Abstract
Bacterial gliding motility is the smooth movement of cells on solid surfaces unaided by flagella or pili. Many diverse groups of bacteria exhibit gliding, but the mechanism of gliding motility has remained a mystery since it was first observed more than a century ago. Recent studies on the motility of Myxococcus xanthus, a soil myxobacterium, suggest a likely mechanism for gliding in this organism. About forty M. xanthus genes were shown to be involved in gliding motility, and some of their protein products were labeled and localized within cells. These studies suggest that gliding motility in M. xanthus involves large multiprotein structural complexes, regulatory proteins, and cytoskeletal filaments. In this review, we summarize recent experiments that provide the basis for this emerging view of M. xanthus motility. We also discuss alternative models for gliding.
Keywords: Myxococcus xanthus, proton motive force, cytoskeleton, protein localization, model
INTRODUCTION
Bacterial motility facilitates colony growth and expansion, as well as a wide range of behaviors such as chemotaxis, virulence, predation, biofilm formation, and development (3, 39, 58, 83). Bacteria have evolved different types of motility patterns, including swimming, swarming, twitching, gliding, and sliding (39). These divergent systems provide specific benefits that help bacteria adapt to their natural habitats.
One of the least understood bacterial motility systems is gliding motility. Gliding motility has been defined as active surface translocation along the long cell axis without the aid of flagella or pili (7, 39). This definition tells us what does not power gliding motility, but fails to tell us what does. Indeed, gliding motility is common among highly divergent bacterial groups, including the myxobacteria, the cyanobacteria, the cytophaga-flavobacteria, and the mycoplasmas. However, in these bacterial groups, more than one motility mechanism may be involved (50). Gliding mechanisms have remained elusive because gliding motility does not cause changes in cell morphology, and the organelles or motility apparatuses associated with gliding motility are not easily observable, except for the legs and the terminal organelles in mycoplasmas (58). In this review, we focus on gliding motility in Myxococcus xanthus, a fruiting myxobacterium. A series of recent experiments suggest how this bacterium might move in the absence of flagella or pili.
Myxococcus xanthus Contains Two Motility Systems
Myxobacteria are Gram-negative δ-proteobacteria. Most species in this family are soil inhabitants that display complex life cycles, including swarming, fruiting body formation, and sporulation (67). M. xanthus, the best studied of the myxobacteria, lacks flagella and is unable to swim in liquid culture. However, cells exhibit gliding motility on solid surfaces usually in coordinated groups but also as isolated adventurous individuals. Motility allows this bacterium to feed on soil detritus and prey on other microorganisms (46, 83). When the availability of nutrients or prey decreases in the environment, most cells exhibit behaviors that include aggregation into fruiting bodies and conversion of individual cells into spores (83). In M. xanthus, motility is essential for swarming, predation, fruiting body formation, and sporulation.
Motility mutants in M. xanthus were first studied by Burchard in 1970 after he observed spontaneous semimotile mutants in his cultures (6). These mutants were nonmotile as individual cells on a surface but became transiently motile when in apposition to other cells. From this strain, he isolated a completely nonmotile mutant that “demonstrated no gliding movement as single cells nor [sic] as swarms” (6). The perplexing phenotypes of these mutants were clarified several years later in an extensive mutagenesis study conducted by Hodgkin & Kaiser (24). In this study, a large number of motility mutants were isolated, and genetic experiments revealed that motility in M. xanthus is controlled by two sets of genes. One set of genes controlled adventurous (A-) motility, the movement of individual cells, whereas the other set of genes was required for social (S-) motility, the movement of cells in groups or rafts (24). This work identified only one locus common to both motility systems, mglA (24). Kaiser and coworkers later showed that cells exhibiting Smotility had type IV pili and that most of the S-motility genes were similar to the twitching motility genes of Pseudomonas and Neisseria (36, 45). S-motility, like twitching motility, is powered by the extension and retraction of type IV pili (36, 78). A-motility does not require surface pili or other external structures for locomotion and is therefore the motility system that best fits the definition of gliding motility in M. xanthus (39, 46).
Both gliding and twitching motilities feature periodic cellular reversals (66). These reversals are coordinated and presumably allow cells to redirect their movements to respond to attractants and repellents (70). Wild-type cells reverse their direction of movements approximately every 7–8 minutes, thus net movement is achieved by biasing the intervals between reversals (83). The reversal frequency is regulated by the frizzy (Frz) chemotaxis system, which features a cytoplasmic methyl-accepting chemotaxis protein (MCP), FrzCD.
It is curious that M. xanthus utilizes two different motility systems, neither of which moves cells much faster than approximately 2–4 μm min−1, a velocity that is approximately one thousand-fold slower than that of most flagellated bacteria (35). We speculate that M. xanthus purposefully utilizes slow-moving motility systems so that it does not outrun its endogenously secreted antibiotics and enzymes. Alternatively, the bacteria may disperse efficiently as fruiting body spores utilizing insect, bird, or bat vectors for rapid transport and thus does not require rapid motility. Each of the two motility systems exhibits selective advantages on different surfaces: In the laboratory, twitching motility works best on moist, soft surfaces (e.g., 0.3–0.5% agar), whereas gliding motility requires a relatively dry, hard surface (e.g., 1.0–2.0% agar) (71). External flagella, which require a moist medium, might not be of much benefit to these bacteria in their relatively dry soil environment.
SEARCHING FOR THE GLIDING MOTILITY ENGINE
Identification of Gliding Motility Genes
More than thirty years ago, Hodgkin & Kaiser identified a large collection of M. xanthus mutants that were nonmotile as isolated cells but still able to move in groups (twitching) (24, 25). From these studies and additional screens, especially from the Hartzell laboratory (for review, see 21), approximately 40 genes were identified as being required for gliding motility, although the functions of these genes were largely unknown. Based on homology predictions, these genes could be classified into several categories:
Genes that encode components of Tol/Ton complexes, usually involved in macromolecule transport and the maintenance of membrane integrity (80). In Escherichia coli, TolQ/TolR proteins share homology with the flagella motor (stator) proteins MotA/MotB (10). Interestingly, AglR/AglS and AglX/AglV, two pairs of M. xanthus genes that encode TolQ/TolR homologs were identified as A-motility genes, suggesting that M. xanthus may assemble these Tol complexes into gliding engines (59, 80).
Genes that encode enzymes for the biosynthesis of polysaccharides (80). These genes have long been speculated to contribute to slime secretion, which potentially could power gliding motility (77, 81). However, none of the mutations in these genes resulted in a slime-free phenotype. If slime comprises polysaccharides, it may consist of more than one kind.
Other genes. This category contains genes that encode proteases, metabolism-related enzymes, and proteins of unknown function (80). The identification of gliding genes with extremely diverse putative functions suggests that gliding motility is a complex process requiring interacting proteins of different function and from different cellular compartments.
Additionally, Hodgkin & Kaiser identified a group of cgl (contact-transient gliding) genes. cgl mutations at five loci restored gliding motility when mixed with wild-type cells or cells with mutations from a different cgl complementation group. The extracellular rescue of motility by members of different complementation groups suggests that surface proteins important for gliding may be freely exchanged between cells (25). For example, the cglB locus encodes a lipoprotein with a high cysteine content that can be exchanged between cells. This protein is required for A-motility, but its specific function is unknown (68). Extracellular complementation of motility proteins is also observed in twitching motility, in which Tgl, an outer membrane lipoprotein, is transferred between cells (62).
Multiprotein Complexes Required for Gliding
Analysis of the large group of A-motility genes and their predicted products suggests that the gliding engines of M. xanthus may be composed of multiprotein complexes. For example, many of the gliding proteins contain TPR (tetratricopeptide repeat) domains that often mediate protein-protein interactions, whereas others, such as the Tol or Ton proteins, are typically found in multiprotein complexes (14).
