Gram-positive pathogenic bacteria display proteins on their surface that may interact with host cells and tissues and play a role in virulence. How these proteins are targeted through the unique cytoplasmic membrane and the thick peptidoglycan to the cell surface and how they remain associated with the cell surface are challenging issues of both clinical and economic relevance. Indeed, a knowledge of the molecular sorting mechanisms may help to identify targets for new therapeutic agents, so urgently needed for Gram-positive bacteria. In this issue of PNAS, Schneewind and colleagues (1) have brought definitive evidence that sortase, an enzyme involved in the covalent linkage of some surface proteins of Staphylococcus aureus to the peptidoglycan, plays a key role in the display of surface proteins and in the virulence of this important human pathogen.
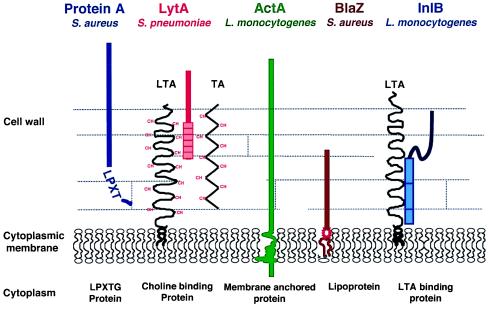
To date, five major mechanisms for displaying proteins at the surface of Gram-positive bacteria have been described (Fig. 1). Each mechanism is characterized by specific structural features that can be identified in the sequence of the proteins and are involved in their specific properties. The LPXTG proteins, exemplified by protein A of S. aureus (2), are the only surface proteins known to be covalently linked to the cell wall. The covalent linkage is dictated by a sorting signal made of a LPXTG motif followed by a hydrophobic domain made of about 20 amino acids and a tail of positively charged amino acids (for a review see ref. 3 and Fig. 2). This mechanism has been reported in many Gram-positive bacteria. In contrast, Streptococcus pneumoniae has evolved a mechanism unique to this organism to associate proteins with the cell wall (4): some S. pneumoniae proteins such as LytA bind to choline-substituted teichoic acids (TA), or lipoteichoic acids (LTA) by means of 20-amino acid repeats (TA and LTA are polymers that traverse the cell wall of Gram-positive bacteria). This mechanism is totally different from that of InlB and other proteins of Listeria monocytogenes that associate directly with LTA. Association of these latter proteins to LTA is mediated by 80-amino acid tandem repeats starting with the dipeptide GW (GW repeats) present in the C-terminal part of the protein (5, 6). Finally, some surface proteins are membrane proteins. Anchoring to the membrane can be achieved, as in ActA, by the presence of a hydrophobic stretch of about 20 amino acids followed by positively charged amino acids acting as a stop-transfer signal, at the C terminus of the protein (7). Alternatively, membrane proteins can be lipoproteins (8). In that case, as in Gram-negative bacteria, cleavage of a characteristic signal peptide generates a N-terminal cysteinyl residue that becomes lipoylated. The lipid moiety tethers the protein to the bacterial membrane.
Figure 1.
Major types of surface proteins in Gram-positive bacteria. Protein A, an immunoglobulin-binding protein, is covalently linked to the cell wall and exposed on the cell surface. The amidase LytA is loosely attached to choline (CH) residues decorating teichoic acid (TA) and lipoteichoic acid (LTA) in Streptococcus pneumoniae (4). The actin-polymerizing protein ActA of Listeria monocytogenes is membrane-anchored and exposed to the medium (7). The β-lactamase BlaZ encoded by the resistance plasmid PI258 of Staphylococcus aureus is associated to the membrane and partially released into the medium (8). InlB of L. monocytogenes is loosely attached to LTA (5, 6). It is weakly exposed on the cell surface.
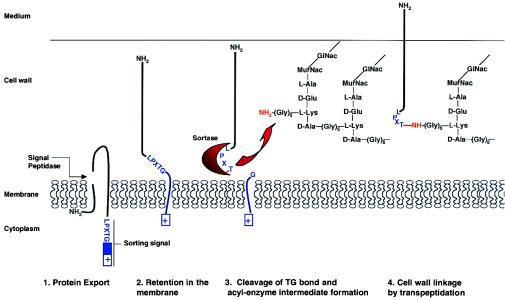
Figure 2.
