Abstract
ProExC expression has been shown to perform similarly to p16 as an aid in the diagnosis of cervical dysplasia but has not been well characterized in head and neck squamous cell carcinomas (SCC). The purpose of this study is to determine if ProExC performs similarly to p16 as a prognostic marker in oropharyngeal SCC and to evaluate the threshold of ProExC and p16 staining that correlates with survival. ProExC, p16 and HPV DNA in situ hybridization (ISH) were performed on tissue microarray (TMA) cores and whole sections from 62 patients with oropharyngeal SCC. Sensitivity and specificity for high-risk HPV and correlation with overall survival (OS), cancer-specific survival (CSS) and time to distant metastasis (TDM) were calculated for ProExC and p16 at different thresholds. ProExC did not prove to be a robust marker. It showed strong correlation with OS at a 66% threshold on TMA cores but correlation with OS was lost on whole sections. It also exhibited low sensitivity (53.7%) on TMA cores and low specificity on whole sections (65%). ProExC at a 33% threshold exhibited unacceptably low specificity and did not correlate with OS, CSS or TDM. Sensitivity and specificity of p16 varied predictably with threshold: higher sensitivity and lower specificity with lower thresholds and vice versa for higher thresholds. p16 at a 50% threshold offers a balance between sensitivity and specificity, and correlates with OS, CSS and TDM on whole sections; correlation with TDM is lost on TMA cores. These findings indicate that ProExC does not perform well enough to be used as a prognostic marker in oropharyngeal SCC. p16 should be used and scored as positive when at least half the tumor is strongly staining.
Keywords: head and neck, squamous cell carcinoma, oropharynx, ProExC, p16, HPV, in situ hybridization, PCR
INTRODUCTION
It is now commonly accepted that there are different pathways for developing head and neck squamous cell carcinomas (HNSCC). The HPV-related pathway is associated with infection by high-risk HPV, predominantly type 16, and most commonly affects the oropharynx. In contrast, non-HPV related HNSCC are associated with tobacco smoking, alcohol use, and poor dentition, and occur more commonly in mucosal sites outside the oropharynx. There has been a significant increase in the incidence of oropharyngeal SCC from 1973–2001 in younger whites (ages 20–44 years), especially in men, but an overall decline in the incidence of non-HPV related HNSCC from 1983–2004. (7, 34) While the published estimates of HPV-related oropharyngeal SCC vary from 8–100%, the International Agency for Research on Cancer (IARC) has determined an overall HPV prevalence of 25.9%. This estimate is based on review of the literature through February 2004 and encompasses 5,035 HNSCC cases from 60 studies and 26 countries.(25)
HPV-related HNSCC have a significantly better prognosis and a higher response to chemotherapy and radiation therapy as compared with their HPV-negative counterparts. (1, 10, 12, 13) In a prospective study of 96 patients with stage III or IV HNSCC (oropharynx or larynx) and median follow-up of 39.1 months, patients with HPV-related tumors showed a significantly better response to induction chemotherapy (82% vs. 55%) and chemoradiation (84% vs. 57%), as well as higher overall survival (95% vs. 62%). (13) Ancillary markers for HPV also play a role in evaluating metastatic carcinomas of unknown primary involving cervical lymph nodes. The presence of high-risk HPV often points to an oropharyngeal (base of tongue or palatine tonsil) primary. (5, 11, 17, 23)
Given the clinical and prognostic implications of HPV in HNSCC, reliable detection of HPV is critical. Immunohistochemical stain for p16 has been shown to be a surrogate marker for HPV that correlates strongly with survival and response to therapy, (12, 13, 26) as well as oropharyngeal origin. (4) However, overexpression of p16 can be observed in a small subset of HPV negative cases and the definition of a positive p16 stain has not been well-delineated, varying from study to study. (35)
ProExC is an immunohistochemical surrogate marker for HPV that targets cell cycle proteins minichromosome maintenance protein-2 (MCM2) and topoisomerase II-alpha (TOP2A). Recent work has shown that TOP2A and MCM2 are among the S-phase proteins induced by viral DNA integration into the host genome, leading to increased levels of E6 and E7 and aberrant S-phase induction. (14, 19, 29) ProExC was first used as a surrogate marker for high-risk HPV in cytology specimens from the uterine cervix and has been shown to perform similarly to p16 as an aid in the histologic diagnosis of uterine cervical dysplasia. (2, 3, 6, 8, 21, 33, 36) Compared with the proliferation marker Ki67, ProExC has been shown to result in a lower false positive rate in the diagnosis of cervical squamous dysplasia. (31) Essentially all cervical squamous cell carcinomas are related to high-risk HPV and ancillary markers are used as an aid in the morphologic diagnosis of precursor lesions. However, immunohistochemical markers for HPV play a different role in HNSCC where only a subset of HNSCC is HPV-related and ancillary markers are used for determining prognosis. To our knowledge, no study has evaluated the utility of ProExC as a prognostic indicator in HNSCC.