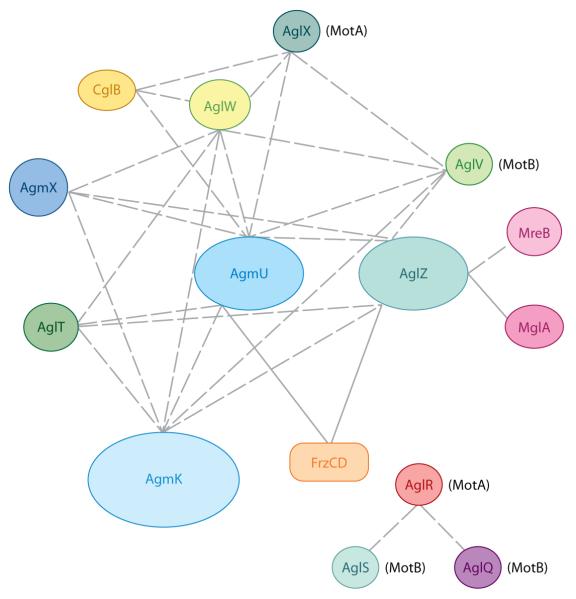
The search for protein-protein interactions involved in M. xanthus gliding motility began with MglA, the only protein known to be essential for both gliding and twitching motilities (24). Yeast two-hybrid screens by the Hartzell laboratory showed that MglA directly interacts with AglZ, a previously unidentified A-motility protein (79). This interaction was confirmed with genetic assays (48). To identify additional interactions between A-motility proteins, Nan et al. used affinity pull-down experiments. For example, GST (glutathione-S-transferase)-tagged proteins or protein fragments were used as baits on affinity columns, and proteins interacting with those baits were eluted and identified by MS/MS mass spectrometry. With the high sensitivity of mass spectrometry, direct and indirect interactions among a wide range of gliding motility proteins were revealed (60). Figure 1 shows that AglZ, AgmU, AglT, AglW, AgmK, AgmX, and CglB, as well as several additional proteins, constitute a large multiprotein complex required for A-motility. Among those proteins:
AgmU, AglT, AgmK and AgmX are encoded by genes in the same operon, suggesting that they share common functions. In-frame deletions in the other genes of this operon, (i.e., cglF, cglE, pglI, agmV, and mxan_4864) also cause gliding defects and/or altered localization of AglZ, indicating that they are also part of the gliding machinery (38, 60, 80, 81).
aglX and aglV, which encode a TolQ/TolR pair, are immediately upstream of aglW in the M. xanthus genome. (80). AglW shows high similarity with the E. coli lipoprotein TolB, known to anchor the membrane embedded TolQ/TolR complex to peptidoglycan (5). AglW contains Trp-Asp (WD) repeats that usually form β-propeller structures that bind TPR domains; this suggests that it may interact with other proteins in the gliding machinery (61). In fact, AglW was found to interact with the gliding proteins, AglX, AglV, AgmU, AgmK, AgmX, and CglB (Figure 1) (59).
TolQ/TolR proteins have been proposed to be homologs of MotA/MotB, the bacterial flagella stator proteins that harness proton motive force (PMF) to power flagella rotation (10, 57). In M. xanthus, two TolQ/TolR systems have been identified related to gliding motility, AglX/AglV and AglR/AglS/AglQ (59, 75, 80). The residues important for proton transport and force generation of MotA/MotB are well conserved in both of the two TolQ/TolR systems (59, 75). In M. xanthus, channels are formed between AglX and AglV, and among AglR, AglS, and AglQ (59, 75). GST-tagged soluble fragments of AglX and AglV were also used in interaction screens. In this case, besides AglW, gliding motility proteins AglZ, AgmU, and AgmK were shown to interact with both baits (Figure 1) (59). This result suggests AglX/AglV and AglR/AglS/AglQ as possible candidates for the proteins that energize the gliding motors.
GST-tagged screening and in vitro cross-linking results showed that the gliding motility proteins AgmU and AglZ directly interact with FrzCD, the MCP of Frz pathway, serving as a possible signal input in the regulation of reversal frequency (49, 60). AglZ was also reported to interact directly with MreB, the actin-like cytoskeleton of M. xanthus (Figure 1) (48).
Figure 1.
Interactions observed for proteins involved in gliding motility. Interactions identified from affinity pull-down and cross-linking experiments are shown with dashed lines, and interactions demonstrated by both biochemistry and in vivo genetic studies are shown with solid lines.
The gliding motility complex identified above may only represent a small fraction of the entire gliding engine or only one type of multiple motility engines. More interactions remain to be explored. Taken together, the protein-protein interaction studies suggest that gliding motility involves large multiprotein complexes that span the cytoplasm (e.g., AglZ), inner membrane (e.g., AglX/AglV), and periplasm (e.g., AglW, CglB), and these complexes interact with the cytoskeleton (MreB) and regulatory proteins (e.g., FrzCD and MglA). For this reason, the activities of proteins in all the different cell compartments must be considered to understand the mechanism of gliding.
The Localization and Dynamics of Gliding Motility Proteins
The dynamic localization of several gliding motility proteins labeled with fluorescence tags were investigated. AglZ and AgmU, arguably the best studied, are discussed below. Tracking the movements of these gliding motility proteins in vivo provided important clues to deciphering the mechanism of gliding motility and the organization of the gliding engines.
AglZ
AglZ is a gliding motility protein that interacts with both MglA (79) and FrzCD (49). AglZ is a large protein (1,395 amino acids) with an N-terminal pseudoreceiver domain and a C-terminal coiled-coil domain similar to myosin class II heavy chain (79). AglZ is probably a regulator of A-motility given that the gliding defect of aglZ deletion mutant can be abrogated by a second mutation in the N-terminal domain of frzCD (49). This suggests that AglZ negatively regulates gliding motility through the Frz path-way (49). Based upon their physical and regulatory interaction, it was predicted that AglZ and FrzCD would colocalize in cells. However, AglZ-mCherry and FrzCD-green fluorescent protein (GFP) were found localized in clusters that occupied different positions in cells, even though these proteins interact directly in vitro through their N-terminal domains. Thus, the interaction between AglZ and FrzCD may be limited to cluster interfaces (49).
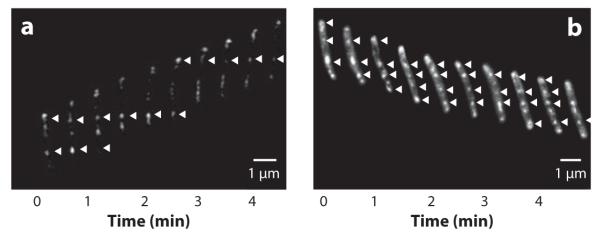
An important breakthrough in our understanding of the mechanism of gliding motility in M. xanthus came from studies of the localization and dynamics of AglZ in moving cells. Mignot et al. (56) showed that AglZ-yellow fluorescent protein (YFP) was organized as an array of clusters that spanned the length of cells. Surprisingly, when cells moved forward, the AglZ-YFP clusters, with the exception of those at the front of cells, did not move along with the cells but remained at fixed positions with respect to the agar surface (Figure 2a) (56). Thus, the cells appeared to move through the clusters. It was hypothesized that new clusters of AglZ were assembled at the leading cell poles and were transported at the same velocity as the cells themselves toward the lagging cell poles, where they were disassembled. The behavior of the AglZ clusters suggested that gliding motility in M. xanthus may have similarities to the focal adhesion complexes seen in some eukaryotic cells (Figure 2a) (56).
Figure 2.
AglZ and AgmU clusters (indicated by white arrows) appear stationary as cells move forward. Gliding cells expressing AglZ-YFP or AgmU-mCherry were imaged by fluorescence microscopy at frequent intervals. (a) AglZ-YFP clusters appear to remain stationary with respect to the substratum as cells glide forward; this behavior is reminiscent of focal adhesion proteins in eukaryotic cells (adapted from Reference 56). (b) Cytoplasmic clusters of AgmU-mCherry appear similar to AglZ clusters (adapted from Reference 60).
AglZ behaves as though it were a protein cargo that is carried by gliding engines toward the lagging cell poles. For example, when cells are not moving or when the gliding engines are disrupted by deletion of other A-motility proteins, AglZ-YFP appears diffuse or forms a single cluster at one cell pole instead of localizing into distributed clusters along the cell body (60). In contrast, when the regulatory N-terminal domain of FrzCD (amino acids 6–130) is deleted, cells retain their ability to glide and AglZ still localizes as distributed clusters, but these clusters now frequently colocalize with FrzCD (49). These observations suggest that instead of being the motor, AglZ is in fact a regulatory protein carried by motor proteins.