Steps involved in the mechanism of covalent anchoring to the cell wall (adapted from refs. 13 and 14). MurNac, N-acetylmuramic acid.
Two main observations pointed to the role of the LPXTG motif in anchoring proteins to the cell surface. First, treatment of Streptococcus pyogenes with trypsin and then muramidase released a trypsin-resistant, cell-wall-embedded fragment of the M protein. Amino acid analysis revealed that this fragment lacked the C-terminal 19 hydrophobic amino acids and the charged tail encoded by the corresponding emm gene (9, 10). These results demonstrated that a posttranslational event generates the mature form of M protein. Second, a conserved LPXTGX hexapeptide preceding the hydrophobic domain and charged tail and present in 11 surface proteins of streptococci and staphylococci was detected by Fischetti and colleagues (11). This list has since dramatically expanded, and the now so-called LPXTG motif has been detected in more than 60 published protein sequences from various Gram-positive bacteria, including lactococci, enterococci, and Listeria (3).
The model that has been most used to study anchoring of LPXTG proteins to the cell wall is protein A of Staphylococcus aureus. Fractionation experiments established that mature protein A, like M protein, is in the cell wall fraction (2). Deletion experiments revealed that both the complete charged tail and the LPXTG motif are necessary for cell wall anchoring. As early as 1992, Olaf Schneewind and colleagues proposed that “Protein A is first led into the export pathway by its signal sequence. The charged tail is required to prevent secretion of the polypeptide into the medium. The retention of protein from the secretory pathway allows proteolytic cleavage near the C-terminal end of the polypeptide chain and a concerted cell wall linkage possibly at the LPXTG motif” (2). Amazingly, over the years, the Schneewind group has elucidated key events and components supporting their hypothesis. Swapping experiments using sorting domains from different bacteria demonstrated the universality and thus the evolutionary conservation of the cell wall anchoring motif, and also showed that this mechanism results in the most efficient attachment of proteins to the bacterial surface (12). Schneewind's group then showed that cleavage occurs between the threonine (T) and the glycine (G) residues in the LPXTG motif (13). The next breakthrough in the field and the definitive establishment of the role of the LPXTG motif was the determination of the fine structure of the cell wall anchor by using a combination of molecular biology and mass spectrometry techniques. After cleavage between threonine and glycine residues of the conserved LPXTG motif, the carboxyl of threonine is amide-linked to the free amino group of pentaglycine cross-bridges in the staphylococcal cell wall (14). These pentaglycine bridges are covalently linked to the ɛ-amino group of lysine residues. Note that other Gram-positive bacteria do not have the same cell wall structure. For example, Streptococcus pyogenes has two alanine residues in place of the pentaglycine bridge, and the T of LPXTG proteins is predicted to be linked to a two-alanine bridge. Listeria monocytogenes has a meso-diaminopimelic acid (m-Dpm) in place of the lysine residue in the peptidoglycan and no amino acid in the bridge. As recently demonstrated, LPXTG proteins are indeed directly attached to m-Dpm (15).
After establishment of the anchor structure, the next issue to address was the identification of the enzyme(s) involved. A so-called “sortase” had long been predicted (14). Would this enzyme be able to perform the peptide cleavage and the transpeptidation? Would these enzymatic activities be harbored by a single protein or a multimeric complex? By using a genetic approach, a small ORF, srtA (surface protein sorting A, or sortase) that complemented a mutant unable to mature a reporter LPXTG protein was identified (16). Sortase is a 206-amino acid protein with a potential N-terminal signal peptide that could act as a membrane anchor. It also has a unique cysteine at position 184, consistent with the observation that the cell wall sorting reaction is sensitive to reagents that modify sulfhydryl groups (17) and later shown to be critical for sortase activity (18). srtA homologs have been detected in several Gram-positive bacteria, including Bacillus subtilis, although no LPXTG protein has been characterized in this organism. All srtA homologs display absolute conservation of the cysteine codon at position 184. Sortase has been purified and shown to catalyze the formation of an hydroxylamine-sensitive acyl enzyme intermediate which, in the presence of peptidoglycan precursors, allows a transpeptidation reaction to proceed (18, 19). Sortase thus possesses both protease and transpeptidase activities. Fractionation experiments and Western blotting experiments using antibodies to recombinant sortase antibodies have shown that sortase is a membrane-associated protein (1).
