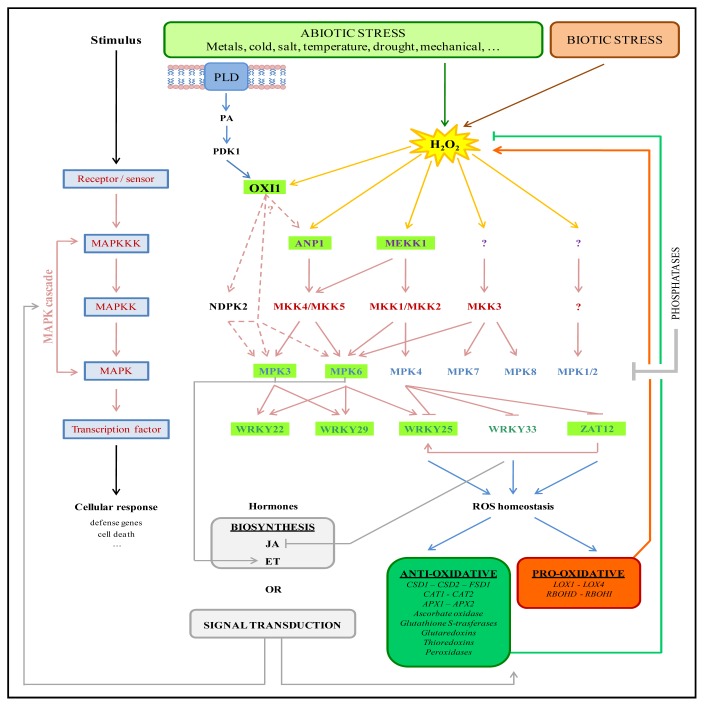
Interplay between the MAPK pathways activated by metal stress and the ones used by other stresses probably exists because ROS generation, which is known to induce MAPK signaling, is common to other abiotic and biotic stress responses (Figure 1).
Figure 1.
Overview of the different MAPK cascades known to function in stress responses as well as their regulation and possible downstream outcomes (signaling components involved in metal stress are highlighted in green). Production of ROS is a common phenomenon between different biotic and abiotic stresses. In particular H2O2 can act as an intra- and intercellular signaling molecule activating MAPK cascades. Via lipid signaling or phosphorylation of OXI1, ROS production can be linked to MAPK activation. MAPK signaling pathways induce gene expression of stress-responsive genes through the activation of transcription factors. On one hand, MAPK cascades influence the cellular redox status by activating gene expression of antioxidative or pro-oxidative enzymes. In this way, MAPK signaling can attenuate or amplify the initial ROS signal. On the other hand, MAPKs interfere with hormone signaling and biosynthesis leading to activation of downstream stress responses. Inactivation of MAPK pathways occurs through phosphatases, which dephosphorylate MAPKs, resulting in the disruption of the MAPK signal and are therefore important in the regulation of downstream responses.
4.2. Comparison to MAPK Pathways Involved in Other Stress Responses
In Arabidopsis protoplasts, Kovtun et al. [63] showed that the MAPKKK ANP1, induced by H2O2, activated the downstream MAPKs MPK3/6. The MAPKKs involved in the activation of MPK3/6 by ANP1 could be MKK4 and MKK5. Ren et al. [79] reported that transgenic Arabidopsis plants, expressing MKK4 and MKK5 under the control of a steroid-inducible promoter, were able to activate MPK3/6, resulting in cell death. The protein kinase OXI1 is, as already mentioned above, involved in H2O2-dependent activation of MPK3/6 [51]. Moreover, oxi1 knockout mutants showed defects in ROS-dependent developmental processes such as root-hair elongation, and in ROS-dependent basal resistance to the fungal pathogen Peronospora parasitica. In what way OXI1 activates MPK3 and MPK6 remains to be addressed, although it is suggested that this activation may be modulated by NDP kinase 2 (NDPK2) [55,59]. Exposure of 2-week-old Arabidopsis plants to 4 mM H2O2 strongly increased gene expression of NDPK2 within 30 min and up to 12h, suggesting that NDPK2 functions in ROS homeostasis. Furthermore, overexpression of NDPK2 resulted in lower levels of ROS as compared to wild type plants and conferred enhanced tolerance to environmental stresses that induce ROS generation, such as freezing during 1h or exposure to 50 mM NaCl for 3 weeks. Specific interaction between NDPK2 and MPK3/6 was discovered using yeast two-hybrid and in vitro protein pull-down assays. NDPK2 was also shown to enhance the myelin basic protein phosphorylation activity of MPK3 in vitro [55].
Nakagami et al. [80] reported on another Arabidopsis MAPKKK, MEKK1, which is also regulated by H2O2 and was found to activate the MAPK MPK4 in response to treatment of Arabidopsis protoplasts with 2 mM H2O2 during 5 min. Ten-day-old mekk1 knockout plants showed a deregulated expression of genes involved in cellular redox control, like glutathione S-transferases, NADPH oxidases and ascorbate peroxidases, and accumulated ROS, suggesting that MEKK1 is necessary for normal redox homeostasis of the cell. The MAPK cascade MEKK1-MKK1/MKK2-MPK4 was earlier identified using yeast two-hybrid and complementation analysis of yeast mutants [81,82]. Later, it was shown that MKK1 phosphorylates MPK4 in vitro as well as in vivo and that this cascade is operating in different environmental stresses such as low temperature, low humidity, hyper-osmolarity and mechanical stress [83–85]. A microarray study with 14-day-old mekk1, mkk1/2 and mpk4 knockout Arabidopsis plants performed by Pitzschke et al. [86] showed that the MEKK1-MKK1/2-MPK4 cascade is a key regulator of ROS- and SA-initiated stress signaling. However, they also suggest that MEKK1 can activate another pathway, independent of MKK1/2 and MPK4, and that MKK1/2 is not only regulating MPK4, but most likely also other MAPKs like MPK3/6. Asai et al. [87] showed that in protoplasts treated with 100 mM of a bacterial flagellin peptide (flg22), MEKK1 activated MPK3/6 through MKK4/MKK5, leading to the expression of early-defense response genes. Activation of the MEKK1-signaling cascade by flg22 is mediated by receptor-like kinases (RLKs), which are also reported to be regulated by Cd stress [88]. Recently, MKK4 was also identified to be involved in abiotic salt stress responses, as a regulator of MPK3 activity [89]. In contrast, studies performed with mekk1 knockout plants instead of the protoplast system used by Asai et al. [87] reported that 14-day-old mekk1 knockout plants did activate MPK3 and MPK6 activity within 10 min after treatment with flg22, but failed to induce MPK4 activity. These results indicate that probably more alternative pathways exist to activate MPK3/6 in the absence of MEKK1 [90]. MKK2, activated by MEKK1, was observed to directly target MPK4 and MPK6 in cold and salt stress [91]. Plants overexpressing MKK2 exhibited constitutive MPK4 and MPK6 activity, showing increased freezing and salt tolerance while mkk2 knockout plants were impaired in MPK4 and MPK6 activation, showing hypersensitivity to salt and cold stress. These studies demonstrate that MEKK1 can integrate different stress signals and ensures stress-specific responses by activating different downstream MAPKs.