We had previously examined the relationship of p16 and HPV DNA ISH with survival but did not study the effect of varying thresholds of p16 reactivity or the performance of ProExC. (24) The aim of the current study is to determine the appropriate threshold for defining p16 and ProExC as positive in oropharyngeal SCC and to determine if ProExC is interchangeable with p16 as a prognostic marker. The performance of the antibodies is correlated with survival and evaluated on whole sections, as well as TMA cores. Since ancillary markers are commonly performed on cell block material from fine needle aspiration (FNA) biopsies, TMA cores were used to mimic the scant material present in FNA specimens.
MATERIALS AND METHODS
Case selection
62 patients with oropharyngeal squamous cell carcinoma were identified from the Stanford Department of Radiation Oncology clinical database. All patients had paraffin-embedded pathology specimens and clinical follow-up, including overall survival (OS, time from starting treatment to death due to any cause), cancer-specific survival (CSS, time from starting treatment until death due to oropharyngeal SCC) and time to distant metastasis (time from starting treatment until metastasis). The mean follow-up was 37.5 months (range: 1.5–122.9 months).
Tissue microarray
The tissue microarray (TMA) was constructed by using a tissue arrayer (Beecher Instruments, Silver Spring, MD) to create a new paraffin block from representative 0.6 mm cores taken in duplicate from 64 paraffin blocks of oropharyngeal SCC.(28) Two of the patients had cores taken from two separate paraffin blocks. The microarray also included control cores of skin, placenta, and benign lymph node.
Immunohistochemistry
4 uM thick sections were cut from the TMA blocks and routine paraffin-embedded blocks of tumor. Routine whole sections were available for all patients (n=62) represented in the tissue microarray. The slides were deparaffinized in xylene, hydrated in graded alcohols, and stained for ProExC (pre-dilute, EDTA; BD, Franklin Lakes, NJ) and p16 (clone E6H4, pre-dilute, Tris pH 9.0; mtm, Westborough, MA). The slides were independently scored by two pathologists (CSK, AMM) who were blinded to the outcome data. Discrepant scores were resolved by dual-headed review of the slides by the two pathologists. Of the 62 patients, all were evaluable for p16 and ProExC on whole sections. 60/62 cases had at least one TMA core evaluable for p16 and 61/62, for ProExC.
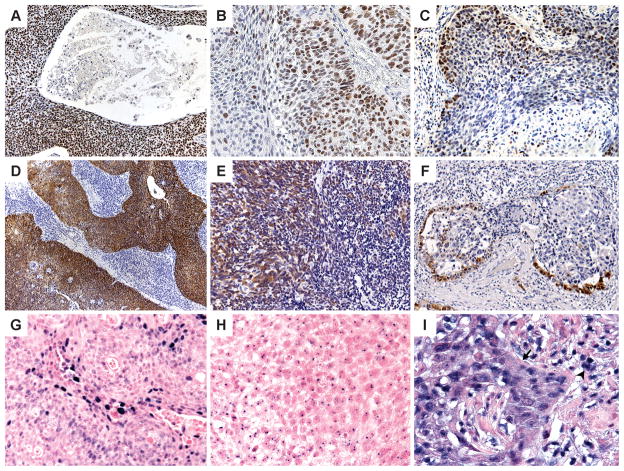
Nuclear staining with ProExC was scored as negative (<33%), focal (33–66%), or strong (>66%). (Figure 1A–C) These thresholds were chosen based on their use in studies of ProExC in the uterine cervix.(3) Since ProExC stains the basal layer of normal squamous mucosa, the mucosal thickness involved has to be evaluated when interpreting ProExC in cervical squamous dysplasia. The difference with evaluating ProExC in SCC is that invasive SCC lacks a basal cell layer. The percent tumor stained with ProExC was evaluated, not distribution of staining.
Figure 1.
ProExC immunohistochemistry: Staining of >95% (A) and 50% (B) of the tumor; staining of less than 33% of the tumor cells (C) is defined as negative. p16 immunohistochemistry: 90% (D), 50% (E) and 10% (F) strong nuclear and cytoplasmic staining of the tumor. HPV in situ hybridization: Positive HPV ISH can appear as diffuse (F) and/or punctate (G) nuclear reactivity; high background (H) can lead to staining of lymphocytes (arrowhead) and punctate staining that extends beyond the tumor nucleus and into the cytoplasm.
Strong nuclear or nuclear and cytoplasmic staining with p16 staining was scored on whole sections and TMA as negative (0–<5%), focal (5–50%), moderate (50–80%) or diffuse (>80%). (Figure 1D–F) On whole sections, a specific percent positive was also determined.
HPV DNA in situ hybridization
The INFORM® HPV ISH assay (Ventana) was performed on 4 μM-thick sections of the TMA and whole sections with the Benchmark ™ Automated Slide Stainer, utilizing the HPV III probe that demonstrates positive hybridization to HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 66. Punctate or diffuse nuclear staining was scored as positive. (Figure 1G–H) High background with staining of non-tumor cells or punctate staining that extended beyond the nucleus was interpreted as equivocal. (Figure 1I) HPV ISH results were available for 61/62 cases on whole sections and 62/62 cases on TMA (at least one core evaluable).