Since several LPXTG proteins of staphylococci (e.g., protein A, clumping factor, fibronectin-binding proteins) contribute to the virulence of S. aureus (20), it was of the utmost interest to test whether incorrect surface presentation due to inactivation of the sortase gene would have an effect on virulence. These experiments are presented in the paper of this issue of PNAS (1). Display of known LPXTG proteins was evaluated by immunofluorescence or other techniques. Clearly, the sortase mutation abrogates exposure on the cell surface, predicting a strong effect of the sortase mutation on the virulence of the bacteria. A mouse model was used to test the effect of the mutation. After peritoneal infection, the LD50 (lethal dose 50, i.e., the dose at which half of the inoculated animals die) of mutants is 100 times higher than that of the wild type. These data provided convincing evidence that cell wall anchoring of surface proteins is an important parameter in the development of a staphylococcal infection. Staphylococci are extracellular pathogens that can lead to severe human infections, in particular endocarditis. However, the animal model and the inoculation protocol chosen by Schneewind and colleagues were not appropriate to test for a role of sortase in such infections. Further work will be needed to really appreciate the role of sortase in the various infections generated by staphylococci. It will also be very interesting to test the role of sortase in infections generated by Streptococcus pneumoniae and other Gram-positive bacteria for which excellent animal models exist. It is possible that even higher effects of the sortase mutation may be observed in the case of bacteria that have fewer toxins and secreted factors compared with staphylococci. Sortase appears to be a very promising target for identifying inhibitors that could be of general use in therapeutics against Gram-positive bacteria.
Acknowledgments
We thank Patrick Stragier for critical reading of this manuscript.
Footnotes
See companion article on page 5510.
References
- 1.Mazmanian S K, Liu G, Jensen E R, Lenoy E, Schneewind O. Proc Natl Acad Sci USA. 2000;97:5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneewind O, Model P, Fischetti V A. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 3.Navarre W W, Schneewind O. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Höltje J V, Tomasz A. J Biol Chem. 1975;250:6072–6076. [PubMed] [Google Scholar]
- 5.Braun L, Dramsi S, Dehoux P, Bierne H, Lindhal G, Cossart P. Mol Microbiol. 1997;25:285–294. doi: 10.1046/j.1365-2958.1997.4621825.x. [DOI] [PubMed] [Google Scholar]
- 6.Jonquières R, Bierne H, Fiedler F, Gounon P, Cossart P. Mol Microbiol. 1999;34:902–914. doi: 10.1046/j.1365-2958.1999.01652.x. [DOI] [PubMed] [Google Scholar]
- 7.Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- 8.Wang P-Z, Novick R P. J Bacteriol. 1987;169:1763–1766. doi: 10.1128/jb.169.4.1763-1766.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pancholi V, Fischetti V A. J Bacteriol. 1988;170:2618–2624. doi: 10.1128/jb.170.6.2618-2624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pancholi V, Fischetti V A. J Exp Med. 1989;170:2119–2133. doi: 10.1084/jem.170.6.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischetti V A, Pancholi V, Schneewind O. Mol Microbiol. 1990;4:1603–1605. doi: 10.1111/j.1365-2958.1990.tb02072.x. [DOI] [PubMed] [Google Scholar]
- 12.Schneewind O, Mihaylova-Petkov D, Model P. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navarre W W, Schneewind O. Mol Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- 14.Schneewind O, Fowler A, Faull K F. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 15.Dhar G, Faull K F, Schneewind O. Biochemistry. 2000;39:3725–3733. doi: 10.1021/bi992347o. [DOI] [PubMed] [Google Scholar]
- 16.Mazmanian S K, Liu G, Ton-That H, Schneewind O. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 17.Ton-That H, Schneewind O. J Biol Chem. 1999;274:24316–24320. doi: 10.1074/jbc.274.34.24316. [DOI] [PubMed] [Google Scholar]
- 18.Ton-That H, Liu G, Mazmanian S K, Faull K F, Schneewind O. Proc Natl Acad Sci USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ton-That H, Mazmanian S K, Faull K F, Schneewind O. J Biol Chem. 2000;275:9876–9880. doi: 10.1074/jbc.275.13.9876. [DOI] [PubMed] [Google Scholar]
- 20.Foster T J, McDevitt D. FEMS Microbiol Lett. 1994;118:199–206. doi: 10.1111/j.1574-6968.1994.tb06828.x. [DOI] [PubMed] [Google Scholar]