Recently, another MAPK component, able to integrate different stress signals, was identified. MKK3 acts as upstream activator of MPK7, which induces target genes such as PR1 in the defense response against Pseudomonas syringae. In Arabidopsis protoplasts, transiently expressing MKK3 and MPK7, MKK3-mediated activity of MPK7 was only induced after application of 4 mM H2O2 during 5 or 15 min, whereas treatment with 1 μM flg22 had no effect on the MPK7 activity. These data led to the observation that the MKK3-MPK7 cascade is induced by H2O2-mediated inhibition of the proteasome-dependent degradation of MKK3 [92]. In contrast, the MKK3-MPK6 pathway functions in jasmonic acid (JA) signaling. In 2-week-old wild type plants, MPK6 activity was enhanced directly after treatment with 50 μM JA. This activity was reduced in mkk3 knockout plants and higher in MKK3 overexpressing plants exposed to JA. In addition, JA-dependent inhibition of root growth and induction of PDF1.2 and VSP2 expression was regulated by MKK3-MPK6 [93]. In the case of mechanical stress, MKK3 is, together with Ca/CaM, responsible for the full activation of MPK8, which negatively regulates the expression of RBOHD (NADPH-oxidase). RBOHD plays an important role in ROS generation and the ROS signal provided by RBOHD is involved in the induction of wound-inducible marker genes, such as OXI1 and ZAT12. Therefore, the negative regulation of RBOHD by MPK8 is required for appropriate production of ROS during mechanical stress responses [94]. In addition, Ortiz-Masia et al. [95] showed a rapid (within 15 min) activation in 4-week-old Arabidopsis plants upon treatment with 5 mM H2O2 as well as by other stress signals such as mechanical stress or application of 50 μM JA or 100 μM abscisic acid (ABA).
5. MAPK Cascades Regulate Stress Responses by Activation of Gene Transcription
5.1. Transcription Factors
MAPK cascades have the possibility to regulate gene transcription by activation or repression of transcription factors. Popescu et al. [96] used high-density Arabidopsis protein microarrays to identify in vitro novel MPK targets. They observed that the largest group of possible MPK targets identified in their screen represent transcription factors. Coexpression of WRKY and TGA transcription factors with specific MKK/MPK modules showed that these transcription factors are also phosphorylated in vivo.
WRKY proteins bind to W-box DNA elements (containing a TGAC core sequence) and act both as positive and negative regulators of target gene expression. WRKY family members are subdivided into three groups based on the number of WRKY domains and certain features of the zinc finger-like motifs (for a review, see [97] and references therein). WRKY transcription factors are known to be involved in diverse biotic and abiotic stresses (Figure 1). Short-term exposure of 3-week-old Arabidopsis plants to 2 μM Cu, induced gene expression of WRKY22, WRKY25 and WRKY29 already after 2h of exposure in leaves and roots. In contrast, after 24 h of exposure to 5 μM Cd only the gene expression of WRKY25 and WRKY29 was affected. These data suggest that these transcription factors play an essential role in regulation of the stress response upon metal exposure [21]. Involvement of these WRKY transcription factors was also reported in other stress conditions confirming the hypothesis that use of these transcription factors is not specific for metal stress signaling but is common between different biotic and abiotic stress responses. Investigation of transcriptome data generated from ROS-related microarray experiments, showed that induction of WRKY22 gene expression is also related to singlet oxygen, ozone and superoxide production [98]. More recently, WRKY22 gene expression was reported to be induced by H2O2 in leaf senescence [99]. Transcript levels of WRKY22 were induced 1h after application of 3% H2O2 or by dark treatment already after 1 day. The involvement of WRKY22 in dark-induced leaf senescence was investigated by comparing wrky22 knockout plants and WRKY22 overexpressing plants with wild type plants in relation to cell death, chlorophyll content and expression of senescence-associated genes [99]. The flagellin induced MAPK cascade MEKK1-MKK4/MKK5-MPK3/MPK6 is known to activate WRKY22 and his close homolog WRKY29. They positively regulate gene expression of disease resistance genes to confer resistance to both bacterial and fungal pathogens [87].
In vitro and in vivo interaction studies revealed a MPK4 substrate MKS1 (MAP kinase 4 substrate 1), which functions in coupling MPK4 to WRKY25 and WRKY33. In this way, WRKY25/33 function in the regulation of pathogen defense responses by repression of SA-dependent resistance. Indeed, Arabidopsis plants overexpressing WRKY25 showed increased disease symptoms together with an invasive bacterial growth after inoculation with Pseudomonas syringae during 3 and 4 days. This was due to reduced expression of PR1, a molecular marker for SA-mediated defense signaling [100,101]. In addition, studies pointed out a role for WRKY25 in the modulation of gene transcription during heat and salt stress [102,103]. Transcript levels of WRKY25 were induced within 30 min in Arabidopsis plants exposed to 42 °C. Five-day-old wrky25 knockout plants showed an increased inhibition of root elongation when exposed to 45 °C for 4 h, whereas 3-week-old wrky25 knockout displayed greater electrolyte leakage after 4h exposure to 42 °C, indicating the positive role of WRKY25 in thermotolerance [102]. Treatment of 3-week-old Arabidopsis plants with 150 mM NaCl during 6 and 24 h induced gene expression of WRKY25 as well as WRKY33 strongly. Root growth was stimulated in WRKY25 and WRKY33 overexpressing plants in the presence of 100 mM NaCl, suggesting the involvement of these transcription factors in tolerance against salt stress [103].
The zinc finger transcription factor (C2H2-type) ZAT12 is also involved during metal stress responses. As was also observed for the WRKY transcription factors, transcript levels of ZAT12 were elevated after 2 h in roots of 3-week-old Arabidopsis plants exposed to 2 μM Cu, whereas ZAT12 expression in 5 μM Cd-exposed roots was not increased before 24 h. In leaves, gene expression of ZAT12 was increased after 24h in both Cu- and Cd-exposed plants [21]. Besides its involvement in metal stress, ZAT12 was also found to respond at the transcriptional level to other abiotic and biotic stresses. A comparison of microarray profiles of 6-week-old wild type and catalase-deficient (20% residual catalase activity) Arabidopsis plants exposed to high light for at least 3 h, identified ZAT12 as a H2O2-responsive transcription factor [24,104]. This observation was confirmed by the results of a microarray study performed on Arabidopsis cell cultures exposed to 20 mM H2O2 during 1.5 and 3 h [105]. Mechanically stressed leaves of 4-week-old Arabidopsis seedlings showed significantly increased transcript levels of ZAT12 after 30 min. This increase in gene expression was still visible after 6h of stress [106]. Activation of ZAT12 transcription was also reported to occur in 3-week-old Arabidopsis plants in response to cold (4 °C, 2 h), heat (38 °C, 1 h), salt (150 mM NaCl, 4 h) and drought (75% relative water content) [107].
5.2. Regulation of the Cellular Redox Status by MAPK Cascades
Metal stress, as well as other biotic and abiotic stresses, is known to disrupt the cellular redox status by stimulating the production of ROS or affecting the antioxidative defense system of the cell. Signaling via MAPK cascades can influence both sides of the redox balance (Figure 1).