HPV DNA PCR/Pyrosequencing
PCR (DNA) for the L1 region of HPV was performed on paraffin-embedded material using the forward primer GP5+ (5′-TTTGTTACTGTTGTTGATACTAC-3′) and the biotinylated reverse primer GP6+ (5′-GAAAAATAAACTGTAAATCATATTC-3′) as previously described.(15) For all samples, the β-globulin gene was used as an internal amplification control to ensure DNA integrity. Single-stranded DNA samples were prepared semi-automatically. Twelve type-specific primers for high risk HPV-16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58 and -59 were used. Sequencing by pyrosequencing technology as described earlier was performed on an automated plate-based bench-top PSQ™ HS96A System.(15, 16)
58/62 samples had sufficient material for evaluation. 8 samples were considered non-evaluable for further HPV analysis based on unreadable signals. 54 of the 58 samples had sufficient material for further PCR evaluation with E1 specific primers; 4 of the samples with unreadable signals did not have sufficient material for further evaluation.
Statistical analysis
R statistical software was used to analyze the relationship of ProExC and p16 to OS, CSS and TDM. Kaplan Meier survival curves were generated and logrank tests were used to compare survival curves. P values less than 0.05 were considered to be statistically significant.
Sensitivity and specificity of p16 and ProExC for the detection of high-risk HPV were calculated with 95% confidence intervals (CI) based on different staining thresholds. HPV DNA PCR and HPV DNA ISH were used as the gold standards with positive defined as positive for HPV DNA by either PCR or ISH.
RESULTS
Data summary (Table 1)
TABLE 1.
Summary of raw p16 and ProExC data. HPV(+) defined as positive by HPV DNA ISH or PCR.
| WHOLE SECTIONS | TMA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| # HPV(+)cases | p16 ≥80 | p16 ≥50 | p16 ≥5 | ProExC >66 | ProExC >33 | # HPV(+)cases | p16 ≥80 | p16 ≥50 | p16 ≥5 | ProExC >66 | ProExC >33 |
| 25 | (+) | (+) | (+) | (+) | (+) | 17 | (+) | (+) | (+) | (+) | (+) |
| 2 | (+) | (+) | (+) | 0 | (+) | 8 | (+) | (+) | (+) | 0 | (+) |
| 3 | 0 | (+) | (+) | (+) | (+) | 3 | (+) | (+) | (+) | 0 | 0 |
| 1 | 0 | (+) | (+) | 0 | 0 | 2 | 0 | (+) | (+) | (+) | (+) |
| 2 | 0 | 0 | (+) | 0 | (+) | 2 | 0 | (+) | (+) | 0 | (+) |
| 6 | 0 | 0 | 0 | (+) | (+) | 2 | 0 | 0 | (+) | (+) | (+) |
| 2 | 0 | 0 | 0 | 0 | (+) | 1 | 0 | 0 | (+) | 0 | (+) |
| 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | (+) | (+) |
| 4 | 0 | 0 | 0 | 0 | 0 | ||||||
| 1 | 0 | (+) | (+) | N/A | N/A | ||||||
| 1 | N/A | N/A | N/A | 0 | 0 | ||||||
| # HPV(−) cases | p16 ≥80 | p16 ≥50 | p16 ≥5 | ProExC >66 | ProExC >33 | # HPV(+) cases | p16 ≥80 | p16 ≥50 | p16 ≥5 | ProExC >66 | ProExC >33 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 0 | 0 | 0 | (+) | 12 | 0 | 0 | 0 | 0 | (+) |
| 5 | 0 | 0 | 0 | (+) | (+) | 1 | 0 | 0 | 0 | (+) | (+) |
| 1 | 0 | 0 | (+) | 0 | 0 | 1 | 0 | 0 | (+) | 0 | (+) |
| 1 | (+) | (+) | (+) | 0 | (+) | 1 | (+) | (+) | (+) | (+) | (+) |
| 2 | (+) | (+) | (+) | (+) | (+) | 1 | N/A | N/A | N/A | (+) | (+) |
(TMA tissue microarray; N/A not available)
On whole sections (WS), median p16 expression was 70% and average, 46.6%. 37/62 cases were positive for p16 in at least 5% of the tumor cells and 33/37 of these cases were positive for HPV DNA by PCR and/or ISH. 30/37 p16(+) cases exhibited ≥80% reactivity (Figure 1D); 4 cases, 50–79% (Figure 1E); and 3 cases, 5–49% (Figure 1F). Of the 4 cases with 50–79% reactivity, 3 (50%, 60%, and 70% p16) were positive for HPV DNA by ISH or PCR and 1 (70% p16) was negative by PCR and ISH. Focal high power fields with extensive (>80%) p16 reactivity could be found in each of the cases with 50–79% reactivity. Of the 3 cases with <50% staining, 2 (10% and 20% p16) were HPV DNA PCR positive and 1 (5% p16) was negative; 1 was HPV DNA ISH positive and 2 were negative. The p16 reactivity in these cases was patchy (Figure 1F); no high power field with extensive staining was seen. For ProExC, 41/62 cases showed staining at a 66% threshold (Figure 1A) and 34 of these cases were HPV DNA positive by PCR and/or ISH. At a 33% threshold (Figure 1B), 55/62 cases were positive for ProExC; 40/55 were HPV DNA positive.
