Seasonal influenza causes hundreds of thousands of deaths annually, and the potential for vastly higher mortality from an influenza pandemic remains an ever-present threat to global health. Two recent papers in Cell1,2 and two in Nature3,4 describe a significant advance in the search for new influenza drugs. The studies reveal hundreds of human genes that are required for the influenza replicative cycle, at least some of which may emerge as novel drug targets.
Like almost all mammalian viruses, influenza virus has evolved strategies for exploiting host cell factors to promote its replication and suppress antiviral immune responses. Identification of these host factors would expand the number of potential drug targets far beyond the 11 proteins encoded in the viral genome. To discover and validate human genes implicated in the influenza life cycle, all four research teams1–4 transfected cultured human cells with pools of short interfering (si)RNAs designed to silence the majority of human genes and monitored the effects of knocking down individual genes on viral activity. Initial hits from each study were used to search databases of protein-protein interactions, allowing prediction of host-cell pathways that are likely to be needed either for the viral replicative cycle or for the immune response to viral infection.
High-throughput siRNA screens of human cells have been used previously to identify host genes important for infection by the HIV5–7, hepatitis C8,9 and West Nile10 viruses. For influenza virus, however, this approach was applied only in Drosophila melanogaster cells11 (Table 1). The relevance of the 110 factors identified in the Drosophila study to infection in humans remains unclear. Because flies are not a natural host of influenza virus, the authors had to use a modified virus. Moreover, flies do not express all of the proteins required for influenza virion assembly and infectivity.
Table 1.
siRNA-based screens to identify host factors associated with influenza virus infection and replication
| Host cell line | Virus used for screen | Readout | Events in viral life cycle | Networks analyzed | Pathways | Number of genes screened | Number of filtered hits | Ref. |
|---|---|---|---|---|---|---|---|---|
| Drosophila D-Mel2 | Recombinant WSN | Luciferase activity | Early only | NDa | NDa | 13,071 | 110 | 11 |
| Human U2OS | PR8b | HA expression | Early and late | HPRDc, BINDd, BioGRIDe | Endosomal acidification, vesicular trafficking, mitochondrial metabolism, RNA splicing, mRNA transport | 17,877 | 312 | 1 |
| Human HBEC | PR8 | Amount of infectious virus | Early and late | BioGRIDe, BINDd, IntActf | WNT, NF-κB, MAPK and p53 signaling,apoptosis | 1,745g | 2 | |
| Human A549 | Recombinant WSN | Luciferase activity | Early only | Reactomeh, BINDd, MINTi, HPRDc | Kinase-regulated signaling, ubiquitination, phosphatase activity, endosomal acidification, actin organization and function, sumoylation, autophagy, nucleocytoplas mic transport | 19,628 | 295 | 3 |
| Human A549 | WSN | NP expression and luciferase activity | Early and late | STRINGj, Reactomeh | Endosome trafficking, RNA splicing, CLK1 and p27 signaling | 22,843 | 287 | 4 |
ND, not determined.
A subset of the results were evaluated with seasonal or pandemic influenza viruses.
Human Protein Reference database (http://www.hprd.org/).
Biomolecular Interaction Network database (http://www.bind.ca/).
1,745 candidate genes identified from yeast two-hybrid and genome wide microarray analyses were subjected to an siRNA screen for validation.
Molecular Interactions database (http://mint.bio.uniroma2.it/mint/).