Pitzschke et al. [86] showed that the MEKK1-MKK1/MMK2-MPK4 pathway negatively controlled the gene expression of WRKY25, WRKY33 and ZAT12. Expression of oxidative stress responsive genes like the Cu/Zn superoxide dismutase CSD1, the catalase CAT2, the NADPH oxidase RBOHI, and certain glutaredoxins and thioredoxins, was also altered in mpk4 knockout plants. This suggests that the MEKK1-MMK1/MMK2-MPK4 pathway regulates ROS homeostasis via the transcription factors WRKY25/33 and ZAT12. Additionally, in the cases of heat or salt stress, WRKY25 and WRKY33 were reported to influence the gene expression of ROS responsive genes [102,103]. Exposure of 21-day-old wrky25 knockout plants to 42 °C for 30, 60 and 120 min showed lower transcript levels of the ascorbate peroxidases APX1 and 2 as compared to wild type plants. These data indicate that WRKY25 can positively regulate the expression of two oxidative stress-responsive genes APX1 and APX2 [102]. Microarray studies on salt-exposed (150 mM, 6 h) wrky33 knockout plants revealed glutathione-S-transferases, class III peroxidases and the lipoxygenase LOX1 as possible targets of WRKY33 transcriptional activity [103]. Because WRKY25 and WRKY33 share very similar protein structures [97], it is possible that they are involved in the regulation of the same genes. For example, Li et al. [108] showed that WRKY25 and WRKY33, together with WRKY26, work coordinately to induce thermotolerance in plants.
Studies with ZAT12 overexpressing as well as knockout plants revealed that ZAT12 is responsible for the induction of oxidative stress-related transcripts, like APX1, CSD1, CSD2, RBOHD, LOX4 and several glutathione S-transferases, while reducing transcript levels of the iron superoxide dismutase FSD1, an L-ascorbate oxidase, several peroxidases and glutaredoxins [107,109–111]. These data suggest that ZAT12 is important in facilitating plants to cope with oxidative stress. ZAT12 was also identified as inducer of WRKY25 during oxidative stress, meaning that WRKY25 acts downstream of ZAT12 to control its target genes [102,109]. Nakagami et al. [80] suggested a role for the MEKK1-MPK4 pathway in the negative regulation of ZAT12 under oxidative stress conditions. Exposure of mekk1 and mpk4 knockout plants to 10 mM H2O2 during 1h resulted in increased ZAT12 transcript levels as compared to wild type plants. Gene expression of CAT1, not CAT2 nor CAT3, is regulated by MKK1-mediated H2O2 production during different types of abiotic stress, such as drought and salt stress, and is related to ABA-signaling [112]. In addition, transcript levels of CAT1, not CAT2 nor CAT3, were reduced in 2-week-old mkk1 knockout plants and increased in MKK1 overexpressing plants as compared to wild type plants after 4h exposure to 300 mM NaCl, drought stress (dehydration of plants to 80% of their original fresh weight followed by incubation at 100% relative humidity at 25 °C) or 0.1 mM ABA. Furthermore, production of H2O2 was abolished in mkk1 knockout plants and higher in MKK1 overexpressors treated with 300 mM NaCl, drought or 100 μM ABA. MKK1 mediates ABA-induced CAT1 expression via MPK6 [113]. As shown for mkk1 knockout and MKK1 overexpressing plants, CAT1 transcript levels were reduced in 2-week-old mpk6 knockout plants and elevated in MPK6 overexpressors exposed to 100 μM ABA. ABA treatment also inhibited H2O2 accumulation in mpk6 knockout plants.
5.3. MAPK Cascades Interfere with Hormone Signaling
Besides their role in physiological processes, ethylene and JA were originally identified as stress hormones essential for plant defense against a variety of abiotic and biotic stresses, such as ozone, UV radiation, mechanical stress, chemicals, metals, drought, extreme temperatures, insect and pathogen infection [114,115]. In the past years, more and more evidence was presented to indicate that MAPK cascades can play a role in ethylene and JA signaling (Figure 1).
5.3.1. Ethylene
MAPkinases have been reported to be involved in ethylene biosynthesis as well as in ethylene signaling. Several data indicate that MPK6 is involved in ethylene biosynthesis. It was reported in twelve-day-old Arabidopsis plants that MPK6, induced by flg22 or mechanical stress, is responsible for the phosphorylation of two ACC synthases, ACS2 and ACS6. Conversion of S-adenosyl-L-methionine to ACC by ACC synthases is the rate-limiting and major regulatory step in stress-induced ethylene production. Phosphorylation of ACS2 and ACS6 stabilizes the ACS proteins, leading to an elevated ACS activity and consequently an elevated ACC and ethylene production [116]. In addition, the MKK9-MPK3/6 pathway was shown to function in ethylene biosynthesis. Constitutive expression of MKK9 induced accumulation of ethylene through activation of MPK3/6 and consecutive positive regulation of ACS2 and ACS6 gene expression [117].
In contrast, other studies report that MAPKinases are involved in ethylene signal transduction rather than in ethylene biosynthesis. The function of the different components in the ethylene signaling pathway was recently reviewed by Shan et al. [118] and will be discussed here briefly. Binding of ethylene to its receptor, ETR1 (Ethylene Response 1), releases CTR1 (Constitutive Triple Response 1), which in the absence of ethylene leads to degradation of EIN2 (Ethylene Insensitive 2), inhibiting downstream signal transduction. Therefore, binding of ethylene leads to accumulation of EIN2 activating a transcriptional cascade, initiated by the transcription factor EIN3 (Ethylene Insensitive 3), regulating transcription of ethylene responsive genes [118]. The first evidence for the involvement of a MAPK cascade in plant ethylene signaling came with the discovery of the first gene in the ethylene signal transduction pathway, CTR1, a negative regulator of ethylene responses. Indeed, the predicted protein sequence of CTR1 showed similarity to the mammalian Raf kinase, a MAPKKK [119]. More recently, Novikova et al. [120] showed that a protein with similarities to a MAPK was activated by exogenous treatment of 6-week-old Arabidopsis plants with 1 μL L−1 ethylene during 1h. Treatment (10 min) of Arabidopsis wild type plants with 1mM ACC, the immediate precursor of ethylene in the biosynthesis pathway, identified the activation of MPK6. Mutant screenings showed that this activation of MPK6 by ACC is mediated by ETR1 and CTR1, but not by EIN2 or EIN3. These results place MPK6, as a positive regulator of ethylene responses, downstream of CTR1 and upstream of EIN2 in the ethylene signal transduction pathway [121]. A study by Yoo et al. [122] reported that the MAPK cascade MKK9-MPK3/6 functions downstream of CTR1, is activated upon binding of ethylene to ETR1 and is able to phosphorylate and stabilize EIN3 leading to transcription of ethylene responsive genes. Yeast-2-hybrid and fluorescence resonance energy transfer identified the in vitro as well in vivo interaction of MPK6 with an ethylene response factor ERF104. The MPK6/ERF104 complex is disrupted by flg22-induced ethylene production, leading to phosphorylation and activation of ERF104. Microarray analysis of ERF104 overexpressing plants identified the ERF104 stimulated targets as pathogenesis related (PDF1.2) or involved in further signal amplification of defense signaling (MKK4, RBOHD, WRKY33) [123].