On TMA cores, 29 cases showed ≥80% p16 staining; 6 cases, 50–79%; and 4 cases, 5–49%. 5/6 cases at the 50–79% threshold were positive for HPV DNA by PCR. 3/4 cases at a threshold of 5–49% were positive for HPV DNA by PCR and/or ISH. For ProExC, 25 cases were positive at a 66% threshold and 50 cases at a 33% threshold. 22/25 cases at the 66% threshold and 33/50 cases at the 33% threshold were positive for HPV DNA by PCR and/or ISH.
Four p16 negative cases (TMA and WS) were positive for HPV DNA by PCR. 2/4 were non-keratinizing SCC (neck lymph node, tonsil) and 2/4 were keratinizing SCC (base of tongue, tonsil).
Correlation with survival (Table 2)
TABLE 2.
Correlation of p16, ProExC, HPV DNA ISH and HPV DNA PCR with survival.
| OVERALL SURVIVAL | CANCER SPECIFIC SURVIVAL | TIME TO DISTANT METASTASIS | |||||
|---|---|---|---|---|---|---|---|
| HR | P-Value | HR | P-Value | HR | P-Value | ||
| WHOLE SECTIONS | p16≥5% | 0.17 (0.07–0.43) | <0.0001 | 0.24 (0.08–0.68) | 0.005 | 0.29 (0.05–1.61) | 0.14 |
| p16 ≥50% | 0.18 (0.07–0.48) | 0.0001 | 0.23 (0.07–0.72) | 0.006 | 0.14 (0.02–1.21) | 0.03 | |
| p16 ≥80% | 0.27 (0.12–0.59) | 0.005 | 0.34 (0.13–0.90) | 0.05 | 0.26 (0.05–1.28) | 0.14 | |
| ProExC >33% | 0.68 (0.23–2.00) | 0.5 | 0.38 (0.12–1.20) | 0.13 | 0.89 (0.08–9.64) | 0.92 | |
| ProExC>66 | 0.57 (0.25–1.27) | 0.17 | 0.33 (0.12–0.90) | 0.03 | 0.12 (0.01–1.03) | 0.03 | |
| HPV ISH | 0.37 (0.17–0.83) | 0.013 | 0.40 (0.15–1.04) | 0.06 | 0.26 (0.05–1.41) | 0.1 | |
| TMA | p16 ≥5% | 0.17 (0.07–0.41) | <0.0001 | 0.25 (0.09–0.73) | 0.004 | 0.49 (0.09–2.69) | 0.37 |
| p16≥50% | 0.21 (0.09–0.47) | 0.0002 | 0.28 (0.10–0.77) | 0.01 | 0.32 (0.06–1.65) | 0.16 | |
| p16 ≥80% | 0.31 (0.14–0.69) | 0.009 | 0.44 (0.16–1.16) | 0.11 | 0.47 (0.10–2.36) | 0.38 | |
| ProExC >33% | 0.88 (0.33–2.36) | 0.8 | 1.82 (0.45–7.29) | 0.38 | 3.20 (0.29–35.69) | 0.33 | |
| ProExC>66 | 0.32 (0.13–0.80) | 0.008 | 0.44 (0.16–1.26) | 0.11 | 0.77 (0.14–4.27) | 0.76 | |
| HPV ISH | 0.33 (0.13–0.84) | 0.01 | 0.45 (0.16–1.30) | 0.13 | 0.49 (0.09–2.70) | 0.4 | |
| PCR | HPV.PCR | 0.72 (0.38–1.38) | 0.32 | 0.42 (0.18–0.98) | 0.04 | 0.08 (0.01–0.65) | 0.003 |
| HPV PCR+ISH | 0.19 (0.08–0.48) | <0.0001 | 0.18 (0.06–0.60) | 0.0003 | 0.19 (0.03–1.14) | 0.03 | |
HR hazard ratio; ISH in situ hybridization; TMA tissue microarray
ProExC at a 66% threshold showed a statistically significant correlation with overall survival (OS) on TMA cores but not whole sections. The converse was true for cancer specific survival (CSS) and time to distant metastasis (TDM): ProExC 66% showed a positive correlation on whole sections but not TMA. ProExC at a 33% threshold (WS, TMA) did not correlate with OS, CSS or TDM.
On whole sections, p16 at a 5%, 50% and 80% threshold on whole sections showed a statistically significant correlation with OS and CSS but only p16 at a 50% threshold correlated with TDM. On TMA cores, p16 at the various thresholds correlated with OS but not TDM. Correlation with CSS was seen with p16 at a 5% or 50% threshold but not 80% threshold.
HPV DNA ISH (WS, TMA) showed a significant correlation with OS but not CSS or TDM. HPV DNA PCR showed the reverse: significant correlation with CSS and TDM but not OS. Combined HPV DNA ISH and PCR results (either ISH or PCR positive) showed a statistically significant correlation with OS, CSS and TDM.
Sensitivity and specificity (Table 3)
TABLE 3.