AgmU
AgmU is a central component of the A-motility gliding complex, as it interacts with many A-motility proteins (see Figure 1) as well as the N-terminal domain of FrzCD (60). It is also a very large protein (1,218 amino acids) with multiple TPR motifs, consistent with its ability to interact with many proteins (60, 80). An AgmU-mCherry fusion that is fully functional localizes in two distinct patterns: distributed cytoplasmic clusters that colocalize with AglZ (and show the same pattern as AglZ clusters when cells are moving) and membrane/periplasm-associated patterns (Figure 2b) (59, 60). Further investigation revealed that these two localization patterns correspond to two subpopulations of AgmU (60). The switch between these two localization patterns is sensitive to the hardness or the viscosity of the gliding surface (60).
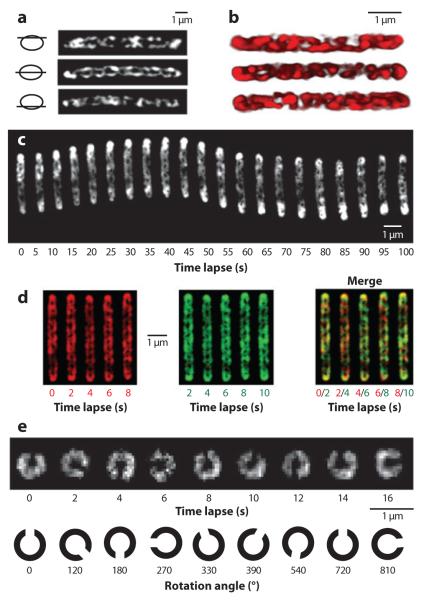
Using high-resolution deconvolution microscopy, the membrane/periplasm-associated population of AgmU-mCherry was found to decorate a closed helical loop with a periodicity of approximately 1 μm (Figure 3a,b) (59). Time-lapse movies taken at 2-s intervals showed that these helical structures rotate as cells move forward on an agar surface (Figure 3c). When cells were suspended in methylcellulose or liquid culture they cannot glide but helix rotation continued (Figure 3d,e) (59). Gliding motility and helix rotation appear to be correlated: (a) Rotation speed, ~7 rpm, correlates with the maximum gliding speed of M. xanthus (~4 μm min−1), considering the helical periodicity (~1 μm) and maximum linear motion that can result from this rotation (~7 μm min−1); (b) rotation direction reverses when cells reverse, as the helices only rotate clockwise if viewed from the lagging cell poles; (c) inhibitors or antibiotics that stop helix rotation [such as cyanide-m-chlorophenylhydrazone (CCCP) or A22] abolish gliding motility (see below) (59).
Figure 3.
Membrane/periplasm-associated AgmU decorates a closed helical loop that rotates as cells glide forward. (a) Deconvolved images of one fixed cell expressing AgmU::mCherry show that AgmU localizes along a looped helical track. (b) 3D reconstructions of the AgmU helix from three individual cells. (c) Time-lapse images of the rotating AgmU-mCherry helix in a cell moving on agar surface. (d ) agmU-mCherry pilA cells were suspended in 1% methylcellulose and the rotation was visualized by merging one frame (red; left panel ) with a frame recorded 2 s later ( green; middle panel ). In the merged images, neither the conformation nor the helical pitch of the AgmU helix changed, but a clear color shift is evident, indicating a rotational movement. (e) Polar view of the rotation of AgmU helix in 1% methylcellulose. All panels are adapted from Reference 59.
The Energy Source for Gliding Motility
Motility in bacteria is usually powered by either PMF or ATP (33). To identify the energy source for gliding motility and AgmU helix rotation, M. xanthus cells were treated with inhibitors that interfere with these processes. For example, when sodium azide was added to cells, both gliding motility and the rotation of the AgmU helices continued. Given that sodium azide disrupts ATP synthesis, these processes are unlikely to be powered by ATP hydrolysis (40). In contrast, when PMF was disrupted by CCCP, both gliding motility and AgmU helix rotation stopped immediately, indicating that these processes are powered by PMF. This inhibition was also observed when AgmU-mCherry-labeled cells failed to show fluorescence recovery after photobleaching (FRAP) after being illuminated in the presence of CCCP. (59). These results indicate that PMF is the major energy source for gliding in M. xanthus. Further investigations identified the pH gradient, rather than membrane potential, as the major component of PMF that powers gliding (59, 75).
In a parallel study, Sun et al. (75) tracked the movement of polystyrene beads that were placed, using laser tweezers, on the outer surface of immobilized cells. In their observations, the beads were transported along the length of the cells with a velocity that was consistent with the gliding speed of the cells. This transport was abolished by CCCP and recovered when CCCP was washed out (75).
The identification of PMF, the energy source for powering flagellar motors in E. coli, as the energy source for M. xanthus gliding suggested that proteins similar to the E. coli flagella motor (stator), MotAB, may be involved in gliding motility. Indeed, Youdarian et al. identified AglX/AglV and AglR/AglS as M. xanthus MotAB homologs and proposed that they could be gliding motor proteins (59, 80). To test this hypothesis, Nan et al. demonstrated that the MotAB homologs AglX and AglV interact with AgmU and other proteins in the A-motility complex (59, 80). Furthermore, AglX-mCherry and AglR-mCherry showed helical localization and rotation similar to that of AgmU-mCherry; rotation of these proteins was strictly dependent on PMF (59). AglQ, another MotB homolog encoded by an open reading frame between aglR and aglS, colocalizes with AglZ clusters. A mutant that expresses a D-to-N mutation that is predicted to disrupt the proton binding channel of AglQ showed no gliding motility, suggesting the direct involvement of AglQ as part of the gliding motor (75). At this time, we do not know how these various proteins are organized in the cell and how they might generate force. We hypothesize that AgmU, AglX/AglV, and AglR/AglS interact in the periplasm and decorate the same helical structure. This structure may also contain AglQ given that it forms periodic clusters similar to those of AgmU and AglZ (75). According to this model, the MotAB homologs AglX/AglV and AglR/AglS/AglQ, instead of being anchored around flagellar basal bodies (57), move along a cytoskeletal filament that spans the whole cell body and twists into a helical structure (see Helical Rotor Model, below).
Cytoskeleton Filaments Play a Role in Gliding Motility
In the not so distant past, bacteria were generally considered to lack significant cytoskeletal structures. However, it is now known that MreB, an actin-like structural homolog, polymerizes into helical filaments that provide the major scaffold for many cellular processes in bacteria (16, 34, 76). The MreB filaments participate in a wide range of functions, such as maintaining cell shape and polarity, guiding the assembly of new peptidoglycan, participating in chromosome segregation, and regulating twitching motility (13, 15-17, 34). That MreB may also be involved in gliding motility was suggested by the helical organization of the A-motility proteins AgmU, AglX, and AglR, and the remarkable similarities observed in the pitch and periodicities of the respective helices (48). Significantly, the cytoplasmic clusters formed by AglZ and AgmU matched the helical pitch of MreB (56, 60).
Evidence for the involvement of MreB in gliding motility came from studies using the MreB-specific perturbing compound A22 (30), which interferes with the dynamic polymerization of MreB and causes M. xanthus cells to slowly round up and lyse (48): (a) A22 blocks gliding motility within several minutes and disrupts the localization of AglZ in vivo (48); (b) MreB and the C-terminal coiled-coil do-main of AglZ were shown to interact in vitro by cross-linking (48); (c) addition of A22 stops AgmU-mCherry helix rotation in wild-type cells but not in a mutant strain that expresses an A22-insensitive MreB mutant (48, 59); (d) AgmU-mCherry helix shows movements from a shielded region to the irradiated regions in FRAP experiments; this movement is blocked by A22 (59). These experiments, in total, indicate a direct connection between an intact MreB cytoskeleton, gliding motility, and the rotation of the helices formed by AgmU, AglX, and AglR.
Interestingly, treatment with A22 does not immediately disperse the established AgmU-mCherry helices as it blocks helix rotation and gliding motility (59). A possible explanation for this observation might be that the interaction between MreB filaments and the gliding engines is transient. In this case, MreB is required for protein localization and rotation, but not for the maintenance of the AgmU-containing helices. Similar observations were reported for RodZ in Caulobacter crescentus. In that case, the localization of RodZ is dependent on MreB, but A22 treatment does not disrupt the ring structure of RodZ immediately (1).
