Abstract
Native human DNA polymerase η, DNA and dATP were crystallized at pH 6.0 without Mg2+. The polymerization reaction was initiated by exposing crystals to 1 mM Mg2+ at pH 7.0, and stopped at various time points by freezing at 77 K for crystallographic analysis. The substrates and two Mg2+ are aligned for reaction within 40 seconds, but the new bond begins to form at 80 seconds. Structures at reaction times from 80s to 300s reveal a mixture of decreasing substrate and increasing product of the nucleotidyl-transfer reaction. In sequence, the nucleophile 3′-OH is deprotonated, the deoxyribose at the primer end converts from C2′-endo to C3′-endo to avoid steric clashes, and the nucleophile and the α-phosphate of dATP approach each other to form the new bond. A third Mg2+ ion, which arrives with the new bond and stabilizes the intermediate state, may be an unappreciated feature of the two-metal-ion mechanism.
Keywords: nucleotidyl transfer, DNA polymerase, Mg2+, deprotonation, sugar pucker
Formation and breakage of chemical bonds underlie all life processes. DNA replication, which is essential for cell proliferation, is but one example 1,2. In each reaction cycle, a dNTP complementary to the templating base is incorporated into DNA in a nucleotidyl-transfer reaction catalyzed by a polymerase, during which a new bond is formed between the 3′-OH of the primer strand and the α-phosphate of the dNTP and the phosphodiester bond between α and β-phosphates of dNTP is broken (Fig. 1a). As a result the primer strand is extended by one nucleotide and a pyrophosphate is released. This reaction has been shown to require two Mg2+ ions and inferred to be the associative SN2 type with a pentacovalent phosphate intermediate 3. A similar two-metal-ion dependent mechanism is believed to be shared by all DNA and RNA polymerases and many nucleases 4,5.
Fig 1.
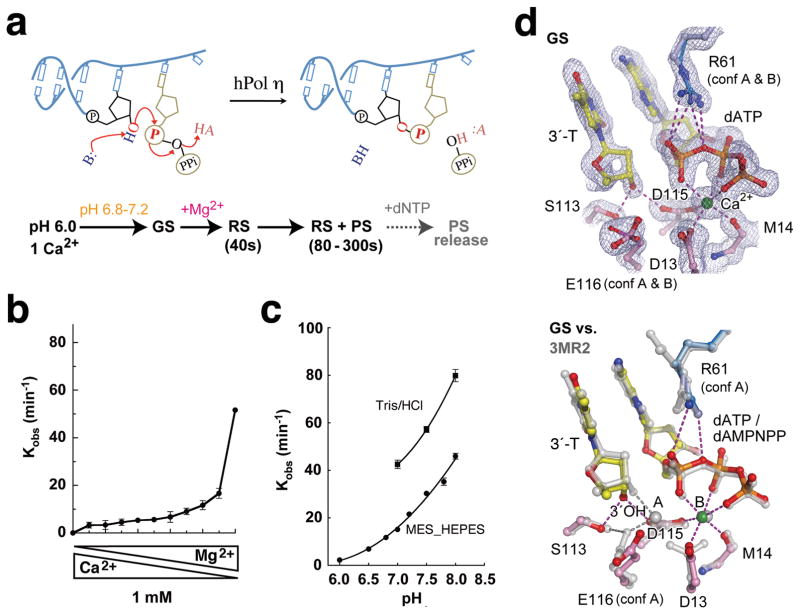
pH and metal ion dependence of hPol η. (a) Diagrams of DNA synthesis and the procedure of the in crystallo reaction. AH and B stands for general acid and base, respectively. (b) Reaction rate (kobs) is plotted against mixed Mg2+ and Ca2+ at the total of 1mM. (c) The pH and buffer dependent reaction rates. (d) The GS structure. The active site is superimposed with the 2Fo-Fc electron density contoured at 1.2σ (upper panel) or with the non-reactive dAMPNPP ternary complex (PDB: 3MR2, silver, lower panel). Hydrogen bonds and metal ion coordination are shown as dashed lines.
DNA synthesis has been analyzed by kinetic measurements, dynamic simulations, and structural studies 2,6,7. Crystal structures of a number of DNA polymerases in substrate-bound forms have been determined using non-reactive substrate analogs or Ca2+ instead of Mg2+ to prevent the nucleotidyl transfer reaction 5,8–18. These structures reveal active-site configurations in an array of pre-reaction ground states. Exquisite pre-steady state kinetic studies of several DNA polymerases indicate that after large conformational changes induced by substrate binding, unspecified subtle conformational changes in the active site are the rate-limiting step 19–27. The actual process of DNA synthesis has never been visualized.