CLK1, CDC-like kinase 1; HA, hemagglutinin; HBEC, human bronchial epithelial cells; ; NP, influenza virus nucleoprotein; PR8, A/PR/8/34; WSN, A/WSN/33
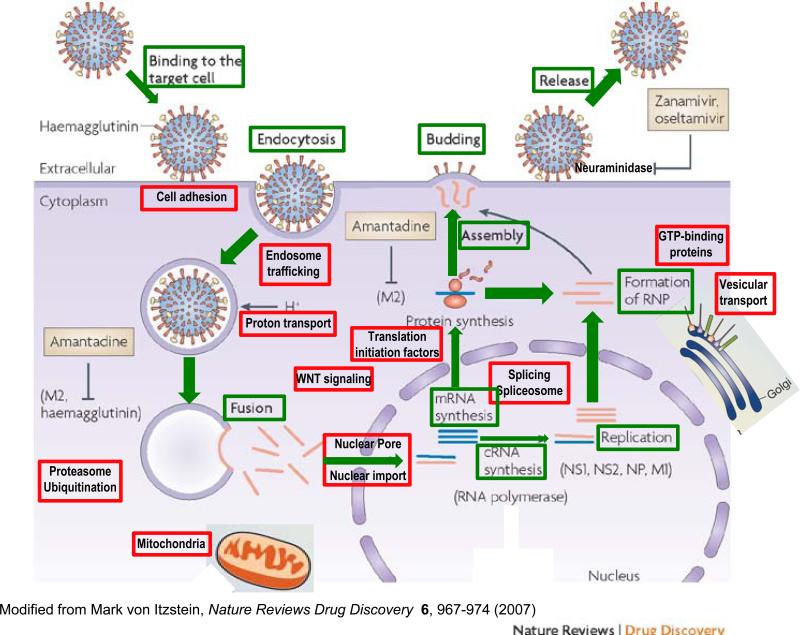
The four recent studies1–4 identified new genes and pathways associated with virtually every step in the influenza virus life cycle (Fig. 1). The early steps include attachment to the appropriate receptor on the surface of the host cell, virus entry through endocytosis, fusion of the endosomal and viral membranes to allow uncoating of the virus particle, and transport of the viral components to the nucleus, where transcription and replication take place. Thereafter, the viral ribonucleoprotein complex and RNA are exported into the cytoplasm, where translation and viral assembly occur, and progeny virions are released from the cell surface; these represent late events.
Figure 1.
Cellular targets for anti-influenza drugs in the context of the replication cycle of influenza virus. Stages of influenza A virus replication are in green. Cellular pathways shown by siRNA screens to be essential for completion of the viral replication cycle are shown in red. The influenza A virus protein hemagglutinin binds to sialylated glycoprotein receptors on the host-cell surface and enters the cell by receptor-mediated endocytosis. Following internalization and endosomal acidification, which permits fusion of the host and viral membranes by altering the conformation of hemagglutinin, viral ribonucleoproteins (RNPs; dark blue) are released in the cytoplasm. In the nucleus of infected cells, the viral RNAs are transcribed into mRNAs and replicated by the viral RNA–dependent RNA polymerase complex. The newly synthesized viral RNPs are exported into the cytoplasm and, after assembly, mature virions bud from the cell surface. Currently, the viral M2 ion channel protein and neuraminidase are the only two targets of influenza antiviral drugs (gray boxes) licensed by the US Food and Drug Administration. Adamantane drugs, which include amantadine and rimantadine, block the action of the viral M2 protein during uncoating of the virus. Zanamivir and oseltamivir target neuraminidase, which is required for release of progeny virus from the cell surface. Adapted from ref. 13.
Brass et al.1 used fluorescence microscopy to score the abundance of the influenza envelope glycoprotein hemagglutinin as a marker of infection with influenza A/PR/8/34 (PR8) virus in siRNA-treated osteosarcoma cells and identified 312 human genes that regulated susceptibility or resistance to influenza PR8 virus infection. Notably, they demonstrated that interferon-inducible transmembrane proteins inhibit an early step of influenza replication.
König et al.3 infected siRNA-transfected cells with a recombinant influenza A/WSN/33 (WSN) virus in which the coding region of hemagglutinin was replaced by a luciferase reporter gene. The absence of functional hemagglutinin protein in this screen limited it to identifying genes involved in early stages of infection (viral entry, uncoating and nuclear import), as well as viral RNA transcription and translation. Late events, such as virus assembly, budding and release, could not be scored. These authors found 295 genes whose knockdown by at least two siRNAs reduced luciferase expression, and knockdown of 219 of these genes inhibited WSN virus replication in multiple-cycle growth. Some of the identified host factors were shown to be important for replication of the swine-origin 2009 pandemic H1N1 virus. Moreover, the authors demonstrated that small-molecule inhibitors of several factors, including the vacuolar ATPase and calcium/calmodulin-dependent protein kinase 2β, inhibited influenza virus replication.