Sensitivity and specificity of p16 and ProExC for the detection of high-risk HPV (gold standard – positive for HPV DNA by PCR and/or ISH) at different thresholds on whole sections and TMA cores
| SENSITIVITY | SPECIFICITY | ||
|---|---|---|---|
| WHOLE SECTIONS | p16 ≥5% | 78.6 (33/42) (CI 62.8–89.2) | 80 (16/20) (CI 55.7–93.3) |
| p16 ≥50% | 73.8 (31/42) (CI 57.7–85.6) | 85 (17/20) (CI 61.1–96.0) | |
| p16 ≥80% | 65.1 (28/43) (CI 49.0–78.5) | 89.5 (17/19) (CI 65.4–98.1) | |
| ProExC >33% | 95.2 (40/42) (CI 82.6–99.2) | 25 (5/20) (CI 10.0–49.4) | |
| ProExC>66 | 78.6 (33/42) (CI 62.8–89.2) | 65 (13/20) (CI 40.9–83.7) | |
| TMA | p16 ≥5% | 87.8 (36/41) (CI 72.9–95.4) | 84.2 (16/19) (CI 59.5–95.8) |
| p16 ≥50% | 80.5 (33/41) (CI 64.6–90.6) | 89.5 (17/19) (CI 65.5–98.2) | |
| p16 ≥80% | 68.3 (28/42) (CI 51.2–81.4) | 94.7 (18/19) (CI 71.9–99.7) | |
| ProExC >33% | 80.5 (33/41) (CI 64.6–90.6) | 15 (3/20) (CI 4.0–38.9) | |
| ProExC>66 | 53.7 (22/41) (CI 37.6–69.0) | 85 (17/20) (CI 61.1–96.0) |
ProExC at a 33% threshold exhibited unacceptably low specificity (15–25%) on TMA and whole sections. At a 66% threshold, ProExC on whole sections showed improved but still low specificity (65%); on TMA, specificity increases to 85% but with a significant decrease in sensitivity (53.7%).
p16 showed higher sensitivity and specificity on TMA than on whole sections but the difference was not statistically significant. Sensitivity and specificity for the detection of HPV predictably varied with the threshold used to define positive staining – higher sensitivity with lower thresholds (78.6–87.8% at 5% threshold, 73.8–80.5% at 50%, 65.1–68.3% at 80%) and higher specificity with higher thresholds (80–84.2% at 5% threshold, 85–89.5% at 50%, 89.5–94.7% at 80%).
DISCUSSION
Our study found that ProExC does not perform well enough to be substituted for p16 as a prognostic marker in oropharyngeal SCC. ProExC at a 66% threshold correlates with OS on TMA cores but not whole sections. ProExC at a 33% threshold shows no correlation with survival. In contrast, p16 at each of the different thresholds (5%, 50%, 80%) on TMA and whole sections strongly correlates with OS, and p16 at a 50% threshold on whole sections also correlates with CSS and TDM.
ProExC is routinely positive in the basal and parabasal layers of benign squamous mucosa. In cervical biopsies, a positive stain is defined as nuclear staining of greater than half of the mucosal thickness. (3, 9, 33) However, with invasive squamous cell carcinomas, the typical organization of squamous mucosa is frequently lost. In HPV-negative tumors, we found staining of at least 33% of the tumor to be common which led to very low specificity unless a threshold of 66% staining was used. Jannapureddy et al reported similar problems with specificity in a study evaluating ProExC vs. p16 and HPV ISH in cell block material from fine needle aspiration biopsies of cervical lymph nodes involved by metastatic squamous cell carcinoma. (20) With positive ProExC defined as any nuclear staining, specificity was reported as 13.8%, similar to our finding of 15.0% for TMA cores at a threshold of 33%. Although specificity is markedly improved using a 66% threshold, specificity of 65% on whole sections is still significantly lower than specificity (85%) for p16 at a 50% threshold. On TMA cores, specificity increases to 85% but at the expense of sensitivity (53.7%).
p16 expression is well-established as an independent prognostic marker that is predictive of improved prognosis in head and neck squamous cell carcinomas. It is generally accepted that weak p16 reactivity or cytoplasmic only reactivity should be interpreted as negative. However, the definition of what constitutes a positive p16 stain varies across studies and ranges from weak reactivity (18) to strong reactivity in any (5), >25% (4, 22), >60% (32), or >80% of tumor cells.(30) We evaluated the impact of varying threshold – 5%, 50% and 80% - on correlation with survival, as well as sensitivity and specificity. p16 at each of the thresholds strongly correlates with OS (TMA and whole sections) but p16 at the 50% threshold on whole sections performed best in terms of correlating with other survival parameters such as CSS and TDM. On TMA cores, p16 at a 50% threshold also correlated with OS and CSS but not TDM. Cases with at least 50% p16 reactivity were notable for high power fields exhibiting extensive (>80%) p16 staining. Cases with less than 50% staining were infrequent – 3/37 cases in this study – and exhibited a patchy pattern of reactivity without any high power fields showing extensive p16 staining. Tumors with less than 50% p16 reactivity are best interpreted as equivocal. Further evaluation with HPV ISH or PCR may be helpful in determining if the tumor is HPV-related.