Human DNA polymerase η(hPol η) is specialized in lesion bypass and has a preformed catalytic center that undergoes limited conformational change during DNA synthesis 28. Crystal structures of the catalytic domain of hPol η complexed with DNA and a non-reactive dNTP analog poised for catalysis have been determined at 1.8 Å resolution 29,30. Taking advantage of the slow reaction rate and hindered post-reaction DNA translocation in crystallo, we follow the course of DNA synthesis by X-ray crystallography and report here structural changes and transient elements that are associated with the reaction.
The Ground State of hPol η
The hPol η activity in a reaction buffer containing 0.9 mM Mg2+ and 0.1 mM Ca2+ was only one-thirds of that with 1mM Mg2+ (Fig. 1b), suggesting that both active-site metal ions have to be Mg2+ for the catalysis. In accordance with the bell-shaped pH-dependence curve characteristic for the acid-base catalysis 17,31, the catalytic rate of hPol η is extremely low at pH 6.0 and rises with increasing pH from 6.0 to 8.0 (Fig. 1c). To prevent nucleotidyl-transfer reaction, crystals of native hPol η-substrate complexes were grown at pH 6.0 with only one Ca2+ per protein-DNA-dATP complex (Methods). After soaking in MES (pH 6.0–7.2) or HEPES buffer (pH 7.0–7.5) with Na+ or K+ but no divalent cation for 5 to 30 min, these crystals maintained excellent diffraction up to pH 7.0, but decayed at pH 7.5 with reduced resolution and increased diffuse scattering.
The structure of hPol η crystal equilibrated at pH 6.8 was refined at 1.50 Å. A Ca2+ ion clearly occupies the B metal-ion binding site and is coordinated in the octahedral geometry by oxygen atoms of the dATP and the active-site residues (Fig. 1d). The A site has low occupancy of a monovalent cation. The occupancy of Na+ or K+ in the A-site increases with the pH (Supplementary Fig. 1a,b), which is probably correlated with deprotonation of the active-site carboxylates. hPol η binds Na+ more readily than K+, particularly below pH 7.0, probably because Na+ is similar to Mg2+ in size and smaller than K+. As expected, there is no nucleotidyl-transfer reaction, and the structure is termed the ground-state ternary complex (GS).
The GS structure is overall similar to the hPol η ternary complex with a non-reactive dATP-analog (dAMPNPP) and two Mg2+ ions (PDB 3MR2) 29. However, without an A-site divalent cation, the 3′-OH of the primer strand shifts away from the dATP and forms hydrogen bonds with the sidechains of S113 and D115, and the active-site carboxylates D13 and E116 adopt different rotamer conformations (Fig. 1d). Replacement of dAMPNPP with dATP leads to stabilization of R61, which makes bidentate hydrogen bonds with the α-phosphate.
Catalysis in crystallo
The nucleotidyl transfer was initiated by transferring hPol η crystals to a pH 6.8 or 7.0 reaction buffer containing 1 mM Mg2+ but no dATP (Methods). After incubation at 293 K for 40s to 300s, the reaction was terminated in ~40s intervals by freezing crystals in liquid nitrogen at 77 K. Diffraction data were collected to 1.50 – 1.95 Å Bragg spacings (Supplementary Table 1). The reaction process is monitored by the electron density corresponding to the new chemical bond in the Fo-Fc map compared with the refined GS structure (Supplementary Fig. 2, Fig. 2a). The reaction time courses at pH 6.8 and 7.0 in crystallo (Fig. 2b) are ~20–100 fold slower than in solution (Fig. 1c), probably due to the reduced thermal motion. By 40s, the A-site was fully occupied with Mg2+ (Fig. 2a, Supplementary Fig. 1c,d). About 50% of Ca2+ in the B site was replaced by Mg2+, and exchange of the remaining Ca2+ took place slowly (Supplementary Fig. 3). Binding of two Mg2+ ions leads to the alignment of the 3′-OH and dATP 18,32. The refined structure is nearly identical to that with dAMPNPP (PDB: 3MR2) except for tighter dATP coordination by R61 and closer of the Mg2+ ions (3.4 Å apart versus 3.6 Å). Since there is no sign of bond formation, the structure is termed the reactant state (RS).
Fig. 2.