5.3.2. Jasmonate
Similar to ethylene, MAPKs are indicated to play a role in the biosynthesis of JA as well as in JA signaling. In case of salt stress, transcripts of two genes involved in JA biosynthesis, OPR1 and OPR2, were shown to be more abundant in wrky33 knockout plants, suggesting that MAPK cascades involving WRKY33 can downregulate JA biosynthesis during heat stress [103]. Earlier, we mentioned that activation of transcription factors WRKY33 or ZAT12 by MAPK cascades induced gene expression of lipoxygenases, LOX1 and LOX4 respectively. Involvement of lipoxygenases in metal stress was also reported. Transcript levels of LOX2 were significantly induced in leaves of Arabidopsis plants after short term exposure to 2 μM Cu or 5 μM Cd. In the roots, a metal-specific upregulation of LOX3 and LOX4 was observed after Cu exposure, whereas transcript levels of LOX5 were specifically downregulated. In contrast, LOX1 and LOX6 gene expression was responsive to both Cd and Cu [21,124]. Further, expression of LOX1 and LOX2 is compromised in roots and leaves of non-exposed mpk6 knockout plants [125]. Lipoxygenases catalyze the first step in JA biosynthesis, namely the oxygenation of α-linolenic acid to hydroperoxides [115]. Earlier, LOX2 was identified as a lipoxygenase responsible for initiating JA biosynthesis upon wounding [126]. Taken together, these data suggest that MAPK cascades can initiate JA biosynthesis via transcriptional control of LOXes.
On the other hand, studies reported that MAPKs would function in JA signal transduction. Leon et al. [127] stated that reversible protein phosphorylation is involved in JA signaling. JA-dependent induction of wound-inducible genes was stimulated by treatment of Arabidopsis plants with the protein kinase inhibitor staurosporin while treatment with the protein phosphatase inhibitor okadaic acid repressed this gene expression. These results suggest a negative regulation of the JA downstream pathway by protein kinase cascades. Petersen et al. [128] showed that mpk4 knockout plants are impaired in expression of PDF1.2 and THI1.2, two JA-responsive genes. Moreover, treatment of 4-week-old mpk4 knockout plants with 50 μM methyl-JA for 48 h failed to induce PDF1.2 or THI1.2 transcript levels, indicating that MPK4 is involved in JA signaling. In addition, the MKK3-MPK6 pathway functions in JA signaling in Arabidopsis as a negative regulator of the downstream transcription factor MYC2. Treatment of mkk3 and mpk6 knockout as well as MKK3 and MPK6 overexpressing plants with 50 μM JA during 12 h, showed that the MKK3/MPK6 pathway induced or reduced the transcript levels of MYC2 respectively [93].
6. Conclusions
MAPK signaling plays a central role in plant metal stress responses. MAPkinases are activated by ROS production, induced upon metal stress, and convert the perception of metals to intracellular signals to the nucleus, where appropriate responses are initiated. However, MAPK cascades are not specific for a single stress condition. One MAPK cascade can be used by different biotic and abiotic stresses and interplay between different pathways is possible. In metal stress, the function of MAPK cascades is poorly understood while knowledge about metal signaling and more specific their downstream targets is essential for understanding plant responses to metal stress. Therefore, in future research it is important to focus on the functional analysis of MAPkinases in plant metal stress. For this purpose, mutants (knock-outs, overexpressors) of the different MAPKKKs, MAPKKs and MAPKs should be investigated under metal exposure in a single and multipollution context. Interaction between the different MAPKinase modules and the possible transcription factors activated by MAPKs can be identified by the use of functional protein microarrays [96] or phosphoproteomics [129]. Specific genes targeted by these transcription factors in their turn can be resolved using different molecular strategies. Better insight into plant metal stress responses and their regulation opens future perspectives to investigate the complexity of signaling modules in plant responses facing a globally changing environment.
Acknowledgments
The authors apologize to any researcher whose work is not cited here due to limitations of space and scope. This work was supported by the research fund of Hasselt University (BOFN0705) with a PhD grant for Kelly Opdenakker. Additional funding was obtained from the Fund for Scientific Research-Flanders (FWO-Belgium) (project G.0807.09), the Methusalem project (08M03VGRJ) and tUL-Impuls financing.
References
- 1.Nriagu J.O., Pacyna J.M. Quantitative assessment of worldwide contamination of air, water and soils by trace metals. Nature. 1988;333:134–139. doi: 10.1038/333134a0. [DOI] [PubMed] [Google Scholar]
- 2.Ruttens A., Boulet J., Weyens N., Smeets K., Adriaensen K., Meers E., van Slycken S., Tack F., Meiresonne L., Thewys T., Witters N., Carleer R., Dupae J., Vangronsveld J. Short rotation coppice culture of willow and poplar as energy crops on metal contaminated agricultural soils. Int. J. Phytoremediat. 2011;13:194–207. doi: 10.1080/15226514.2011.568543. [DOI] [PubMed] [Google Scholar]
- 3.Kirkham M.B. Cadmium in plants on polluted soils: Effects of soil factors, hyperaccumulation, and amendments. Geoderma. 2006;137:19–32. [Google Scholar]
- 4.Chary N.S., Kamala C.T., Raj D.S.S. Assessing risk of heavy metals from consuming food grown on sewage irrigated soils and food chain transfer. Ecotoxicol. Environ. Saf. 2008;69:513–524. doi: 10.1016/j.ecoenv.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 5.Marschner H. Functions of Mineral Nutrients: Micronutrients. In: Marschner H., editor. Mineral Nutrition of Higher Plants. 2nd ed. Academic Press; London, UK: 1995. pp. 313–404. [Google Scholar]
- 6.Broadley M.R., White P.J., Hammond J.P., Zelko I., Lux A. Zinc in plants. New Phytol. 2007;173:677–702. doi: 10.1111/j.1469-8137.2007.01996.x. [DOI] [PubMed] [Google Scholar]
- 7.Cuypers A., Smeets K., Vangronsveld J. Heavy Metal Stress in Plants. In: Hirt H., editor. Plant Stress Biology: From Genomics to Systems Biology. 1st ed. Wiley-VCH Verlag; Weinheim, Germany: 2009. pp. 161–178. [Google Scholar]
- 8.Leonard S.S., Bower J.J., Shi X. Metal-induced toxicity, carcinogenesis, mechanisms and cellular responses. Mol. Cell. Biochem. 2004;255:3–10. doi: 10.1023/b:mcbi.0000007255.72746.a6. [DOI] [PubMed] [Google Scholar]
- 9.Hogervorst J., Plusquin M., Vangronsveld J., Nawrot T., Cuypers A., van Hecke E., Roels H.A., Carleer R., Staessen J.A. House dust as possible route of environmental exposure to cadmium and lead in the adult general population. Environ. Res. 2007;103:30–37. doi: 10.1016/j.envres.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Nawrot T., Plusquin M., Hogervorst J., Roels H.A., Celis H., Thijs L., Vangronsveld J., van Hecke E., Staessen J.A. Environmental exposure to cadmium and risk of cancer: A prospective population-based study. Lancet Oncol. 2006;7:119–226. doi: 10.1016/S1470-2045(06)70545-9. [DOI] [PubMed] [Google Scholar]