THE REGULATION OF GLIDING MOTILITY
The Frz Pathway
M. xanthus cells are very flexible and frequently bend and change direction in response to the unevenness of the substrate or interactions with other cells. Because these rod-shaped cells move by gliding in the direction of their long axis, periodic cell reversals are required for cells to redirect their movements. During reversals, the polarity of the cells is inverted so that old lagging cell poles become new leading cell poles. Cellular reversals are complex, requiring many changes in protein localization: For example, polar localized proteins, like type IV pili, must be relocalized to the leading cell poles. Directed movements are required for vegetative swarming, predation, and fruiting body formation.
The Frz chemosensory system controls reversals for both pilus-mediated twitching motility and gliding motility (4). Mutants defective in the Frz pathway almost never reverse and are therefore defective in swarming and developmental aggregation. In contrast, some frz mutants hyper-reverse, giving rise to non-spreading colonies, as individual cells reverse before they achieve net translocation (4). The Frz system is composed of bacterial chemotaxis homologs: FrzCD, a cytoplasmic MCP; FrzA and FrzB, CheW-like coupling proteins; FrzE, a CheA-CheY hybrid histidine kinase-response regulator protein; FrzZ, a dual-response regulator protein; FrzF, a methyltransferase; and FrzG, a methylesterase (52). According to the current view, chemotaxis signals or repellents are sensed by FrzCD, which activates FrzE, a histidine protein kinase. To trigger cell reversals, FrzE autophosphorylates its kinase domain and transfers the phosphoryl group to the CheY domains of FrzZ (9, 28, 29). Reversible methylation of the FrzCD chemoreceptor allows the bacteria to adapt to changes in environmental stimuli. Site-specific methylation appears to have differential effects on the overall activity of FrzCD (2, 51, 53, 54, 69).
FrzCD is an unusual bacterial chemoreceptor in that it lacks the periplasmic and transmembrane domains that are usually involved in ligand recognition in other chemoreceptors. However, it contains an N-terminal domain that interacts with the gliding proteins AglZ and AgmU (as discussed above) (49, 60). Those interactions put the Frz pathway into a regulatory feedback loop in which the output protein, FrzZ-P, regulates gliding motility according to the current gliding status sensed by FrzCD (29, 49). The localization pattern of FrzCD is consequently very different from that of the enteric MCPs, which typically cluster in patches or arrays near cell poles (19, 23). In contrast, FrzCD is localized in helically arranged clusters along the length of the cell body. The distribution of these clusters is highly dynamic, continuously changing in number and location (47). Surprisingly, when M. xanthus cells make side-by-side contact, the FrzCD clusters in adjacent cells transiently align; this alignment is usually followed by the reversal of one of the contacting cells. These alignments depend on a functional Frz pathway (47). These interesting observations suggest that FrzCD senses cell-cell contacts or an opposing surface. Indeed, the gliding machineries may serve as contact sensors, suggested by the following: (a) Multiple components of the gliding engines are periplasmic or membrane-embedded proteins and could therefore be expected to be sensitive to pressure from cell contacts (59, 60); (b) the gliding proteins AglZ and AgmU interact with FrzCD directly as indicated by affinity pulldown experiments and in vitro cross-linking (49, 60); (c) the localizations of AglZ and AgmU are sensitive to surface conditions and vary according to whether cells are utilizing twitching or gliding motility; for example, the protein clusters are less defined under conditions that favor twitching motility (49, 60).
FrzCD, when viewed by deconvolution microscopy, appears as a continuous helical structure, with a pitch similar to that of MreB and the gliding motility proteins AgmU, AglX, and AglR (47, 59). However, no evidence has been found that FrzCD associates with MreB or the other gliding motility proteins in vivo. To the contrary, the clusters of FrzCD appear to localize at different sites from these proteins, suggesting that another cytoskeletal filament may be involved in the organization of FrzCD or, alternatively, that another totally different mechanism may be involved (49, 60).
MglA
MglA is a regulatory protein that is required for both gliding and twitching motilities (20, 22, 24). The sequence of MglA shares similarity with Ras family GTPases that are involved in a wide range of functions, including signal transduction and cell migration in eukaryotic cells (12, 22). In M. xanthus, MglA binds and hydrolyzes GTP, and its GTPase activity is activated by MglB, its cognate guanine nucleotide hydrolysis activating protein (GAP) (43, 48, 64, 82). The ΔmglAB mutant was initially thought to be completely nonmotile, but more careful analyses showed that it hyper-reverses at such high frequency that displacements are very small and cause almost no net translocation (73).
Interestingly, although MglA and MglB interact as part of a regulatory cycle, they are localized at different sites: MglA is primarily localized at the leading cell pole, whereas MglB is localized at the lagging cell pole. During cell reversals, the polar clusters of MglA and MglB oscillate inversely and synchronously, defining the new leading and lagging poles (43, 82). Cross-correlation analysis confirmed that the oscillations of MglA are coupled with cell reversals (43, 82). MglA mutants that mimic the stable GTP-bound state either localize to both cell poles (82) or lose their ability to reside at the leading pole (43); in the absence of MglB, MglA localizes to both cell poles (43, 82). Taken together, the data suggest that the function of MglB is to inhibit reversals by activating the GTPase activity of MglA and preventing MglA-GTP from localizing at the lagging cell poles. This hypothesis is supported by in vitro data that purified MglA does not hydrolyze GTP efficiently and that MglB specifically recognizes and binds to the GTP-bound conformation of MglA (43, 82).
Genetic evidence suggests that MglB works downstream of FrzZ, the output module of the Frz pathway, and upstream of MglA (43, 82). MglB switches to the new lagging pole less frequently in the frz null mutants and more frequently in the hyper-reversing frz mutant (82). It will be important to investigate the connection between MglB and the Frz pathway, especially with FrzZ, the dual response regulator. On the other hand, signaling from the Frz pathway may be branched: In the absence of MglB, the reversal frequency of cells is still partially controlled by the Frz pathway, suggesting the existence of additional interactions between MglA and the Frz pathway (82).
It is still unknown how the GTP cycle of MglA triggers the change in localization of so many motility proteins. It has been established that MglA directly interacts with AglZ and FrzS, components of the gliding and twitching machineries, respectively (48, 79). In the absence of MglA, AglZ appears diffuse or localizes into poorly defined clusters (48, 82).
MglA shows localization patterns and behaviors strikingly similar to those of other gliding motility proteins. Under some experimental conditions, MglA appears as clusters that share the same localization pattern as AglZ and AgmU (48, 56, 60). Observed by immunofluorescence, MglA also decorates a helical structure that resembles that of MreB, AgmU, AglX, and AglR (48, 59, 64). Treatment of cells with A22 caused the lateral clusters of MglA to disperse, suggesting a connection with MreB (48). Significantly, the helically arranged localization of MglA is enhanced on soft surfaces, whereas the lateral clusters are more predominant on harder surfaces; this localization pattern is similar to that of AgmU (64). The switch between helices and lateral clusters may reflect the working status of the gliding engines. Given that the twitching engines are assembled at the poles and the gliding engines are distributed and helically arranged, it is possible that MglA regulates twitching and gliding motilities through the polar and lateral clusters, respectively.
RomR
RomR is another protein required for gliding motility that has an interesting localization pattern (42). RomR-GFP localizes to both cell poles, but the brighter cluster resides at the lagging cell pole. When cells reverse, the fluorescence of the brighter lagging cell pole decreases and the fluorescence of the new lagging pole increases. MglA regulates the localization of RomR. In mglA mutants, RomR localizes either predominantly at the leading pole or at both poles simultaneously (43). RomR contains an N-terminal response regulator domain and a C-terminal proline-rich output domain (42). The C-terminal domain of RomR is required for polar localization and the N-terminal domain for dynamic distribution. The RomR protein may indeed undergo phosphorylation cycles in vivo because the D to N mutant that mimics the dephosphorylated form of RomR can no longer show the asymmetric localization associated with reversals, and the cells carrying a D to E mutation in RomR that mimics the phosphorylated form reverse more frequently than the wild type (42). The conjugate protein kinase that presumably phosphorylates RomR still remains to be identified.