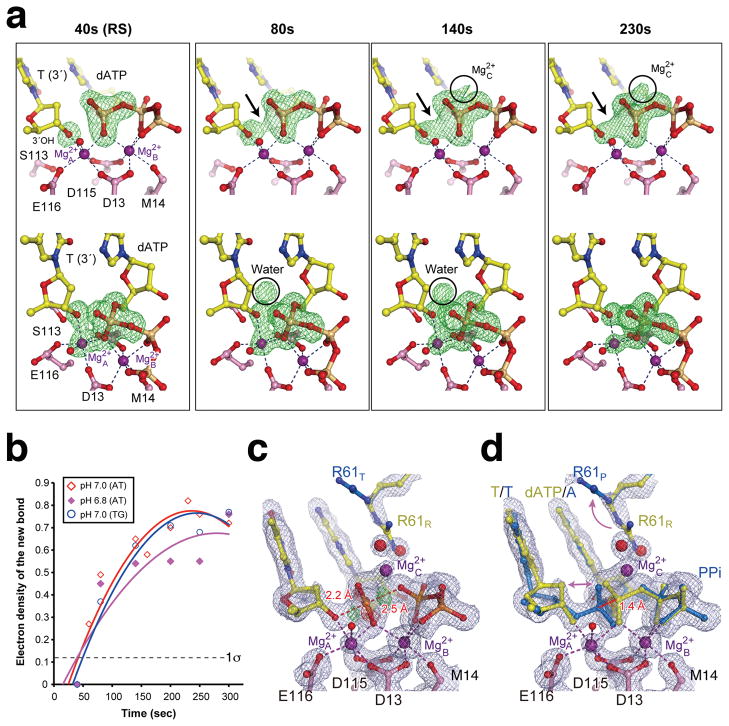
Reaction time course. (a) Two views (upper and lower panels) of omit Fo40-230s - Fc40s maps (4.0σ) superimposed onto the 40s structure (pH 7.0). The emerging densities are pointed out or circled. (b) A plot of the absolute peak height of the new bond density versus reaction time at pH 6.8 and 7.0 in AT and TG crystals (Supplementary Table 1a,b). The noise level (1σ) is indicated. (c) The 230s structure refined as the pentacovalent transition state (TS), or (d) the RS (yellow) and PS (blue) mixture. The 2Fo-Fc (1σ, grey) and Fo-Fc map (±3.0σ green and red) are superimposed.
Electron density corresponding to a new bond between the 3′-OH and the α-phosphorus of dATP begins to emerge at 80 seconds, increases quickly in the following 60s, and reaches its maximum after 200s when the reaction is 60–70% complete (Fig. 2b). The seemingly reduced rate after 140s is likely due to the reverse reaction in crystallo. The slight decline of product after 250s (Fig. 2b) is due to a sideway product translocation, which is clear with longer incubation (Supplementary Fig 4a). To alleviate an impediment to proper DNA translocation by the crystal lattice, we replaced an AT basepair with a mismatch at the DNA end that formed lattice contacts (Methods, Supplementary Table 2). One DNA with a TG mismatch led to isomorphous crystals and proper translocation of the DNA product (Supplementary Fig 4b). Interestingly, the time courses of nucleotidyl transfer in the AT and TG crystals are nearly identical and unaffected by the lattice contacts (Fig. 2b).
At the peak of chemical bond formation between 200 and 250s, the scissile phosphate can be refined in a penta-covalent transition state without restraints (Fig. 2c). But the bond distances between the phosphorus and the attacking or leaving oxygen atoms are 2.2 to 2.5 Å (Fig. 2c), much longer than the expected 2.0 Å observed with transition-state mimics like AlF4 or MgF3 33–35. Moreover, in the Fo-Fc map residual electron densities are observed around the new and scissile phosphodiester bonds (Fig. 2c). The same diffraction data, however, can be well fitted as a mixture of the reactant (RS) and the product state (PS) immediately before and after the nucleotidyl transfer (Fig 2d). Because the transition state is transient and unstable, the structures obtained between 80s and 300s are refined as a mixture of the RS and PS at different ratios (Fig. 2b, Supplementary Table 1a). For instance the 1.52 Å structure at 230s contains 40% substrate and 60% product complexes.
Between the RS and PS, the protein, DNA and dATP are superimposable except for atoms in the reaction center. Most notably, the α-phosphorus of the dATP moves 1.4 Å along a straight line between the attacking and leaving oxygen atoms (separated by 4.6 Å) (Fig. 2d). The shift of the α-phosphate is accompanied by alteration of R61 of hPolη, which flips away from the scissile phosphate and is replaced by a new metal ion and water molecules (Fig. 2a,d, see details below). On the primer strand, changes are confined to the 3′ nucleotide. The 3′-OH together with the deoxyribose moves towards the α-phosphate by 0.5 Å, and the sugar pucker changes from C2′ endo in the RS to C3′ endo in the PS (Fig. 2d). With the loss of the nucleophile and α-phosphate as ligands, the A-site Mg2+ dissociates in the PS as evidenced by the declining occupancy (Supplementary Fig 1c,d). Concomitantly, D13 assumes a second conformation and forms an H-bond with K224.