- 11.Nawrot T., Staessen J.A., Roels H., Munters E., Cuypers A., Richart T., Ruttens A., Smeets K., Clijsters H., Vangronsveld J. Cadmium exposure in the population: from risks to measures of prevention. Biometals. 2010;23:769–782. doi: 10.1007/s10534-010-9343-z. [DOI] [PubMed] [Google Scholar]
- 12.Thijssen S., Cuypers A., Maringwa J., Smeets K., Horemans N., Lambrichts I., van Kerkhove E. Low cadmium exposure triggers a biphasic oxidative stress response in mice kidneys. Toxicology. 2007;236:29–41. doi: 10.1016/j.tox.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 13.DalCorso G., Farinati S., Maistri S., Furini A. How plants cope with cadmium: Staking all on metabolism and gene expression. J. Integr. Plant Biol. 2008;50:1268–1280. doi: 10.1111/j.1744-7909.2008.00737.x. [DOI] [PubMed] [Google Scholar]
- 14.Yruela I. Copper in plants: Acquisition, transport and interactions. Funct. Plant Biol. 2009;36:409–430. doi: 10.1071/FP08288. [DOI] [PubMed] [Google Scholar]
- 15.Smeets K., Opdenakker K., Remans T., van Sanden S., van Belleghem F., Semane B., Horemans N., Guisez Y., Vangronsveld J., Cuypers A. Oxidative stress-related responses at transcriptional and enzymatic levels after exposure to Cd or Cu in a multipollution context. J. Plant Physiol. 2009;166:1982–1992. doi: 10.1016/j.jplph.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 16.Cuypers A., Smeets K., Ruytinx J., Opdenakker K., Keunen E., Remans T., Horemans N., Vanhoudt N., van Sanden S., van Belleghem F., Guisez Y., Colpaert J., Vangronsveld J. The cellular redox state as a modulator in cadmium and copper responses in Arabidopsis thaliana seedlings. J. Plant Physiol. 2011;168:309–316. doi: 10.1016/j.jplph.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Cuypers A., Keunen E., Bohler S., Jozefczak M., Opdenakker K., Gielen H., Vercampt H., Bielen A., Schellingen K., Vangronsveld J., Remans T. Cadmium and Copper Stress Induce a Cellular Oxidative Challenge Leading to Damage versus Signalling. In: Gupta D.K.G., Sandalio L.M., editors. Metal Toxicity in Plants: Perception, Signalling and Remediation. 1st ed. Springer-Verlag GmbH; Berlin, Germany: 2011. pp. 65–90. [Google Scholar]
- 18.Gratao P.L., Polle A., Lea P.J., Azevedo R.A. Making the life of heavy metal-stressed plants a little easier. Funct. Plant Biol. 2005;32:481–494. doi: 10.1071/FP05016. [DOI] [PubMed] [Google Scholar]
- 19.Mittler R., Vanderauwera S., Gollery M., van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Pitzschke A., Forzani C., Hirt H. Reactive oxygen species signaling in plants. Antioxid. Redox Sign. 2006;8:1757–1764. doi: 10.1089/ars.2006.8.1757. [DOI] [PubMed] [Google Scholar]
- 21.Opdenakker K., Remans T., Keunen E., Vangronsveld J., Cuypers A. Exposure of Arabidopsis thaliana to Cd or Cu excess leads to oxidative stress mediated alterations in MAPKinase transcript levels. Environ. Exp. Bot. 2012;83:53–61. [Google Scholar]
- 22.Kalbina I., Strid A. The role of NADPH oxidase and MAP kinase phosphatase in UV-B-dependent gene expression in Arabidopsis. Plant Cell Environ. 2006;29:1783–1793. doi: 10.1111/j.1365-3040.2006.01555.x. [DOI] [PubMed] [Google Scholar]
- 23.Miller G., Suzuki N., Ciftci-Yilmaz S., Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010;33:453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- 24.Vandenabeele S., Vanderauwera S., Vuylsteke M., Rombauts S., Langebartels C., Seidlitz H.K., Zabeau M., van Montagu M., Inze D., van Breusegem F. Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 2004;39:45–58. doi: 10.1111/j.1365-313X.2004.02105.x. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki N., Mittler R. Reactive oxygen species and temperature stresses: A delicate balance between signaling and destruction. Physiol. Plant. 2006;126:45–51. [Google Scholar]
- 26.Torres M.A., Jones J.D.G., Dangl J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006;141:373–378. doi: 10.1104/pp.106.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers H.J. Is there an important role for reactive oxygen species and redox regulation during floral senescence? Plant Cell Environ. 2012;35:217–233. doi: 10.1111/j.1365-3040.2011.02373.x. [DOI] [PubMed] [Google Scholar]
- 28.Bright J., Desikan R., Hancock J.T., Weir I.S., Neill S.J. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 2006;45:113–122. doi: 10.1111/j.1365-313X.2005.02615.x. [DOI] [PubMed] [Google Scholar]
- 29.De Pinto M.C., Locato V., de Gara L. Redox regulation in plant programmed cell death. Plant Cell Environ. 2012;35:234–244. doi: 10.1111/j.1365-3040.2011.02387.x. [DOI] [PubMed] [Google Scholar]
- 30.Foreman J., Demidchik V., Bothwell J.H.F., Mylona P., Miedema H., Torres M.A., Linstead P., Costa S., Brownlee C., Jones J.D.G., Davies J.M., Dolan L. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature. 2003;422:442–446. doi: 10.1038/nature01485. [DOI] [PubMed] [Google Scholar]
- 31.Apel K., Hirt H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 32.Miller G., Shulaev V., Mittler R. Reactive oxygen signaling and abiotic stress. Physiol. Plant. 2008;133:481–489. doi: 10.1111/j.1399-3054.2008.01090.x. [DOI] [PubMed] [Google Scholar]
- 33.Quan L.J., Zhang B., Shi W.W., Li H.Y. Hydrogen peroxide in plants: A versatile molecule of the reactive oxygen species network. J. Integr. Plant Biol. 2008;50:2–18. doi: 10.1111/j.1744-7909.2007.00599.x. [DOI] [PubMed] [Google Scholar]
- 34.Mishra N.S., Tuteja R., Tuteja N. Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 2006;452:55–68. doi: 10.1016/j.abb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Ichimura K., Shinozaki K., Tena G., Sheen J., Henry Y., Champion A., Kreis M., Zhang S.Q., Hirt H., Wilson C., Heberle-Bors E., Ellis B.E., Morris P.C., Innes R.W., Ecker J.R., Scheel D., Klessig D.F., Machida Y., Mundy J., Ohashi Y., Walker J.C. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 2002;7:301–308. doi: 10.1016/s1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- 36.Jonak C., Okresz L., Bogre L., Hirt H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 2002;5:415–424. doi: 10.1016/s1369-5266(02)00285-6. [DOI] [PubMed] [Google Scholar]
- 37.Morrison D.K., Davis R.J. Regulation of map kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 38.Nakagami H., Kiegerl S., Hirt H. OMTK1, a novel MAPKKK, channels oxidative stress signaling through direct MAPK interaction. J. Biol. Chem. 2004;279:26959–26966. doi: 10.1074/jbc.M312662200. [DOI] [PubMed] [Google Scholar]