MODELS FOR GLIDING MOTILITY
The identification of genes required for gliding motility and the study of the localization and dynamics of gliding motility proteins have inspired several molecular models to explain this type of M. xanthus motility. The most popular models for the mechanism of gliding motility are described below.
The Slime Secretion Model
The slime secretion model for gliding motility was first proposed in 1924 by Jahn, who thought that the extrusion and hydration of slime might push cells forward (31). This model was inspired in part by the observation that extracellular slime secretion is characteristic of most gliding bacteria (7). This model was ignored for many years but received renewed interest in 1998 as a result of the work of Hoiczyk & Baumeister (26, 27). These investigators were studying gliding in the cyanobacteria Phormidium and Anabaena. In these species, multicellular filaments glide on solid surfaces, leaving mucilaginous slime trials behind. The slime ribbons originated from cellular junctions where junction pore complexes are found (26, 27). The observation that slime trails are only found behind moving cells and that the elongation rate of slime trails is consistent with the gliding velocity suggested to them the possibility that cells might be pushed forward by the slime (26, 27).
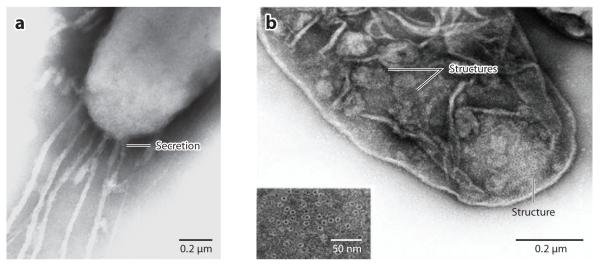
In M. xanthus, the slime secretion model is supported by several observations. (a) Gliding myxobacteria, e.g., cyanobacteria, routinely leave slime trails in their wake and the rate of slime deposition is consistent with gliding velocities (Figure 4a) (8, 81). (b) Nozzle structures, similar in appearance to the junction pore complexes of cyanobacteria, were observed by electron microscopy on the surface of M. xanthus cells near the cell poles (Figure 4b). Ribbons of slime appeared to be exiting from these structures (Figure 4a) (77). (c) Some genes found to be required for gliding motility encode enzymes for polysaccharide biosynthesis (e.g., sugar transferases and polymerases) and transport (e.g., Tol/Ton complexes) (80). (d) Some mutants defective in gliding motility were observed to leave slime trails at both ends. Additionally, some mutants that showed long pauses between reversals were defective in genes encoding sugar transferases and polymerases (81). (e) Calculations based on physical principles suggested that as few as 50 slime nozzles could generate sufficient thrust to account for the slow gliding velocity observed for M. xanthus (77).
Figure 4.
Nozzle structures and slime secretion in Myxococcus xanthus. (a) Electron micrograph of ribbon-like materials secreted from the cell pole. (b) Electron micrograph of the nozzle structures near the cell pole. Both panels are adapted from Reference 77, with the permission from Elsevier.
However, despite this evidence, none of these findings rule out the possibility that the deposition of slime, rather than being the driving force, is indeed a consequence of gliding. Most importantly, the components of the slime gun have never been identified. Furthermore, slime is complex and may consist of multiple components that have never been properly characterized. Slime could be the extracellular matrix material (fibrils or exopolysaccharides) that functions in twitching motility, or even the extracellular vesicles and tubes that protrude from cells, which might appear as slime trails under the light microscope (41, 63). The slime could have functions not directly related to motility, such as the adhesive that attaches cells to the substratum, as a capsule that protects cells from proteolysis, or as a matrix that immobilizes extracellular enzymes (7).
Indeed, evidence has been accumulating that does not directly support the slime secretion model. (a) Filamentous cells (snakes) created by cephalexin treatment glide at the same rate as shorter cells (74). Given that there is no evidence for increased slime secretion at the lagging cell poles of the elongated cells, gliding speeds would be expected to be inversely proportional to cell length to overcome the resistance of the larger cells. However, this is not observed. Furthermore, in some cephalexin-treated cells, the anterior portion of the elongated cells move forward, whereas the posterior portions lag behind or appear to not move at all (72). In the latter case, one might argue that secretion nozzles located in the vicinity of undeveloped division sites might extrude slime, pushing the anterior regions of cells forward (37). However, if this were occurring, lateral movements as observed in Phormidium should have been observed but were not reported. (b) M. xanthus cells continue to glide for more than 30 minutes after treated with NaN3, a reagent that dramatically reduces ATP synthesis. Gliding motility powered by slime secretion would be expected to be ATP dependent, as ATP is required for the synthesis and transport of polysaccharides. Indeed, in both myxobacteria and cyanobacteria the energy source of gliding motility is PMF (18, 59). (c) Localization studies of most gliding motility proteins and regulators show that these factors are distributed along the cell length rather than being concentrated at the lagging pole of moving cells, as might be expected for slime secretion nozzles localized at the lagging cell pole (56, 59, 60, 64). Computational simulations indicate that because of cell flexibility, force generated only at the rear of the cells should not be sufficient to power gliding motility along the long axis of cells (32).
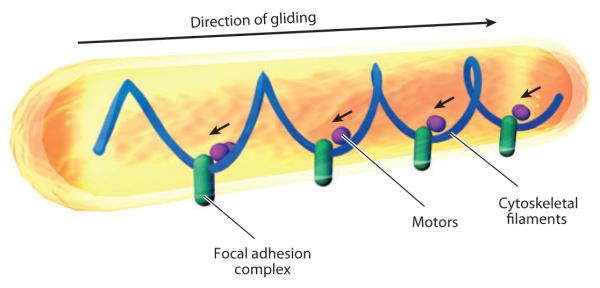
The Focal Adhesion Model
The focal adhesion model was proposed to explain the unexpected localization pattern of AglZ, an important A-motility protein. AglZ appears to be localized as an array of distributed stationary clusters in moving cells, i.e., AglZ clusters appeared to remain stationary with respect to the substratum rather than moving forward with the cells (56). This localization pattern is also observed for AgmU (60). To explain these results, Mignot et al. (56) proposed that gliding motility in M. xanthus is powered by distributed engines. According to this model, motor proteins that connect to cytoskeleton and envelope-spanning focal adhesion complexes assemble at the leading cell pole and move toward the lagging cell pole. This movement drives the cells forward by pushing against the focal adhesion complexes. Thus, the distributed focal adhesion complexes appear stationary as cells move forward (Figure 5) (55).
Figure 5.
The focal adhesion model for gliding motility. In this model, large focal adhesion complexes penetrate the cell envelope, stick to the substratum at one end, and connect to cytoskeletal filaments at the other end. Motor proteins push backward (marked by small arrows) against those focal adhesion complexes, pushing the cells forward.
The focal adhesion model is supported by the following experimental observations: (a) The gliding velocity of filamentous cells created by cephalexin treatment is proportional to the number of AglZ clusters and independent of cell length (56). This observation favors the hypothesis that the gliding engines are distributed. Given that the AglZ clusters always appear to be assembled at the leading cell poles, directional movements could be encoded by the regulators that reside at the leading pole, such as MglA, and countered by regulators at the lagging pole, like RomR and MglB (42, 43, 48, 56, 82). (b) M. xanthus cells are very flexible; cells frequently turn, bend, or even do U-turns. When moving cells bend, the clusters of AglZ are always found at the sites of bending, where the pushing force would be concentrated (56). (c) This model connects gliding motility to the MreB cytoskeleton. AglZ and MreB have been shown to interact both in vivo and in vitro (48). The periodicity of the AglZ clusters is consistent with the pitch of the MreB helices.
The focal adhesion model is based on the behavior of one protein, AglZ, and is undoubtedly overly simplistic. To expand this model, observations on more components and regulators of the gliding motility machinery should be included. Specifically, several basic questions remain to be addressed. For example:
What is the nature of the hypothetical focal adhesion complexes? Do they actually exist? Protein-protein interaction studies suggest the existence of protein complexes that contain components in the cytoplasm, inner membrane, and the periplasm (60), but among the identified gliding motility genes, none of them are predicted to encode a structure that dynamically sticks cells to the substratum (21, 80).