p16 at a 50% threshold showed good sensitivity (73.8–80.5%, WS-TMA) and specificity (85–89.5%, WS-TMA). Although the differences were not statistically significant, there was a predictable trend towards higher sensitivity at a 5% threshold (78.6–87.8%, WS-TMA) and higher specificity (89.5–94.7%, WS-TMA) at an 80% threshold, and a trend towards higher sensitivity and specificity for p16 on TMA cores vs. whole sections. Dahlstrand et al reported similar sensitivity (80% at 10% threshold) and specificity that ranged from 76% (threshold 10–50%) to 95% (threshold 50–90%) to 100% (threshold >90%), using HPV PCR (DNA) as a gold standard. (10) Sensitivity can be further affected by the fact that not all tumors with biologically active HPV and positive E6/E7 transcript expression are p16 positive. (33, 37) In our study, there were 4 cases with HPV DNA detected by PCR that were p16 negative. Additionally, specificity is impacted by the small subset of head and neck tumors that strongly express p16 in the absence of detectable HPV DNA. (27, 35)
A recent study by Lewis et al. found that p16(+) tumors correlate with an improved prognosis, regardless of HPV status as detected by HPV DNA ISH or PCR. (27) Although HPV DNA PCR is often regarded as a gold standard, there is wide variability in PCR assays – different primer sets, variable extraction and detection methods, disparate specimen types, etc. - that lead to different test characteristics. For an HPV PCR assay to be clinically valid as a prognostic marker in HNSCC, it must be shown to correlate with survival. The HPV DNA PCR assay used in our study correlates with CSS and TDM but not OS. In contrast, HPV DNA ISH correlates with OS and CSS (whole sections only) but not TDM. This indicates that neither the HPV DNA ISH nor the PCR assay used in this study is optimal for clinical use in determining prognosis. However, by combining HPV DNA PCR and ISH, the results do correlate with OS, CSS and TDM. While this supports the use of HPV DNA PCR plus ISH in lieu of p16 as a prognostic marker, in reality, immunohistochemical stains are more readily available than PCR and easier to perform and interpret than in situ hybridization. With ISH, the presence of HPV DNA is often manifested only as focal, faint, punctuate reactivity, requiring careful high-power examination and limiting the utility in small biopsies where sensitivity is lower.
In summary, these findings indicate that ProExC is not reliable as a prognostic marker for oropharyngeal SCC and should not be used in lieu of p16. For prognostic purposes, p16 is best defined as positive when at least half the tumor exhibits strong nuclear or nuclear and cytoplasmic staining. Patchy staining with less than 50% p16 reactivity should be interpreted as equivocal. These thresholds apply to both whole sections and TMA cores which were evaluated as a surrogate for small samples such as FNA cell block material.
Acknowledgments
This study was supported by the following grants from the National Institute of Health: R01 CA118582-01 (QTL, CSK), PO1- CA67166 (QTL), and HG000205 (NP) and by the Stanford Cancer Council Grant (CSK).
Footnotes
This work was presented in part at the USCAP 99th Annual Meeting in Washington DC, March 2010
References
- 1.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, Westra WH, Chung CH, Jordan RC, Lu C, Kim H, Axelrod R, Silverman CC, Redmond KP, Gillison ML. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aximu D, Azad A, Ni R, Colgan T, Nanji S. A pilot evaluation of a novel immunohistochemical assay for topoisomerase II-alpha and minichromosome maintenance protein 2 expression (ProEx C) in cervical adenocarcinoma in situ, adenocarcinoma, and benign glandular mimics. Int J Gynecol Pathol. 2009;28:114–119. doi: 10.1097/PGP.0b013e3181895573. [DOI] [PubMed] [Google Scholar]
- 3.Badr RE, Walts AE, Chung F, Bose S. BD ProEx C: a sensitive and specific marker of HPV-associated squamous lesions of the cervix. Am J Surg Pathol. 2008;32:899–906. doi: 10.1097/PAS.0b013e31815bbb69. [DOI] [PubMed] [Google Scholar]
- 4.Begum S, Cao D, Gillison M, Zahurak M, Westra WH. Tissue distribution of human papillomavirus 16 DNA integration in patients with tonsillar carcinoma. Clin Cancer Res. 2005;11:5694–5699. doi: 10.1158/1078-0432.CCR-05-0587. [DOI] [PubMed] [Google Scholar]
- 5.Begum S, Gillison ML, Nicol TL, Westra WH. Detection of human papillomavirus-16 in fine-needle aspirates to determine tumor origin in patients with metastatic squamous cell carcinoma of the head and neck. Clin Cancer Res. 2007;13:1186–1191. doi: 10.1158/1078-0432.CCR-06-1690. [DOI] [PubMed] [Google Scholar]