- 39.Bartels S., Besteiro M.A.G., Lang D., Ulm R. Emerging functions for plant MAP kinase phosphatases. Trends Plant Sci. 2010;15:322–329. doi: 10.1016/j.tplants.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Ulm R., Revenkova E., di Sansebastiano G.P., Bechtold N., Paszkowski J. Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Gene Dev. 2001;15:699–709. doi: 10.1101/gad.192601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ulm R., Ichimura K., Mizoguchi T., Peck S.C., Zhu T., Wang X., Shinozaki K., Paszkowski J. Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 2002;21:6483–6493. doi: 10.1093/emboj/cdf646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson J.C., Bartels S., Besteiro M.A.G., Shahollari B., Ulm R., Peck S.C. Arabidopsis MAP kinase phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J. 2011;67:258–268. doi: 10.1111/j.1365-313X.2011.04588.x. [DOI] [PubMed] [Google Scholar]
- 43.Besteiro M.A.G., Bartels S., Albert A., Ulm R. Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV-B stress tolerance, independent of the UVR8 photoreceptor pathway. Plant J. 2011;68:727–737. doi: 10.1111/j.1365-313X.2011.04725.x. [DOI] [PubMed] [Google Scholar]
- 44.Lee K., Song E.H., Kim H.S., Yoo J.H., Han H.J., Jung M.S., Lee S.M., Kim K.E., Kim M.C., Cho M.J., Chung W.S. Regulation of MAPK phosphatase 1 (AtMKP1) by calmodulin in Arabidopsis. J. Biol. Chem. 2008;283:23581–23588. doi: 10.1074/jbc.M801549200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartels S., Anderson J.C., Besteiro M.A.G., Carreri A., Hirt H., Buchala A., Metraux J.P., Peck S.C., Ulm R. MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell. 2009;21:2884–2897. doi: 10.1105/tpc.109.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J.S., Ellis B.E. Arabidopsis MAPK phosphatase 2 (MKP2) positively regulates oxidative stress tolerance and inactivates the MPK3 and MPK6 MAPKs. J. Biol. Chem. 2007;282:25020–25029. doi: 10.1074/jbc.M701888200. [DOI] [PubMed] [Google Scholar]
- 47.Lumbreras V., Vilela B., Irar S., Sole M., Capellades M., Valls M., Coca M., Pages M. MAPK phosphatase MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. Plant J. 2010;63:1017–1030. doi: 10.1111/j.1365-313X.2010.04297.x. [DOI] [PubMed] [Google Scholar]
- 48.Bargmann B.O., Munnik T. The role of phospholipase D in plant stress responses. Curr. Opin. Plant Biol. 2006;9:515–522. doi: 10.1016/j.pbi.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 49.Yu L.J., Nie J.N., Cao C.Y., Jin Y.K., Yan M., Wang F.Z., Liu J., Xiao Y., Liang Y.H., Zhang W.H. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 2010;188:762–773. doi: 10.1111/j.1469-8137.2010.03422.x. [DOI] [PubMed] [Google Scholar]
- 50.Anthony R.G., Henriques R., Helfer A., Meszaros T., Rios G., Testerink C., Munnik T., Deak M., Koncz C., Bogre L. A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 2004;23:572–581. doi: 10.1038/sj.emboj.7600068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rentel M.C., Lecourieux D., Ouaked F., Usher S.L., Petersen L., Okamoto H., Knight H., Peck S.C., Grierson C.S., Hirt H., Knight M.R. OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature. 2004;427:858–861. doi: 10.1038/nature02353. [DOI] [PubMed] [Google Scholar]
- 52.Camehl I., Drzewiecki C., Vadassery J., Shahollari B., Sherameti I., Forzani C., Munnik T., Hirt H., Oelmuller R. The OXI1 kinase pathway mediates Piriformospora indica-induced growth promotion in Arabidopsis. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anthony R.G., Khan S., Costa J., Pais M.S., Bogre L. The Arabidopsis protein kinase PTI1–2 is activated by convergent phosphatidic acid and oxidative stress signaling pathways downstream of PDK1 and OXI1. J. Biol. Chem. 2006;281:37536–37546. doi: 10.1074/jbc.M607341200. [DOI] [PubMed] [Google Scholar]
- 54.Forzani C., Carreri A., van Bentem S.D., Lecourieux D., Lecourieux F., Hirt H. The Arabidopsis protein kinase Pto-interacting 1–4 is a common target of the oxidative signal-inducible 1 and mitogen-activated protein kinases. Febs J. 2011;278:1126–1136. doi: 10.1111/j.1742-4658.2011.08033.x. [DOI] [PubMed] [Google Scholar]
- 55.Moon H., Lee B., Choi G., Shin S., Prasad D.T., Lee O., Kwak S.S., Kim D.H., Nam J., Bahk J., Hong J.C., Lee S.Y., Cho M.J., Lim C.O., Yun D.J. NDP kinase 2 interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proc. Natl. Acad. Sci. USA. 2003;100:358–363. doi: 10.1073/pnas.252641899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang T., Liu Y., Yang T., Zhang L., Xu S., Xue L., An L. Diverse signals converge at MAPK cascades in plant. Plant Physiol. Biochem. 2006;44:274–283. doi: 10.1016/j.plaphy.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 57.Pitzschke A., Hirt H. Disentangling the Complexity of Mitogen-activated protein kinases and reactive oxygen species signaling. Plant Physiol. 2009;149:606–615. doi: 10.1104/pp.108.131557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang S.Q., Klessig D.F. MAPK cascades in plant defense signaling. Trends Plant Sci. 2001;6:520–527. doi: 10.1016/s1360-1385(01)02103-3. [DOI] [PubMed] [Google Scholar]
- 59.Colcombet J., Hirt H. Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 2008;413:217–226. doi: 10.1042/BJ20080625. [DOI] [PubMed] [Google Scholar]
- 60.Liu X.M., Kim K.E., Kim K.C., Nguyen X.C., Han H.J., Jung M.S., Kim H.S., Kim S.H., Park H.C., Yun D.J., Chung W.S. Cadmium activates Arabidopsis MPK3 and MPK6 via accumulation of reactive oxygen species. Phytochemistry. 2010;71:614–618. doi: 10.1016/j.phytochem.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 61.Rao K.P., Vani G., Kumar K., Wankhede D.P., Misra M., Gupta M., Sinha A.K. Arsenic stress activates MAP kinase in rice roots and leaves. Arch. Biochem. Biophys. 2011;506:73–82. doi: 10.1016/j.abb.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 62.Jonak C., Nakagami H., Hirt H. Heavy metal stress. Activation of distinct mitogen-activated protein kinase pathways by copper and cadmium. Plant Physiol. 2004;136:3276–3283. doi: 10.1104/pp.104.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kovtun Y., Chiu W.L., Tena G., Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Opdenakker K. Centre for Environmental Sciences, Hasselt University, Diepenbeek, Belgium. Arabidopsis thaliana oxi1 and mpk6 knockout seedlings are less sensitive to Cu exposure. 2012 Unpublished work. [Google Scholar]
- 65.Roelofs D., Aarts M.G.M., Schat H., van Straalen N.M. Functional ecological genomics to demonstrate general and specific responses to abiotic stress. Funct. Ecol. 2008;22:8–18. [Google Scholar]
- 66.Suzuki N., Koizumi N., Sano H. Screening of cadmium-responsive genes in Arabidopsis thaliana. Plant Cell Environ. 2001;24:1177–1188. [Google Scholar]
- 67.Gupta M., Sharma P., Sarin N.B., Sinha A.K. Differential response of arsenic stress in two varieties of Brassica juncea L. Chemosphere. 2009;74:1201–1208. doi: 10.1016/j.chemosphere.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 68.Agrawal G.K., Rakwal R., Iwahashi H. Isolation of novel rice (Oryza sativa L.) multiple stress responsive MAP kinase gene, OsMSRMK2, whose mRNA accumulates rapidly in response to environmental cues. Biochem. Bioph. Res. Commun. 2002;294:1009–1016. doi: 10.1016/S0006-291X(02)00571-5. [DOI] [PubMed] [Google Scholar]