How do cells maintain their integrity if focal adhesion complexes continuously breach the cell envelope? The focal adhesion model predicts that during gliding, focal adhesion complexes must cut through the cell envelope laterally, breaching the inner membrane, peptidoglycan, and outer membrane. It is difficult to imagine how cells might maintain their integrity under these stressful conditions. Indeed, there is no evidence that moving cells are more osmotically sensitive than stationary cells.
The Helical Rotor Model
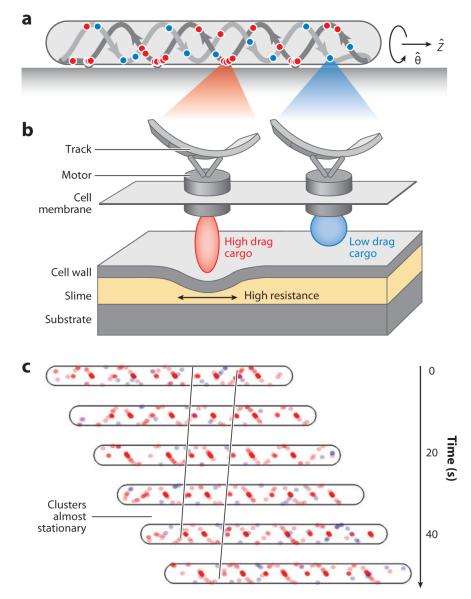
The helical rotor model is a mechanochemical model that attempts to assimilate recent data based on the analysis and localization of several gliding motility proteins. The model proposes that motor proteins move along a helical cytoskeletal track, creating forces that distort the cell surface and generate surface waves that push the cells forward (Figure 6).
Figure 6.
The helical rotor model. In this model, flagella motor homologs carry different protein cargos and ride on a looped helical track. (a,b) The high-drag cargos deform the cell envelope at the ventral side of cells, where the thrust is exerted. (c) The motors carrying high-drag cargos slow down at the sites of surface distortion, collecting in traffic jams that appear as stationary periodic clusters in the observations of AgmU and AglZ. All the panels are adapted from Reference 59.
According to this model, gliding motility is powered by modified flagella motors (MotAB homologs) that harness PMF to facilitate movement. These MotAB homologs, instead of being tethered to the peptidoglycan as in flagellated bacteria, move freely along an endless, looped cytoskeletal filament. Different protein cargos are carried along the helical track by the motors. High-drag cargos, such as motility proteins AglZ and AgmU, are part of a large multiprotein complex (60); this complex exerts force at the ventral side (the side where cells contact with the substratum) of the track and deforms the cell surface, generating wave-like contours on the ventral cell surface (Figure 6) (59). The surface waves push on slime, generating the force that drives cells forward. Because the cytoskeletal track is helical, the cells rotate. Thus, cells are powered by the backward propagation of transverse surface waves (Figure 6b), a similar mechanism also adopted by snails (11). The low-drag cargos could be regulatory proteins, such as FrzCD, MglA, and RomR, which are carried from pole to pole on the same track in an oscillatory manner without generating force (Figure 6b).
How then can we explain the focal adhesion clusters observed for AglZ (56), AgmU (60), and MglA (64)? These proteins are considered to be cargos and motor-associated proteins that travel on the ventral side of the helical track. At these sites, high-drag cargos are forced to slow down and back-up into traffic jams while exerting force on the cell surface. These stalled motor-associated proteins appear as nearly stationary clusters at these sites. Because they only form at the side of the helical track that contacts a surface, the periodicity of these clusters is consistent with the helical pitch of MreB (Figure 6c). This hypothesis is also supported by the observation that the fluorescence of AglZ-YFP is enhanced near moving polystyrene beads, where the traction force is predicted to be generated (75). On soft agar, methylcellulose, or in water, motors and cargos continue to move along the track but cells are not able to glide because very little drag force can be generated between the cell surface and the external substrate. For the same reason, few molecules are forced to slow down and aggregate into the traffic jam sites. That explains the reason that the formation of the clusters by AglZ, AgmU, and MglA is sensitive to the surface hardness and favored by hard surfaces (49, 60, 64). On a glass surface, almost all the motor and cargo molecules slow down and make even stronger aggregates, where bigger and brighter clusters were reported (60).
The helical rotor model attempts to explain many experimental observations:
Multiple gliding motility proteins (AgmU, AglX, and AglV) and regulators (FrzCD and MglA) localize along helical tracks (47, 59, 64).
The helices containing motility proteins rotate when cells are gliding forward and this rotation correlates with gliding motility in both direction and speed (59).
Both the rotation and gliding motility are powered by PMF and independent of ATP; chemicals that disrupt PMF stop gliding (59, 75).
Homologs of the flagella motors were found to be A-motility proteins (59, 75).
The localization and rotation of gliding motility proteins depends on intact MreB cytoskeletal filaments (48, 59).
The gliding motility complexes are composed of components that reside in the cytoplasm, membrane, and periplasm (60).
The polystyrene beads attached to cells can move in opposite directions (75).
The helical rotor model is consistent with additional experimental data. For example, the recovery of fluorescence of bleached AgmU-mCherry cells is bidirectional. This is consistent with AgmU riding on a track that forms a closed loop (59). Additionally, deformation of the cell surface is observed by total internal reflection fluorescence microscopy and electronic microscopy (44, 59, 65). This is consistent with deformations in the cell surface caused by protein cargos.
As with the two other gliding motility models, some details are still absent in this model and remain to be investigated. For example, to generate torque, both rotor and stator are required. In this model, MreB is presumed to be the stator, and the MotAB homologs serve as rotors. Further investigation is required to determine which cellular components are actually moving. Additionally, the net driving force in this model results from the unequal distribution of cargos loaded on the motors running to opposite directions. To ensure directed motility, the high-drag cargos and some of the regulatory proteins should be loaded onto the motors at the leading cell poles, transported backward, and unloaded at the lagging poles, as indicated by the biased transport of polystyrene beads (75), while other regulatory proteins should be loaded, transported, and unloaded in the opposite direction. The determinants of this biased transportation are still unknown. It is also unknown how cargos are transported back to one pole when they are unloaded at the other pole.
CONCLUSIONS
Bacterial cells are highly organized so that proteins and protein complexes are deployed to particular locations to facilitate diverse functions and behaviors. The key to uncovering the mystery of M. xanthus gliding motility requires the understanding of the motility components, their localization, and dynamics. Recent genetic and biochemical studies have identified many but not all of the proteins involved in the gliding machinery. The application of high-resolution fluorescence microscopy, biochemistry, and biophysical approaches has contributed important insights into the dynamics of gliding motility proteins. However, many questions remain. What are the motors? How do the motors propel the gliding engines? How do the gliding engines interact with the cytoskeletal filaments? How do the regulatory proteins change gliding direction? In many ways, gliding motility in the myxobacteria remains a mystery and is a subject of active investigation and a source for insights into the molecular organization and machinery of bacteria.
SUMMARY POINTS.
The machinery of M. xanthus gliding motility comprises multiple protein components, which assemble into large multiprotein complexes that span the cytoplasm, the inner membrane, and the periplasm.
Gliding motility proteins AgmU, AglX, and AglR localize along a closed helical loop that rotates as cells glide forward.
PMF is the energy source of M. xanthus gliding motility and the rotation of the helices formed by motility proteins. MotAB homologs are candidates for the gliding motors.
The rotation of the helices formed by motility proteins depends on actin-like MreB cytoskeletal filaments.
The reversal frequency of M. xanthus gliding motility is regulated by the Frz chemotaxis system, with MglA and RomR as outputs.
FUTURE ISSUES.
What are the motors for M. xanthus gliding motility? How do the motors interact with other components in the gliding machinery?
How is the gliding machinery organized? In addition to the machinery containing AgmU, AglZ, AglX, AglR, etc., are there other complexes involved in gliding motility?
How does the gliding machinery interact with MreB?
How do MglA and RomR interact with the gliding machinery and regulate the reversal frequency? What is the signal input of the Frz pathway?
What is the role of cell-cell contact in the regulation of gliding motility?
How is gliding motility coordinated with twitching motility?
What is the function of slime secretion in gliding motility? What is slime?
ACKNOWLEDGMENTS
We thank Eva Campodonico, James Berleman, Christine Kaimer, Im-Hong Sun and Linda Pan for helpful discussions. Research in our laboratory is funded by the National Institutes of Health (GM020509).