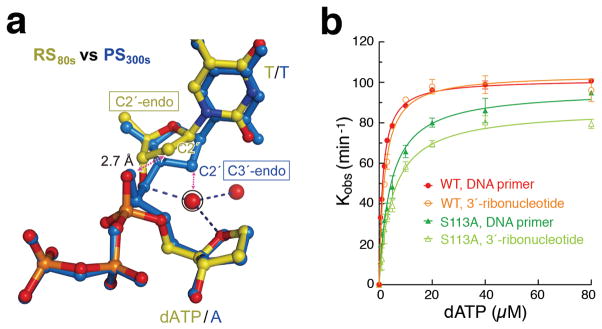
The C3′-endo (A-form) conformation at the 3′ primer end was observed in ternary complexes with the A, B and X-family DNA polymerases and thought to be important for forming a shallow minor-groove for dNTP selection 9,11,12,14,15,17,18,32. Among the Y-family polymerases, the primer end has always been observed as C2′-endo28. In the hPol η GS and RS complexes, the dATP and the nucleotide 5′ to the primer end have the A-form conformation, but only in the PS structure does the primer end adopt the C3′-endo conformation to avoid clashes between its C2′ atom and the non-bridging oxygen of dATP during and immediately after the nucleophilic attack (Fig. 2d, 3a). Since the electron density is weak for the sugar moiety at the 3′-primer end, we tested the effect of the A-form conformation by using a primer with a ribonucleotide at its 3′ end. The catalytic efficiency (kcat/KM) of hPol ηis comparable whether it is rA- or dA (Fig. 3b), as observed for DNA pol β36, indicating that the A-form conformation is probably necessary for DNA synthesis in general.
Fig. 3.
Deprotonation of the 3′-OH. (a) Superposition of the refined RS (80s, yellow) and PS (300s, blue). The transient water molecule, likely deprotonating the 3′-OH, is circled. The sugar pucker at the primer 3′-end changes from C2′-endo in the RS to C3′-endo in the PS. The clashes between the α-phosphate and the C2′ if it is C2′-endo, and between the C2′ and the transient water if the water doesn’t depart, are indicated by dashed double arrowheads. (b) WT and S113A mutant hPol η can both extend primer with a ribonucleotide at the 3′end (lighter color) as with pure DNA (darker color).
A Transient Water Molecule
Two unexpected spheres of electron density appear in the Fo-Fc maps in the course of new bond formation (Fig. 2a). The first is within hydrogen bonding distance of the 3′-OH. Its electron density peaks at 80s and drastically declines after 140s when the PS becomes prominent. Although absent in the GS and the 40s RS structure, the electron density is superimposable with a water molecule observed in the hPol η-dAMPNPP ternary complex (3MR2). This water is hydrogen bonded with the 3′-OH, the O4′ of dATP and another water in the 80s RS structure, but it is incompatible with the C3′-endo conformation in the PS (Fig. 3a). The 3′-OH is also hydrogen bonded to a water molecule in an X-family DNA polymerase crystallized with dUMPNPP 32. A water-mediated and substrate-assisted catalytic mechanism was proposed previously 37,38. Besides this water, S113 is hydrogen bonded with the 3′-OH in the GS and may pick up the proton and passes it to E116 in the RS (Fig. 1d). However, the S113A mutant hPol η retains 95% of kcat with a 3-fold increase of KM (Fig. 3b, Supplementary Table 3). Interestingly, mutations of the S113-equivalent in the A- and B-family DNA polymerases (highly conserved histidine and threonine, respectively) have limited impact on catalytic efficiency 39–41. We therefore propose that 3′-OH is prone to deprotonation when coordinated by the Mg2+ and aligned with the incoming nucleotide 4. The proton can be passed onto the transient water molecule and then to bulk solvent. When the 3′-end of primer is a ribonucleotide, the O2′ may relay the proton out instead. An incoming dNTP most probably participates in deprotonation of the nucleophile because with the nitrogen substitution in dAMPNPP the water molecule is stably bound to the 3′-OH 29,30.