- 6.Boucher J, Anku-Bertholet C, Temmar R, Pelmus M. Evaluation of p16INK4a, minichromosome maintenance protein 2, DNA topoisomerase IIalpha, ProEx C, and p16INK4a/ProEx C in cervical squamous intraepithelial lesions. Hum Pathol. 2009;40:904–905. doi: 10.1016/j.humpath.2009.02.004. author reply 905–906. [DOI] [PubMed] [Google Scholar]
- 7.Chaturvedi AK, Engels EA, Anderson WF, Gillison ML. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J Clin Oncol. 2008;26:612–619. doi: 10.1200/JCO.2007.14.1713. [DOI] [PubMed] [Google Scholar]
- 8.Conesa-Zamora P, Domenech-Peris A, Orantes-Casado FJ, Ortiz-Reina S, Sahuquillo-Frias L, Acosta-Ortega J, Garcia-Solano J, Perez-Guillermo M. Effect of human papillomavirus on cell cycle-related proteins p16, Ki-67, Cyclin D1, p53, and ProEx C in precursor lesions of cervical carcinoma: a tissue microarray study. Am J Clin Pathol. 2009;132:378–390. doi: 10.1309/AJCPO0WY1VIFCYDC. [DOI] [PubMed] [Google Scholar]
- 9.Conesa-Zamora P, Domenech-Peris A, Ortiz-Reina S, Orantes-Casado FJ, Acosta-Ortega J, Garcia-Solano J, Perez-Guillermo M. Immunohistochemical evaluation of ProEx C in human papillomavirus-induced lesions of the cervix. J Clin Pathol. 2009;62:159–162. doi: 10.1136/jcp.2008.061408. [DOI] [PubMed] [Google Scholar]
- 10.Dahlstrand H, Dahlgren L, Lindquist D, Munck-Wikland E, Dalianis T. Presence of human papillomavirus in tonsillar cancer is a favourable prognostic factor for clinical outcome. Anticancer Res. 2004;24:1829–1835. [PubMed] [Google Scholar]
- 11.El-Mofty SK, Lu DW. Prevalence of human papillomavirus type 16 DNA in squamous cell carcinoma of the palatine tonsil, and not the oral cavity, in young patients: a distinct clinicopathologic and molecular disease entity. Am J Surg Pathol. 2003;27:1463–1470. doi: 10.1097/00000478-200311000-00010. [DOI] [PubMed] [Google Scholar]
- 12.Fakhry C, Gillison ML. Clinical implications of human papillomavirus in head and neck cancers. J Clin Oncol. 2006;24:2606–2611. doi: 10.1200/JCO.2006.06.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, Gillison ML. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 14.Freeman A, Morris LS, Mills AD, Stoeber K, Laskey RA, Williams GH, Coleman N. Minichromosome maintenance proteins as biological markers of dysplasia and malignancy. Clin Cancer Res. 1999;5:2121–2132. [PubMed] [Google Scholar]
- 15.Gharizadeh B, Oggionni M, Zheng B, Akom E, Pourmand N, Ahmadian A, Wallin KL, Nyren P. Type-specific multiple sequencing primers: a novel strategy for reliable and rapid genotyping of human papillomaviruses by pyrosequencing technology. J Mol Diagn. 2005;7:198–205. doi: 10.1016/S1525-1578(10)60546-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gharizadeh B, Zheng B, Akhras M, Ghaderi M, Jejelowo O, Strander B, Nyren P, Wallin KL, Pourmand N. Sentinel-base DNA genotyping using multiple sequencing primers for high-risk human papillomaviruses. Mol Cell Probes. 2006;20:230–238. doi: 10.1016/j.mcp.2006.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, Shah KV, Sidransky D. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 18.Hashibe M, Brennan P, Benhamou S, Castellsague X, Chen C, Curado MP, Dal Maso L, Daudt AW, Fabianova E, Fernandez L, Wunsch-Filho V, Franceschi S, Hayes RB, Herrero R, Koifman S, La Vecchia C, Lazarus P, Levi F, Mates D, Matos E, Menezes A, Muscat J, Eluf-Neto J, Olshan AF, Rudnai P, Schwartz SM, Smith E, Sturgis EM, Szeszenia-Dabrowska N, Talamini R, Wei Q, Winn DM, Zaridze D, Zatonski W, Zhang ZF, Berthiller J, Boffetta P. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J Natl Cancer Inst. 2007;99:777–789. doi: 10.1093/jnci/djk179. [DOI] [PubMed] [Google Scholar]
- 19.Ishimi Y, Okayasu I, Kato C, Kwon HJ, Kimura H, Yamada K, Song SY. Enhanced expression of Mcm proteins in cancer cells derived from uterine cervix. Eur J Biochem. 2003;270:1089–1101. doi: 10.1046/j.1432-1033.2003.03440.x. [DOI] [PubMed] [Google Scholar]
- 20.Jannapureddy S, Cohen C, Lau S, Beitler JJ, Siddiqui MT. Assessing for primary oropharyngeal or nasopharyngeal squamous cell carcinoma from fine needle aspiration of cervical lymph node metastases. Diagn Cytopathol. 2010;38:795–800. doi: 10.1002/dc.21293. [DOI] [PubMed] [Google Scholar]
- 21.Kelly D, Kincaid E, Fansler Z, Rosenthal DL, Clark DP. Detection of cervical high-grade squamous intraepithelial lesions from cytologic samples using a novel immunocytochemical assay (ProEx C) Cancer. 2006;108:494–500. doi: 10.1002/cncr.22288. [DOI] [PubMed] [Google Scholar]