Glossary
- Chemotaxis
directed movement toward attractants and away from repellents
- Gliding motility
the active and smooth translocation of cells on a surface without the aid of flagella or pili
- MCP
methyl-accepting chemotaxis protein
- Slime
the material secreted and left behind as cells move. It contains carbohydrate and protein but its specific composition remains to be determined
- PMF
proton motive force
- Cytoskeleton
protein scaffolding or skeletal structures
- FRAP
fluorescence recovery after photobleaching
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacobs-Wagner C. RodZ, a component of the bacterial core morphogenic apparatus. Proc. Natl. Acad. Sci. USA. 2009;106:1239–44. doi: 10.1073/pnas.0810794106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astling DP, Lee JY, Zusman DR. Differential effects of chemoreceptor methylation-domain mutations on swarming and development in the social bacterium Myxococcus xanthus. Mol. Microbiol. 2006;59:45–55. doi: 10.1111/j.1365-2958.2005.04926.x. [DOI] [PubMed] [Google Scholar]
- 3.Berleman JE, Kirby JR. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol. Rev. 2009;33:942–57. doi: 10.1111/j.1574-6976.2009.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackhart BD, Zusman DR. “Frizzy” genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc. Natl. Acad. Sci. USA. 1985;82:8767–70. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonsor DA, Hecht O, Vankemmelbeke M, Sharma A, Krachler AM, et al. Allosteric beta-propeller signaling in TolB and its manipulation by translocating colicins. Embo J. 2009;28:2846–57. doi: 10.1038/emboj.2009.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burchard RP. Gliding motility mutants of Myxococcus xanthus. J. Bacteriol. 1970;104:940–47. doi: 10.1128/jb.104.2.940-947.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burchard RP. Gliding motility of prokaryotes: ultrastructure, physiology, and genetics. Annu. Rev. Microbiol. 1981;35:497–529. doi: 10.1146/annurev.mi.35.100181.002433. [DOI] [PubMed] [Google Scholar]
- 8.Burchard RP. Trail following by gliding bacteria. J. Bacteriol. 1982;152:495–501. doi: 10.1128/jb.152.1.495-501.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bustamante VH, Martinez-Flores I, Vlamakis HC, Zusman DR. Analysis of the Frz signal transduction system of Myxococcus xanthus shows the importance of the conserved C-terminal region of the cytoplasmic chemoreceptor FrzCD in sensing signals. Mol. Microbiol. 2004;53:1501–13. doi: 10.1111/j.1365-2958.2004.04221.x. [DOI] [PubMed] [Google Scholar]
- 10.Cascales E, Lloubes R, Sturgis JN. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol. Microbiol. 2001;42:795–807. doi: 10.1046/j.1365-2958.2001.02673.x. [DOI] [PubMed] [Google Scholar]
- 11.Chan B, Balmforth NJ, Hosoi AE. Building a better snail: lubrication and adhesive locomotion. Phys. Fluids. 2005;17:113101–10. [Google Scholar]
- 12.Charest PG, Firtel RA. Big roles for small GTPases in the control of directed cell movement. Biochem. J. 2007;401:377–90. doi: 10.1042/BJ20061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowles KN, Gitai Z. Surface association and the MreB cytoskeleton regulate pilus production, localization and function in Pseudomonas aeruginosa. Mol. Microbiol. 2010;76:1411–26. doi: 10.1111/j.1365-2958.2010.07132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem. Sci. 2003;28:655–62. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Divakaruni AV, Baida C, White CL, Gober JW. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol. Microbiol. 2007;66:174–88. doi: 10.1111/j.1365-2958.2007.05910.x. [DOI] [PubMed] [Google Scholar]
- 16.Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. USA. 2004;101:8643–48. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–41. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Glagoleva TN, Glagolev AN, Gusev MV, Nikitina KA. Protonmotive force supports gliding in cyanobacteria. FEBS Lett. 1980;117:49–53. [Google Scholar]
- 19.Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, et al. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartzell P, Kaiser D. Function of MglA, a 22-kilodalton protein essential for gliding in Myxococcus xanthus. J. Bacteriol. 1991;173:7615–24. doi: 10.1128/jb.173.23.7615-7624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartzell P, Shi W, Youderian P. Gliding motility of Myxococcus xanthus. In: Whitworth DE, editor. Myxobacteria: Multicellularity and Differentiation. ASM Press; 2008. pp. 103–22. [Google Scholar]
- 22.Hartzell PL. Complementation of sporulation and motility defects in a prokaryote by a eukaryotic GTPase. Proc. Natl. Acad. Sci. USA. 1997;94:9881–86. doi: 10.1073/pnas.94.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hazelbauer GL, Lai WC. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr. Opin. Microbiol. 2010;13:124–32. doi: 10.1016/j.mib.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol. Gen. Genet. 1979;171:177–91. [Google Scholar]
- 25.Hodgkin J, Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. USA. 1977;74:2938–42. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoiczyk E. Gliding motility in cyanobacterial: observations and possible explanations. Arch. Microbiol. 2000;174:11–17. doi: 10.1007/s002030000187. [DOI] [PubMed] [Google Scholar]
- 27.Hoiczyk E, Baumeister W. The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr. Biol. 1998;8:1161–68. doi: 10.1016/s0960-9822(07)00487-3. [DOI] [PubMed] [Google Scholar]
- 28.Inclan YF, Laurent S, Zusman DR. The receiver domain of FrzE, a CheA-CheY fusion protein, regulates the CheA histidine kinase activity and downstream signalling to the A- and S-motility systems of Myxococcus xanthus. Mol. Microbiol. 2008;68:1328–39. doi: 10.1111/j.1365-2958.2008.06238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inclan YF, Vlamakis HC, Zusman DR. FrzZ, a dual CheY-like response regulator, functions as an output for the Frz chemosensory pathway of Myxococcus xanthus. Mol. Microbiol. 2007;65:90–102. doi: 10.1111/j.1365-2958.2007.05774.x. [DOI] [PubMed] [Google Scholar]
- 30.Iwai N, Nagai K, Wachi M. Novel S-benzylisothiourea compound that induces spherical cells in Escherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2. Biosci. Biotechnol. Biochem. 2002;66:2658–62. doi: 10.1271/bbb.66.2658. [DOI] [PubMed] [Google Scholar]
- 31.Jahn E. I. Die Polyangiden. In: Borntraeger G, editor. Beitrage zur botanischen Protistologie. Leipzig; Leipzig, Germany: 1924. [Google Scholar]
- 32.Janulevicius A, van Loosdrecht MC, Simone A, Picioreanu C. Cell flexibility affects the alignment of model myxobacteria. Biophys. J. 2010;99:3129–38. doi: 10.1016/j.bpj.2010.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 2008;6:466–76. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 34.Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–22. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser AD, Crosby C. Cell movement and its coordination in swarms of Myxococcus xanthus. Cell Motil. 1983;3:227–45. [Google Scholar]
- 36.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA. 1979;76:5952–56. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaiser D. Are there lateral as well as polar engines for A-motile gliding in myxobacteria? J. Bacteriol. 2009;191:5336–41. doi: 10.1128/JB.00486-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaiser D, Robinson M, Kroos L. Myxobacteria, polarity, and multicellular morphogenesis. Cold Spring Harb. Perspect. Biol. 2010;2:a000380. doi: 10.1101/cshperspect.a000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearns DB. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010;8:634–44. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi H, Maeda M, Anraku Y. Membrane-bound adenosine triphosphatase of Escherichia coli. III. Effects of sodium azide on the enzyme functions. J. Biochem. 1977;81:1071–7. doi: 10.1093/oxfordjournals.jbchem.a131530. [DOI] [PubMed] [Google Scholar]
- 41.Konovalova A, Petters T, Sogaard-Andersen L. Extracellular biology of Myxococcus xanthus. FEMS Microbiol. Rev. 2010;34:89–106. doi: 10.1111/j.1574-6976.2009.00194.x. [DOI] [PubMed] [Google Scholar]