The Third Metal Ion
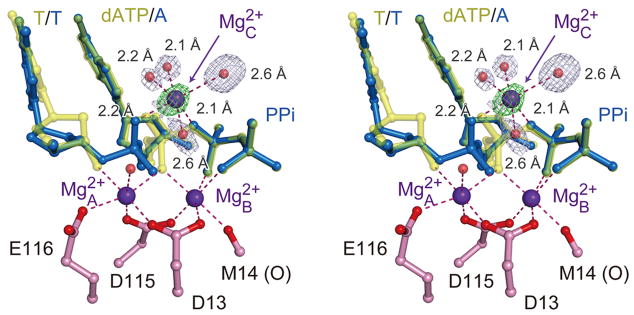
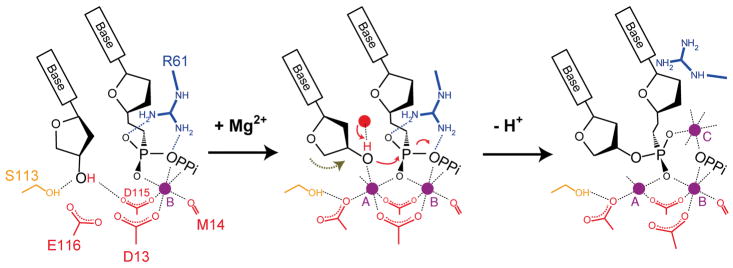
The second emerging sphere of electron density is very close to the α-phosphate on the opposite side of the A- and B-site Mg2+ ions (Fig. 2a). It starts to appear at 140s and intensifies with the reaction time. By 230s with the structure refined at 1.52 Å resolution, the octahedral geometry and the short coordination distances indicate that this is a divalent cation and most likely Mg2+ (Fig. 4). This third metal ion is liganded by four water molecules, two of which occupy the space of the departed R61 (Fig. 2d). Two additional ligands are the leaving oxygen (bridging between the α and β phosphate) and the non-bridging oxygen of the α-phosphate (Fig. 4). This Mg2+ thus bridges the two reaction products destined to separate and prevents DNA translocation. In the TG crystals with 1mM Mg2+ in the reaction buffer, the mixed RS and PS intermediate stays at equilibrium for up to 15 minutes, but with reduced Mg2+ (5μM) and addition of dATP (5μM), product release occurs much faster (Supplemental Information). The physiological role of the third metal ion is likely to neutralize the negative charge built up in the transition state and may also facilitate protonation of the pyrophosphate.
Fig. 4.
The 3rd Mg2+ ion in hPol η catalysis. A stereo view of the three metal ions observed in the intermediate state of 230s after Mg2+ addition, which consists of a mixture of PS (blue) and RS (yellow). The 3rd Mg2+ has six ligands, four of which are water molecules. The 2Fo-Fc map contoured at 1.5 σ (grey) and Fo-Fc map with the 3rd Mg2+ omitted contoured at 4.0 σ (green) are overlaid with the ball-and-stick model.
Three metal ions have been structurally observed in a number of enzymes catalyzing phosphoryl-transfer reactions, including alkaline phosphatase, P1 nuclease and Endo IV 33,42–44. In the case of Endo IV, the three Zn2+ ions observed in both substrate and product state 44 are reminiscent of the Mg2+ with hPol η(Supplementary Fig. 5). The two metal ions essential for catalysis flank the scissile phosphate on one side, and the third metal ion bridges the reaction products on the other. A similar arrangement of three metal ions was proposed for group I introns based on chemical probing 45. However, the third metal ion was absent in crystal structure of a splicing intermediate, likely due to its transient nature 46.
Concluding Remarks
Previously, flash-freeze was used to trap covalent intermediates and conformational changes associated with a chemical reaction 47–49. Here we extend the technology to record DNA synthesis in real time and at atomic resolution (Fig. 5, Supplementary Movie 1). The arrival of a water molecule to deprotonate the 3′-OH occurs only after two-Mg2+ induced substrate alignment and in the presence of a correct dNTP but not a non-reactive dAMPNPP. Mg2+ dependent and water-mediated deprotonation may occur in all DNA polymerases. The third metal ion that replaces the sidechain of R61 during the nucleotidyl transfer likely stabilizes the transition state and facilitates product release. In the one-metal-ion dependent HUH nucleases and topoisomerases a conserved Lys or Arg also occupies the position equivalent to R61 in hPol η50. The involvement of an additional transient metal ion may be a general feature of both one- and two-metal-ion mechanism.
Fig. 5.
A proposed mechanism for metal-ion dependent polymerase reaction. The B-site metal ion is stably associated with the incoming dNTP and the enzyme. Binding of Mg2+ at the A-site aligns the reactants, in particular the 3′-OH, and promotes its deprotonation, which is assisted by the dNTP and the transient water. After the reaction is initiated and before the products are released, a third metal ion replaces R61 to stabilize the reaction intermediates.
Methods
hPol η, DNA and dATP complexes mixed with Ca2+ at 1:1 molar ratio were concentrated to ~3 mg/ml hPol η and crystallized by the hanging-drop vapor diffusion method29. For in crystallo reactions, crystals were first stabilized in 0.1 M MES (pH 6.8–7.2), 5 μM dATP, 20% (w/v) PEG 2K-MME and 1 mM DTT at a specific pH for ~30 min, transferred in a nylon loop to the reaction buffer containing 0.1 M MES (pH 6.8–7.2), 1 mM MgCl2, 20% (w/v) PEG 2K-MME and 1 mM DTT for the desired amount of time, followed by a quick dipping in a cryo-solution supplemented with 20% (w/v) glycerol and flash cooled in liquid nitrogen. The time courses were determined based on the Fo-Fc maps with all data scaled against and compared to the refined GS structures. The intermediate states were refined as a mixture of RS and PS, and their ratio at each time point was determined according to the reaction course (Fig. 2b) and difference (Fo-Fc) electron density maps. Catalytic rates (kobs), kcat and KM were measured using the template/primer listed in Supplementary Table 3.