- 69.Agrawal G.K., Agrawal S.K., Shibato J., Iwahashi H., Rakwal R. Novel rice MAP kinases OsMSRMK3 and OsWJUMK1 involved in encountering diverse environmental stresses and developmental regulation. Biochem. Bioph. Res. Commun. 2003;300:775–783. doi: 10.1016/s0006-291x(02)02868-1. [DOI] [PubMed] [Google Scholar]
- 70.Yeh C.M., Hsiao L.J., Huang H.J. Cadmium activates a mitogen-activated protein kinase gene and MBP kinases in rice. Plant Cell Physiol. 2004;45:1306–1312. doi: 10.1093/pcp/pch135. [DOI] [PubMed] [Google Scholar]
- 71.Yeh C.M., Hung W.C., Huang H.J. Copper treatment activates mitogen-activated protein kinase signalling in rice. Physiol. Plant. 2003;119:392–399. [Google Scholar]
- 72.Hung W.C., Huang D.D., Yeh C.M., Huang H.J. Reactive oxygen species, calcium and serine/threonine phosphatase are required for copper-induced MAP kinase gene OsMAPK2, expression in rice. Plant Growth Regul. 2005;45:233–241. [Google Scholar]
- 73.Yeh C.M., Chien P.S., Huang H.J. Distinct signalling pathways for induction of MAP kinase activities by cadmium and copper in rice roots. J. Exp. Bot. 2007;58:659–671. doi: 10.1093/jxb/erl240. [DOI] [PubMed] [Google Scholar]
- 74.Lin C.W., Chang H.B., Huang H.J. Zinc induces mitogen-activated protein kinase activation mediated by reactive oxygen species in rice roots. Plant Physiol. Biochem. 2005;43:963–968. doi: 10.1016/j.plaphy.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 75.Huang T.L., Huang H.J. ROS and CDPK-like kinase-mediated activation of MAP kinase in rice roots exposed to lead. Chemosphere. 2008;71:1377–1385. doi: 10.1016/j.chemosphere.2007.11.031. [DOI] [PubMed] [Google Scholar]
- 76.Tsai T.M., Huang H.J. Effects of iron excess on cell viability and mitogen-activated protein kinase activation in rice roots. Physiol. Plant. 2006;127:583–592. [Google Scholar]
- 77.Wang J.X., Ding H.D., Zhang A., Ma F.F., Cao J.M., Jiang M.Y. A novel mitogen-activated protein kinase gene in maize (Zea mays), ZmMPK3, is involved in response to diverse environmental cues. J. Integr. Plant Biol. 2010;52:442–452. doi: 10.1111/j.1744-7909.2010.00906.x. [DOI] [PubMed] [Google Scholar]
- 78.Ding H.D., Tan M.P., Zhang C., Zhang Z.W., Zhang A.Y., Kang Y.J. Hexavalent chromium (VI) stress induces mitogen-activated protein kinase activation mediated by distinct signal molecules in roots of Zea mays L. Environ. Exp. Bot. 2009;67:328–334. [Google Scholar]
- 79.Ren D.T., Yang H.P., Zhang S.Q. Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 2002;277:559–565. doi: 10.1074/jbc.M109495200. [DOI] [PubMed] [Google Scholar]
- 80.Nakagami H., Soukupova H., Schikora A., Zarsky V., Hirt H. A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 2006;281:38697–38704. doi: 10.1074/jbc.M605293200. [DOI] [PubMed] [Google Scholar]
- 81.Ichimura K., Mizoguchi T., Irie K., Morris P., Giraudat J., Matsumoto K., Shinozaki K. Isolation of ATMEKK1 (a MAP kinase kinase kinase)—Interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem. Bioph. Res. Commun. 1998;253:532–543. doi: 10.1006/bbrc.1998.9796. [DOI] [PubMed] [Google Scholar]
- 82.Mizoguchi T., Ichimura K., Irie K., Morris P., Giraudat J., Matsumoto K., Shinozaki K. Identification of a possible MAP kinase cascade in Arabidopsis thaliana based on pairwise yeast two-hybrid analysis and functional complementation tests of yeast mutants. FEBS Lett. 1998;437:56–60. doi: 10.1016/s0014-5793(98)01197-1. [DOI] [PubMed] [Google Scholar]
- 83.Ichimura K., Mizoguchi T., Yoshida R., Yuasa T., Shinozaki K. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 2000;24:655–665. doi: 10.1046/j.1365-313x.2000.00913.x. [DOI] [PubMed] [Google Scholar]
- 84.Huang Y.F., Li H., Gupta R., Morris P.C., Luan S., Kieber J.J. ATMPK4, an Arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol. 2000;122:1301–1310. doi: 10.1104/pp.122.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Matsuoka D., Nanmori T., Sato K., Fukami Y., Kikkawa U., Yasuda T. Activation of AtMEK1, an Arabidopsis mitogen-activated protein kinase kinase, in vitro and in vivo: Analysis of active mutants expressed in E-coli and generation of the active form in stress response in seedlings. Plant J. 2002;29:637–647. doi: 10.1046/j.0960-7412.2001.01246.x. [DOI] [PubMed] [Google Scholar]
- 86.Pitzschke A., Djamei A., Bitton F., Hirt H. A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Mol. Plant. 2009;2:120–137. doi: 10.1093/mp/ssn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 88.Ding Y.F., Chen Z., Zhu C. Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa) J. Exp. Bot. 2011;62:3563–3573. doi: 10.1093/jxb/err046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim S.H., Woo D.H., Kim J.M., Lee S.Y., Chung W.S., Moon Y.H. Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem. Bioph. Res. Commun. 2011;412:150–154. doi: 10.1016/j.bbrc.2011.07.064. [DOI] [PubMed] [Google Scholar]
- 90.Suarez-Rodriguez M.C., Adams-Phillips L., Liu Y.D., Wang H.C., Su S.H., Jester P.J., Zhang S.Q., Bent A.F., Krysan P.J. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 2007;143:661–669. doi: 10.1104/pp.106.091389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Teige M., Scheikl E., Eulgem T., Doczi R., Ichimura K., Shinozaki K., Dangl J.L., Hirt H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell. 2004;15:141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 92.Doczi R., Brader G., Pettko-Szandtner A., Rajh I., Djamei A., Pitzschke A., Teige M., Hirt H. The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell. 2007;19:3266–3279. doi: 10.1105/tpc.106.050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takahashi F., Yoshida R., Ichimura K., Mizoguchi T., Seo S., Yonezawa M., Maruyama K., Yamaguchi-Shinozaki K., Shinozaki K. The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell. 2007;19:805–818. doi: 10.1105/tpc.106.046581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takahashi F., Mizoguchi T., Yoshida R., Ichimura K., Shinozaki K. Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis. Mol. Cell. 2011;41:649–660. doi: 10.1016/j.molcel.2011.02.029. [DOI] [PubMed] [Google Scholar]
- 95.Ortiz-Masia D., Perez-Amador M.A., Carbonell J., Marcote M.J. Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. FEBS Lett. 2007;581:1834–1840. doi: 10.1016/j.febslet.2007.03.075. [DOI] [PubMed] [Google Scholar]