- 42.Leonardy S, Freymark G, Hebener S, Ellehauge E, Sogaard-Andersen L. Coupling of protein localization and cell movements by a dynamically localized response regulator in Myxococcus xanthus. Embo J. 2007;26:4433–44. doi: 10.1038/sj.emboj.7601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leonardy S, Miertzschke M, Bulyha I, Sperling E, Wittinghofer A, Sogaard-Andersen L. Regulation of dynamic polarity switching in bacteria by a Ras-like G-protein and its cognate GAP. Embo J. 2010;29:2276–89. doi: 10.1038/emboj.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lunsdorf H, Schairer HU. Frozen motion of gliding bacteria outlines inherent features of the motility apparatus. Microbiology. 2001;147:939–47. doi: 10.1099/00221287-147-4-939. [DOI] [PubMed] [Google Scholar]
- 45.Mattick JS. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 46.Mauriello EM, Mignot T, Yang Z, Zusman DR. Gliding motility revisited: How do the myxobacteria move without flagella? Microbiol. Mol. Biol. Rev. 2010;74:229–49. doi: 10.1128/MMBR.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mauriello EMF, Astling DP, Sliusarenko O, Zusman DR. Localization of a bacterial cytoplasmic receptor is dynamic and changes with cell-cell contacts. Proc. Natl. Acad. Sci. USA. 2009;106:4852–57. doi: 10.1073/pnas.0810583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mauriello EMF, Mouhamar F, Nan B, Ducret A, Dai D, et al. Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. Embo J. 2010;29:315–26. doi: 10.1038/emboj.2009.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mauriello EMF, Nan B, Zusman DR. AglZ regulates adventurous (A-) motility in Myxococcus xanthus through its interaction with the cytoplasmic receptor, FrzCD. Mol. Microbiol. 2009;72:964–77. doi: 10.1111/j.1365-2958.2009.06697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McBride MJ. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 2001;55:49–75. doi: 10.1146/annurev.micro.55.1.49. [DOI] [PubMed] [Google Scholar]
- 51.McBride MJ, Kohler T, Zusman DR. Methylation of FrzCD, a methyl-accepting taxis protein of Myxococcus xanthus, is correlated with factors affecting cell behavior. J. Bacteriol. 1992;174:4246–57. doi: 10.1128/jb.174.13.4246-4257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McBride MJ, Weinberg RA, Zusman DR. “Frizzy” aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc. Natl. Acad. Sci. USA. 1989;86:424–28. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McBride MJ, Zusman DR. FrzCD, a methyl-accepting taxis protein from Myxococcus xanthus, shows modulated methylation during fruiting body formation. J. Bacteriol. 1993;175:4936–40. doi: 10.1128/jb.175.15.4936-4940.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCleary WR, McBride MJ, Zusman DR. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J. Bacteriol. 1990;172:4877–87. doi: 10.1128/jb.172.9.4877-4887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mignot T. The elusive engine in Myxococcus xanthus gliding motility. Cell Mol. Life Sci. 2007;64:2733–45. doi: 10.1007/s00018-007-7176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. Evidence that focal adhesion complexes power bacterial gliding motility. Science. 2007;315:853–56. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minamino T, Imada K, Namba K. Molecular motors of the bacterial flagella. Curr. Opin. Struct. Biol. 2008;18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 58.Miyata M. Unique centipede mechanism of Mycoplasma gliding. Annu. Rev. Microbiol. 2010;64:519–37. doi: 10.1146/annurev.micro.112408.134116. [DOI] [PubMed] [Google Scholar]
- 59.Nan B, Chen J, Neu JC, Berry RM, Oster G, Zusman DR. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc. Natl. Acad. Sci. USA. 2011;108:2498–503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nan B, Mauriello EMF, Sun I, Wong A, Zusman DR. A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol. Microbiol. 2010;76:1539–54. doi: 10.1111/j.1365-2958.2010.07184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 62.Nudleman E, Wall D, Kaiser D. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science. 2005;309:125–27. doi: 10.1126/science.1112440. [DOI] [PubMed] [Google Scholar]
- 63.Palsdottir H, Remis JP, Schaudinn C, O’Toole E, Lux R, et al. Three-dimensional macromolecular organization of cryofixed Myxococcus xanthus biofilms as revealed by electron microscopic tomography. J. Bacteriol. 2009;191:2077–82. doi: 10.1128/JB.01333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patryn J, Allen K, Dziewanowska K, Otto R, Hartzell PL. Localization of MglA, an essential gliding motility protein in Myxococcus xanthus. Cytoskeleton (Hoboken) 2010;67:322–37. doi: 10.1002/cm.20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pelling AE, Li Y, Shi W, Gimzewski JK. Nanoscale visualization and characterization of Myxococcus xanthus cells with atomic force microscopy. Proc. Natl. Acad. Sci. USA. 2005;102:6484–89. doi: 10.1073/pnas.0501207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reichenbach H. Myxococcus spp. (Myxobacterales) Schwarmentwicklung und Bildung von Protocysten. Publikationen zu wissenschaftlichen Filmen; Gottingen, Germany: 1966. pp. 557–78. [Google Scholar]
- 67.Reichenbach H. Biology of myxobacteria: ecology and taxonomy. In: Dworkin M, Kaiser AD, editors. Myxobacteria II. Am. Soc. Microbiol.; Washington DC: 1993. pp. 13–62. [Google Scholar]
- 68.Rodriguez AM, Spormann AM. Genetic and molecular analysis of cglB, a gene essential for single-cell gliding in Myxococcus xanthus. J. Bacteriol. 1999;181:4381–90. doi: 10.1128/jb.181.14.4381-4390.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scott AE, Simon E, Park SK, Andrews P, Zusman DR. Site-specific receptor methylation of FrzCD in Myxococcus xanthus is controlled by a tetra-trico peptide repeat (TPR) containing regulatory domain of the FrzF methyltransferase. Mol. Microbiol. 2008;69:724–35. doi: 10.1111/j.1365-2958.2008.06323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi W, Kohler T, Zusman DR. Chemotaxis plays a role in the social behaviour of Myxococcus xanthus. Mol. Microbiol. 1993;9:601–11. doi: 10.1111/j.1365-2958.1993.tb01720.x. [DOI] [PubMed] [Google Scholar]
- 71.Shi W, Zusman D. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc. Natl. Acad. Sci. USA. 1993;90:3378–82. doi: 10.1073/pnas.90.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sliusarenko O, Zusman D, Oster G. The motors powering A-motility in Myxococcus xanthus are distributed along the cell body. J. Bacteriol. 2007 doi: 10.1128/JB.00923-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spormann A, Kaiser D. Gliding mutants of Myxococcus xanthus with high reversal frequencies and small displacements. J. Bacteriol. 1999;181:2593–601. doi: 10.1128/jb.181.8.2593-2601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun H, Yang Z, Shi W. Effect of cellular filamentation on adventurous and social gliding motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA. 1999;96:15178–83. doi: 10.1073/pnas.96.26.15178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T. Motor-driven intracellular transport powers bacterial gliding motility. Proc. Natl. Acad. Sci. USA. 2011;108:7559–64. doi: 10.1073/pnas.1101101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van den Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 77.Wolgemuth C, Hoiczyk E, Kaiser D, Oster G. How myxobacteria glide. Curr. Biol. 2002;12:369–77. doi: 10.1016/s0960-9822(02)00716-9. [DOI] [PubMed] [Google Scholar]
- 78.Wu SS, Kaiser D. Genetic and functional evidence that Type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 1995;18:547–58. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 79.Yang R, Bartle S, Otto R, Stassinopoulos A, Rogers M, et al. AglZ is a filament-forming coiled-coil protein required for adventurous gliding motility of Myxococcus xanthus. J. Bacteriol. 2004;186:6168–78. doi: 10.1128/JB.186.18.6168-6178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Youderian P, Burke N, White DJ, Hartzell PL. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol. Microbiol. 2003;49:555–70. doi: 10.1046/j.1365-2958.2003.03582.x. [DOI] [PubMed] [Google Scholar]
- 81.Yu R, Kaiser D. Gliding motility and polarized slime secretion. Mol. Microbiol. 2007;63:454–67. doi: 10.1111/j.1365-2958.2006.05536.x. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y, Franco M, Ducret A, Mignot T. A bacterial Ras-like small GTP-binding protein and its cognate GAP establish a dynamic spatial polarity axis to control directed motility. PLoS Biol. 2010;8:e1000430. doi: 10.1371/journal.pbio.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 2007;5:862–72. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]