Online Only Methods
Protein and Crystal Preparation
hPol η mutant S113A was made using QuikChange (Stratagene). Wildtype hPolη(residues 1–432) and S113A mutant proteins were expressed and purified as described29. The ternary complexes of hPolη, DNA and dATP were mixed with Ca2+ at 1:1 molar ratio and concentrated to a protein concentration of ~3 mg/ml. Oligonucleotides used for original (A:T pair at the DNA end) and three “loosely packed” crystals (T:G or G:T mispaired and an unpaired A) are shown in Supplementary Table 2. An unpaired nucleotide at the primer 5′ end was disordered in the original hPol η structures 35, and was therefore removed to improve the diffraction quality. All crystals were obtained using the hanging-drop vapor diffusion method against a reservoir solution containing 0.1 M MES (pH 6.0) and 9-17% (w/v) PEG 2K-MME and streak seeding.
Chemical Reaction in crystallo
Based on diffraction qualities, the complexes containing the original A:T and T:G-end DNA were selected for chemical reaction in crystallo. These crystals were first transferred and incubated in a pre-reaction buffer containing 0.1 M MES (pH 6.8–7.2), 5 μM dATP, 20% (w/v) PEG 2K-MME and 1 mM DTT for ~30 min. For the pH 6.8 time course (Fig. 2b, Table S1b) and translocation in the TG crystal, the pre-reaction buffer also contained 5μM MgCl2. The different pre-reaction buffer might cause the larger variations of the pH 6.8 reactions. Chemical reaction was initiated by transferring the crystals with a Nylon loop into a reaction buffer containing 0.1 M MES (pH 6.8–7.2), 1 mM MgCl2, 20% (w/v) PEG 2K-MME and 1 mM DTT. After incubation for a desired time point (40s and intervals of ~40s), the crystals were quickly dipped in a cryo-solution supplemented with 20% (w/v) glycerol and flash cooled in liquid nitrogen. For in crystallo reactions, MES buffer was titrated by KOH and not NaOH to avoid binding of Na+ ion at the A site. To observe product release, TG crystals were soaked in 5μM dATP and 5μM MgCl2 for 30 min and chased with addition of 1 mM MgCl2 for 1–8 min.
Data Collection and Structure Determination
Diffraction data were collected at 100 K on beamlines 22ID and 22BM of the Advanced Photon Source. Data were processed with HKL2000 51 or XDS 52, and converted to structure factors by TRUNCATE. All data were isomorphic in the P61 space group and processed with identical unit-cell parameters a = b = 98.8 Å and c = 82.4 Å for structure and electron density comparison. All structural figures were drawn using PyMOL (http://www.pymol.org).
To monitor the new bond formation (Fig. 2a), Fo-Fc maps are calculated using the Fo of 40s, 80s, 140s and 230s after addition of 1mM Mg2+ (pH 7.0) and the Fc of the refined RS structure (40s) with the 3′-OH of the primer end, the A site Mg2+ and the α-phosphate of dATP omitted. To determine the reaction time courses (Fig. 2b and Supplementary Fig. 1-3), all data were scaled against and compared to the refined GS structure, and electron densities for the new bond (between the 3′-OH of the primer strand and the α-phosphorus of dATP) and the metal ions were measured in the Fo-Fc map (with the B-site metal ion omitted) at each time point using programs in CCP4 53. After refinement of the GS structure, the intermediate structures were refined as a mixture of RS and PS using PHENIX 54 and COOT 55. The ratio of RS and PS at each time point was determined according to the time course of the new bond formation (Fig. 2b) and difference (Fo-Fc) electron density maps. For high-resolution refinement (~1.5 Å resolution), the translation-libration-screw refinement was applied. Data collection and refinement statistics are summarized in Supplementary Table 1.
Kinetic Measurements
Kinetic measurements including Km and kcat were done using the template/primer shown in supplementary Table 3. The basic reaction mixture contained 2.5 nM hPol η, 5 uM 5′-FAM-labeled primer and template, and 0-80 μM dNTP in 40 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 10 mM dithiothreitol, 100 mM KCl, 0.1 mg/ml bovine serum albumin, and 5% glycerol. For the metal ion competition assay, Mg2+ was increased from 0 to 1.0 mM at 0.1 mM step and Ca2+ was decreased to keep the combined concentration of 1.0 mM. The effects of pH were screened from pH 6.0 to pH 8.0. All reactions took place at room temperature for 4 or 8 min and were stopped by addition of 10X formamide loading buffer to the final concentrations of 8% formamide, 1 mM EDTA (pH 8.0) and 0.1 mg/ml xylene cyanol. After heating to 90 °C for 3 min and immediately placing on ice, products were resolved on 20% poly-acrylamide sequencing gels containing 5.5 M urea. Quantification and curve fitting were carried out as described 20. KM, kcat and (kcat/KM) of WT and S113A hPol ηcatalyzing single nucleotide incorporation using deoxyribonucleotide versus ribonucleotide primer are summarized in Supplementary Table 3 and plotted in Fig. 3b.