- 96.Popescu S.C., Popescu G.V., Bachan S., Zhang Z.M., Gerstein M., Snyder M., Dinesh-Kumar S.P. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Gene Dev. 2009;23:80–92. doi: 10.1101/gad.1740009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 98.Gadjev I., Vanderauwera S., Gechev T.S., Laloi C., Minkov I.N., Shulaev V., Apel K., Inze D., Mittler R., van Breusegem F. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006;141:436–445. doi: 10.1104/pp.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou X., Jiang Y.J., Yu D.Q. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells. 2011;31:303–313. doi: 10.1007/s10059-011-0047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Andreasson E., Jenkins T., Brodersen P., Thorgrimsen S., Petersen N.H.T., Zhu S., Qiu J.L., Micheelsen P., Rocher A., Petersen M., Newman M.A., Nielsen H.B., Hirt H., Somssich I., Mattsson O., Mundy J. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 2005;24:2579–2589. doi: 10.1038/sj.emboj.7600737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zheng Z.Y., Abu Qamar S., Chen Z.X., Mengiste T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006;48:592–605. doi: 10.1111/j.1365-313X.2006.02901.x. [DOI] [PubMed] [Google Scholar]
- 102.Li S.J., Fu Q.T., Huang W.D., Yu D.Q. Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. 2009;28:683–693. doi: 10.1007/s00299-008-0666-y. [DOI] [PubMed] [Google Scholar]
- 103.Jiang Y.Q., Deyholos M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009;69:91–105. doi: 10.1007/s11103-008-9408-3. [DOI] [PubMed] [Google Scholar]
- 104.Vanderauwera S., Zimmermann P., Rombauts S., Vandenabeele S., Langebartels C., Gruissem W., Inze D., van Breusegem F. Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol. 2005;139:806–821. doi: 10.1104/pp.105.065896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Desikan R., Mackerness S.A.H., Hancock J.T., Neill S.J. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 2001;127:159–172. doi: 10.1104/pp.127.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cheong Y.H., Chang H.S., Gupta R., Wang X., Zhu T., Luan S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002;129:661–677. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Davletova S., Schlauch K., Coutu J., Mittler R. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 2005;139:847–856. doi: 10.1104/pp.105.068254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li S.J., Fu Q.T., Chen L.G., Huang W.D., Yu D.Q. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta. 2011;233:1237–1252. doi: 10.1007/s00425-011-1375-2. [DOI] [PubMed] [Google Scholar]
- 109.Rizhsky L., Davletova S., Liang H.J., Mittler R. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 2004;279:11736–11743. doi: 10.1074/jbc.M313350200. [DOI] [PubMed] [Google Scholar]
- 110.Davletova S., Rizhsky L., Liang H.J., Zhong S.Q., Oliver D.J., Coutu J., Shulaev V., Schlauch K., Mittler R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell. 2005;17:268–281. doi: 10.1105/tpc.104.026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vogel J.T., Zarka D.G., Van Buskirk H.A., Fowler S.G., Thomashow M.F. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- 112.Xing Y., Jia W.S., Zhang J.H. AtMEK1 mediates stress-induced gene expression of CAT1 catalase by triggering H2O2 production in Arabidopsis. J. Exp. Bot. 2007;58:2969–2981. doi: 10.1093/jxb/erm144. [DOI] [PubMed] [Google Scholar]
- 113.Xing Y., Jia W.S., Zhangl J.H. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 2008;54:440–451. doi: 10.1111/j.1365-313X.2008.03433.x. [DOI] [PubMed] [Google Scholar]
- 114.Kendrick M.D., Chang C. Ethylene signaling: New levels of complexity and regulation. Curr. Opin. Plant Biol. 2008;11:479–485. doi: 10.1016/j.pbi.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wasternack C., Kombrink E. Jasmonates: Structural requirements for lipid-derived signals active in plant stress responses and development. ACS Chem. Biol. 2010;5:63–77. doi: 10.1021/cb900269u. [DOI] [PubMed] [Google Scholar]
- 116.Liu Y.D., Zhang S.Q. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell. 2004;16:3386–3399. doi: 10.1105/tpc.104.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu J., Li Y., Wang Y., Liu H.X., Lei L., Yang H.L., Liu G.Q., Ren D.T. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 2008;283:26996–27006. doi: 10.1074/jbc.M801392200. [DOI] [PubMed] [Google Scholar]
- 118.Shan X.Y., Yan J.B., Xie D.X. Comparison of phytohormone signaling mechanisms. Curr. Opin. Plant Biol. 2012;15:84–91. doi: 10.1016/j.pbi.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 119.Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. Ctr1, a negative regulator of the ethylene response pathway in Arabidopsis, Encodes a member of the raf family of protein-kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 120.Novikova G.V., Moshkov I.E., Smith A.R., Hall M.A. The effect of ethylene on MAPKinase-like activity in Arabidopsis thaliana. FEBS Lett. 2000;474:29–32. doi: 10.1016/s0014-5793(00)01565-9. [DOI] [PubMed] [Google Scholar]
- 121.Ouaked F., Rozhon W., Lecourieux D., Hirt H. A MAPK pathway mediates ethylene signaling in plants. EMBO J. 2003;22:1282–1288. doi: 10.1093/emboj/cdg131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yoo S.D., Cho Y.H., Tena G., Xiong Y., Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature. 2008;451:789–796. doi: 10.1038/nature06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bethke G., Unthan T., Uhrig J.F., Poschl Y., Gust A.A., Scheel D., Lee J. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA. 2009;106:8067–8072. doi: 10.1073/pnas.0810206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Remans T., Opdenakker K., Smeets K., Mathijsen D., Vangronsveld J., Cuypers A. Metal-specific and NADPH oxidase dependent changes in lipoxygenase and NADPH oxidase gene expression in Arabidopsis thaliana exposed to cadmium or excess copper. Funct. Plant Biol. 2010;37:532–544. [Google Scholar]
- 125.Opdenakker K. Centre for Environmental Sciences, Hasselt University, Diepenbeek, Belgium. MPK6 regulates transcription of lipoxygenases in Arabidopsis thaliana. 2012 Unpublished work. [Google Scholar]
- 126.Bell E., Creelman R.A., Mullet J.E. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1995;92:8675–8679. doi: 10.1073/pnas.92.19.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Leon J., Rojo E., Sanchez-Serrano J.J. Wound signalling in plants. J. Exp. Bot. 2001;52:1–9. doi: 10.1093/jexbot/52.354.1. [DOI] [PubMed] [Google Scholar]
- 128.Petersen M., Brodersen P., Naested H., Andreasson E., Lindhart U., Johansen B., Nielsen H.B., Lacy M., Austin M.J., Parker J.E., Sharma S.B., Klessig D.F., Martienssen R., Mattsson O., Jensen A.B., Mundy J. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/s0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- 129.Van Bentem S.D., Anrather D., Dohnal I., Roitinger E., Csaszar E., Joore J., Buijnink J., Carreri A., Forzani C., Lorkovic Z.J., Barta A., Lecourieux D., Verhounig A., Jonak C., Hirt H. Site-specific phosphorylation profiling of Arabidopsis proteins by mass spectrometry and peptide chip analysis. J. Proteome Res. 2008;7:2458–2470. doi: 10.1021/pr8000173. [DOI] [PubMed] [Google Scholar]