- 22.Klussmann JP, Gultekin E, Weissenborn SJ, Wieland U, Dries V, Dienes HP, Eckel HE, Pfister HJ, Fuchs PG. Expression of p16 protein identifies a distinct entity of tonsillar carcinomas associated with human papillomavirus. Am J Pathol. 2003;162:747–753. doi: 10.1016/S0002-9440(10)63871-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klussmann JP, Weissenborn SJ, Wieland U, Dries V, Kolligs J, Jungehuelsing M, Eckel HE, Dienes HP, Pfister HJ, Fuchs PG. Prevalence, distribution, and viral load of human papillomavirus 16 DNA in tonsillar carcinomas. Cancer. 2001;92:2875–2884. doi: 10.1002/1097-0142(20011201)92:11<2875::aid-cncr10130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 24.Kong CS, Narasimhan B, Cao H, Kwok S, Erickson JP, Koong A, Pourmand N, Le QT. The relationship between human papillomavirus status and other molecular prognostic markers in head and neck squamous cell carcinomas. Int J Radiat Oncol Biol Phys. 2009;74:553–561. doi: 10.1016/j.ijrobp.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14:467–475. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- 26.Lassen P, Eriksen JG, Hamilton-Dutoit S, Tramm T, Alsner J, Overgaard J. Effect of HPV-associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. J Clin Oncol. 2009;27:1992–1998. doi: 10.1200/JCO.2008.20.2853. [DOI] [PubMed] [Google Scholar]
- 27.Lewis JS, Jr, Thorstad WL, Chernock RD, Haughey BH, Yip JH, Zhang Q, El-Mofty SK. p16 positive oropharyngeal squamous cell carcinoma:an entity with a favorable prognosis regardless of tumor HPV status. Am J Surg Pathol. 2010;34:1088–1096. doi: 10.1097/PAS.0b013e3181e84652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu CL, Prapong W, Natkunam Y, Alizadeh A, Montgomery K, Gilks CB, van de Rijn M. Software tools for high-throughput analysis and archiving of immunohistochemistry staining data obtained with tissue microarrays. Am J Pathol. 2002;161:1557–1565. doi: 10.1016/S0002-9440(10)64434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nieh S, Chen SF, Chu TY, Lai HC, Fu E. Expression of p16 INK4A in Papanicolaou smears containing atypical squamous cells of undetermined significance from the uterine cervix. Gynecol Oncol. 2003;91:201–208. doi: 10.1016/s0090-8258(03)00479-7. [DOI] [PubMed] [Google Scholar]
- 30.O’Regan EM, Toner ME, Finn SP, Fan CY, Ring M, Hagmar B, Timon C, Smyth P, Cahill S, Flavin R, Sheils OM, O’Leary JJ. p16(INK4A) genetic and epigenetic profiles differ in relation to age and site in head and neck squamous cell carcinomas. Hum Pathol. 2008;39:452–458. doi: 10.1016/j.humpath.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Pinto AP, Schlecht NF, Woo TY, Crum CP, Cibas ES. Biomarker (ProEx C, p16(INK4A), and MiB-1) distinction of high-grade squamous intraepithelial lesion from its mimics. Mod Pathol. 2008;21:1067–1074. doi: 10.1038/modpathol.2008.101. [DOI] [PubMed] [Google Scholar]
- 32.Reimers N, Kasper HU, Weissenborn SJ, Stutzer H, Preuss SF, Hoffmann TK, Speel EJ, Dienes HP, Pfister HJ, Guntinas-Lichius O, Klussmann JP. Combined analysis of HPV-DNA, p16 and EGFR expression to predict prognosis in oropharyngeal cancer. Int J Cancer. 2007;120:1731–1738. doi: 10.1002/ijc.22355. [DOI] [PubMed] [Google Scholar]
- 33.Shi J, Liu H, Wilkerson M, Huang Y, Meschter S, Dupree W, Schuerch C, Lin F. Evaluation of p16INK4a, minichromosome maintenance protein 2, DNA topoisomerase IIalpha, ProEX C, and p16INK4a/ProEX C in cervical squamous intraepithelial lesions. Hum Pathol. 2007;38:1335–1344. doi: 10.1016/j.humpath.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 34.Shiboski CH, Schmidt BL, Jordan RC. Tongue and tonsil carcinoma: increasing trends in the U.S. population ages 20–44 years. Cancer. 2005;103:1843–1849. doi: 10.1002/cncr.20998. [DOI] [PubMed] [Google Scholar]
- 35.Smeets SJ, Hesselink AT, Speel EJ, Haesevoets A, Snijders PJ, Pawlita M, Meijer CJ, Braakhuis BJ, Leemans CR, Brakenhoff RH. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer. 2007;121:2465–2472. doi: 10.1002/ijc.22980. [DOI] [PubMed] [Google Scholar]
- 36.Walts AE, Bose S. p16, Ki-67, and BD ProExC immunostaining: a practical approach for diagnosis of cervical intraepithelial neoplasia. Hum Pathol. 2009;40:957–964. doi: 10.1016/j.humpath.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 37.Wiest T, Schwarz E, Enders C, Flechtenmacher C, Bosch FX. Involvement of intact HPV16 E6/E7 gene expression in head and neck cancers with unaltered p53 status and perturbed pRb cell cycle control. Oncogene. 2002;21:1510–1517. doi: 10.1038/sj.onc.1205214. [DOI] [PubMed] [Google Scholar]