Supplementary Material
Acknowledgments
We thank D. Leahy, M. Gellert and R. Craigie for editing the manuscript. The research was supported by the intramural research program of NIDDK, NIH (W.Y., T.N. and Y.Z.), the JSPS Institutional Program for Young Researcher Overseas Kumamoto University, and KUMAYAKU Alumni Research Fund (T.N.), Chinese Ministry of Education scholarship (Y.Z.), National Natural Science Foundation of China (Y.H.), and Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Y.Y.).
Footnotes
Author contributions
T.N. and Y.Z. carried out crystallography. Y.Z. did mutagenesis and kinetic analyses. T.N. handled crystals, refined structures and analyzed reaction time courses. W.Y. designed the project. T.N., Y.Z. and W.Y. prepared the manuscript. T.N. and Y.Z. contributed equally to the study. Y.Y. supervised T.N. and Y.H. advised Y.Z.. All authors discussed the results and commented on the manuscript
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank with accession codes from 4ECQ to 4ECZ, 4ED0 to 4ED3, and 4ED6 to 4ED8
The authors declare no competing financial interests.
References
- 1.Echols H, Goodman MF. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 2.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 3.Brody RS, Frey PA. Unambiguous determination of the stereochemistry of nucleotidyl transfer catalyzed by DNA polymerase I from Escherichia coli. Biochemistry. 1981;20:1245–1252. doi: 10.1021/bi00508a030. [DOI] [PubMed] [Google Scholar]
- 4.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Johnson KA. Role of induced fit in enzyme specificity: a molecular forward/reverse switch. J Biol Chem. 2008;283:26297–26301. doi: 10.1074/jbc.R800034200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warshel A, et al. Electrostatic basis for enzyme catalysis. Chem Rev. 2006;106:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 8.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 9.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 10.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Korolev S, Waksman G. Crystal structures of open and closedforms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 13.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 14.Berman AJ, et al. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J. 2007;26:3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc Natl Acad Sci U S A. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans RJ, et al. Structure of PolC reveals unique DNA binding and fidelity determinants. Proc Natl Acad Sci U S A. 2008;105:20695–20700. doi: 10.1073/pnas.0809989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F, Yang W. Structural insight into translesion synthesis by DNA Pol II. Cell. 2009;139:1279–1289. doi: 10.1016/j.cell.2009.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batra VK, et al. Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 20.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 21.Showalter AK, Tsai MD. A reexamination of the nucleotide incorporation fidelity of DNA polymerases. Biochemistry. 2002;41:10571–10576. doi: 10.1021/bi026021i. [DOI] [PubMed] [Google Scholar]
- 22.Shah AM, Li SX, Anderson KS, Sweasy JB. Y265H mutator mutant of DNA polymerase beta. Proper teometric alignment is critical for fidelity. J Biol Chem. 2001;276:10824–10831. doi: 10.1074/jbc.M008680200. [DOI] [PubMed] [Google Scholar]
- 23.Fiala KA, Suo Z. Mechanism of DNA polymerization catalyzed by Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry. 2004;43:2116–2125. doi: 10.1021/bi035746z. [DOI] [PubMed] [Google Scholar]
- 24.Cramer J, Restle T. Pre-steady-state kinetic characterization of the DinB homologue DNA polymerase of Sulfolobus solfataricus. J Biol Chem. 2005;280:40552–40558. doi: 10.1074/jbc.M504481200. [DOI] [PubMed] [Google Scholar]
- 25.Choi JY, Guengerich FP. Adduct size limits efficient and error-free bypass across bulky N2-guanine DNA lesions by human DNA polymerase eta. J Mol Biol. 2005;352:72–90. doi: 10.1016/j.jmb.2005.06.079. [DOI] [PubMed] [Google Scholar]