Karlas et al.4 used a two-step approach in which A549 cells transfected with an siRNA library were infected with the WSN virus and cells were immunostained for viral nucleoprotein expression as a marker of virus infection. In the second step, the culture supernatants from the A549 cells were transferred to 293T human embryonic kidney reporter cells bearing an inducible influenza virus–specific luciferase construct. This approach identified 287 host factors that affected replication of WSN virus. The authors then confirmed the involvement of a majority of these host factors in the replicative cycles of other influenza viruses, including the 2009 pandemic H1N1 virus, by showing that transfection with siRNAs targeting the host factors reduced virus titers. Notably, they provided in vivo evidence for the importance of the p27 gene by studying p27 knockout mice.
In contrast to these fluorescence-based approaches1,3,4, Shapira et al.2 used a combination of yeast two-hybrid and genome-wide microarray analyses to define a physical and regulatory map of host-virus interactions comprising 1,745 candidate genes involved in influenza PR8 infection. Using siRNA technology, they then validated the roles of all 1,745 candidate genes.
Notwithstanding the value of these screens to identify candidate genes for further analysis using molecular biology and virology tools, it must be noted that all siRNA screens have inherent limitations. First, because siRNAs deplete only a single gene at a time, genes whose functions are redundant with other genes might not be found. Second, unannotated genes and genes that are crucial for virus infection but are lethal or toxic to the viability of host cell will probably be excluded. Lastly, the data set gathered from each screen seems to depend strongly on the conditions and readouts of the screen. Indeed, although each of the five studies1–5 had the same goal, they did not identify the same set of host genes; a close comparison of the five studies shows that fewer than 20 genes were recovered more than once.
This discrepancy can likely be explained by differences in the cell lines, assay designs, and siRNA libraries used in the four studies (Table 1). Whereas Brass et al.1 worked with an osteosarcoma cell line (U2OS) and PR8 virus, König et al.3 and Karlas et al.4 both used carcinoma human alveolar basal epithelial cells (A549), and used recombinant and wild-type WSN viruses, respectively. The conditions for siRNA treatment and virus infection also differed between the studies. Brass et al.1 treated cells with siRNA for 72 h before infection and scored viral infectivity 12 h after infection. In contrast, König et al.3 and Karlas et al.4 treated cells with siRNA for 48 h and scored readouts 12 h after infection. Thus, the screens may have identified host genes that affect different stages of infection.
The hundreds of new host factors identified by the four studies1–4 should be welcome news to drug developers. Current influenza drugs target only two proteins, and both are viral gene products: the M2 ion channel (the target of amantadine (Symmetrel) and its derivative rimantadine) and neuraminidase (the target of zanamivir (Relenza) and oseltamivir (Tamiflu)). Unfortunately, resistance to both classes of drugs is now widespread, and new antiviral drugs are urgently needed to combat influenza epidemics and pandemics. Host-cell factors have been successfully targeted in the case of other viral infections (e.g., the CCR5 coreceptor of HIV-1), and they have the distinct advantage of lacking the high mutation rates of influenza virus genes, which have enabled the development of resistance to current drugs. Finally, it is worth noting that viruses from different families may interact with the same cellular factors with widely disparate effects. For example, CCR5 deficiency is a risk factor for early clinical manifestations of West Nile virus infection12. Although the extent to which the data from these screens will influence drug development remains to be seen, they unquestionably extend our understanding of influenza virus biology and suggest potentially fruitful leads for the rational design of broad-spectrum antiviral drugs.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Brass AL, et al. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shapira SD, et al. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.König R, et al. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlas A, et al. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 5.Brass AL, et al. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 6.König R, et al. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H, et al. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Cherry S, et al. Genes Dev. 2005;19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, et al. Proc. Natl. Acad. Sci. USA. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnan MN, et al. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao L, et al. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim JK, et al. J. Infect. Dis. 2010;201:178–185. doi: 10.1086/649426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Itzstein M. Nat. Rev. Drug Discov. 2007;6:967–974. doi: 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]