- 26.Rothwell PJ, Mitaksov V, Waksman G. Motions of the fingers subdomain of klentaq1 are fast and not rate limiting: implications for the molecular basis of fidelity in DNA polymerases. Mol Cell. 2005;19:345–355. doi: 10.1016/j.molcel.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Cao W, Zakharova E, Konigsberg W, De La Cruz EM. Fluorescence of 2-aminopurine reveals rapid conformational changes in the RB69 DNA polymerase-primer/template complexes upon binding and incorporation of matched deoxynucleoside triphosphates. Nucleic Acids Res. 2007;35:6052–6062. doi: 10.1093/nar/gkm587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci U S A. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biertumpfel C, et al. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, et al. Structural basis of human DNA polymerase eta-mediated chemoresistance to cisplatin. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1202681109. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castro C, et al. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc Natl Acad Sci U S A. 2007;104:4267–4272. doi: 10.1073/pnas.0608952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Role of the catalytic metal during polymerization by DNA polymerase lambda. DNA Repair (Amst) 2007;6:1333–1340. doi: 10.1016/j.dnarep.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Du MH, et al. Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase. J Mol Biol. 2002;316:941–953. doi: 10.1006/jmbi.2001.5384. [DOI] [PubMed] [Google Scholar]
- 34.Graham DL, et al. MgF(3)(-) as a transition state analog of phosphoryl transfer. Chem Biol. 2002;9:375–381. doi: 10.1016/s1074-5521(02)00112-6. [DOI] [PubMed] [Google Scholar]
- 35.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavanaugh NA, Beard WA, Wilson SH. DNA polymerase beta ribonucleotide discrimination: insertion, misinsertion, extension, and coding. J Biol Chem. 2010;285:24457–24465. doi: 10.1074/jbc.M110.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Broyde S, Zhang Y. Polymerase-tailored variations in the water-mediated and substrate-assisted mechanism for nucleotidyl transfer: insights from a study of T7 DNA polymerase. J Mol Biol. 2009;389:787–796. doi: 10.1016/j.jmb.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Yu X, Hu P, Broyde S, Zhang Y. A water-mediated and substrate-assisted catalytic mechanism for Sulfolobus solfataricus DNA polymerase IV. J Am Chem Soc. 2007;129:4731–4737. doi: 10.1021/ja068821c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polesky AH, Dahlberg ME, Benkovic SJ, Grindley ND, Joyce CM. Side chains involved in catalysis of the polymerase reaction of DNA polymerase I from Escherichia coli. J Biol Chem. 1992;267:8417–8428. [PubMed] [Google Scholar]
- 40.Copeland WC, Wang TS. Mutational analysis of the human DNA polymerase alpha. The most conserved region in alpha-like DNA polymerases is involved in metal-specific catalysis. J Biol Chem. 1993;268:11028–11040. [PubMed] [Google Scholar]
- 41.Zakharova E, Wang J, Konigsberg W. The activity of selected RB69 DNA polymerase mutants can be restored by manganese ions: the existence of alternative metal ion ligands used during the polymerization cycle. Biochemistry. 2004;43:6587–6595. doi: 10.1021/bi049615p. [DOI] [PubMed] [Google Scholar]
- 42.Kim EE, Wyckoff HW. Reaction mechanism of alkaline phosphatase based on crystal structures. Two-metal ion catalysis. J Mol Biol. 1991;218:449–464. doi: 10.1016/0022-2836(91)90724-k. [DOI] [PubMed] [Google Scholar]
- 43.Romier C, Dominguez R, Lahm A, Dahl O, Suck D. Recognition of single-stranded DNA by nuclease P1: high resolution crystal structures of complexes with substrate analogs. Proteins. 1998;32:414–424. [PubMed] [Google Scholar]
- 44.Garcin ED, et al. DNA apurinic-apyrimidinic site binding and excision by endonuclease IV. Nat Struct Mol Biol. 2008;15:515–522. doi: 10.1038/nsmb.1414. [DOI] [PubMed] [Google Scholar]
- 45.Shan S, Yoshida A, Sun S, Piccirilli JA, Herschlag D. Three metal ions at the active site of the Tetrahymena group I ribozyme. Proc Natl Acad Sci U S A. 1999;96:12299–12304. doi: 10.1073/pnas.96.22.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stahley MR, Strobel SA. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 47.Ding X, Rasmussen BF, Petsko GA, Ringe D. Direct structural observation of an acyl-enzyme intermediate in the hydrolysis of an ester substrate by elastase. Biochemistry. 1994;33:9285–9293. [PubMed] [Google Scholar]
- 48.Scott WG, Murray JB, Arnold JR, Stoddard BL, Klug A. Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science. 1996;274:2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- 49.Murray JB, Szoke H, Szoke A, Scott WG. Capture and visualization of a catalytic RNA enzyme-product complex using crystal lattice trapping and X-ray holographic reconstruction. Mol Cell. 2000;5:279–287. doi: 10.1016/s1097-2765(00)80423-2. [DOI] [PubMed] [Google Scholar]
- 50.Yang W. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 52.Kabsch W. XDS. Acta Crystallographica Section D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 54.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.