Abstract
How chromatin remodelers cooperate to organize nucleosomes around the start and end of genes is not known. We determined the genome-wide binding of remodeler complexes SWI/SNF, RSC, ISW1a, ISW1b, ISW2, and INO80 to individual nucleosomes in Saccharomyces, and determined their functional contributions to nucleosome positioning through deletion analysis. We applied ultra-high resolution ChIP-exo mapping to Isw2 to determine its sub-nucleosomal orientation and organization on a genomic scale. Remodelers interacted with selected nucleosome positions relative to the start and end of genes, and produced net directionality in moving nucleosomes either away or towards nucleosome-free regions at the 5′ and 3′ ends of genes. Isw2 possessed a sub-nucleosomal organization in accord with biochemical and crystallographic-based models that place its linker binding region within promoters and abutted against Reb1-bound locations. Together these findings reveal a coordinated position-specific approach taken by remodelers to organize genic nucleosomes into arrays.
INTRODUCTION
The nucleosome is the basic repeating unit of chromatin. Genome-wide mapping of nucleosome positions reveals that they are organized into uniformly-spaced arrays at the 5′ and to a lesser extent 3′ ends of genes (Jiang and Pugh, 2009b). Most genes are bounded by nucleosome-free promoter and termination regions (NFRs), although a select subset of repressed genes have a nucleosome situated over the promoter. Arrays start with the “+1” nucleosome positioned at a fairly precise distance from the transcription start site (TSS). A “−1” nucleosome, located on the upstream side of the promoter NFR, is positioned to potentially control access to gene regulatory sequences.
While the underlying DNA sequence contributes substantially to the occupancy levels of nucleosomes across a genome (Segal and Widom, 2009), ATP-dependent chromatin remodeling complexes play a key role in guiding their proper positioning at the start and end of genes on a genomic scale (Whitehouse et al., 2007; Badis et al., 2008; Hartley and Madhani, 2009; Tirosh et al., 2010), perhaps by packing them against barriers (Zhang et al., 2011b). Regulation of these barriers and/or nucleosome spacing may control gene expression.
Chromatin-remodeling complexes propagate nucleosome movement via ATP-dependent alterations in histone-DNA contacts (Peterson and Workman, 2000; Gangaraju and Bartholomew, 2007b; Clapier and Cairns, 2009; Narlikar, 2010). They are classified based on the sequence homology of their conserved ATPase subunit into four distinct families: ISWI (ISW1a, ISW1b, and ISW2), INO80/SWR1, CHD, and SWI/SNF (including RSC) (Mellor and Morillon, 2004; Hota and Bartholomew, 2011). However, their biochemical mechanism of nucleosome movement can be thought of in terms of two groups: ISWI and SWI/SNF, based upon requirements for flanking linker DNA sequences.
In the ISWI group, Isw1, Isw2, Chd1, and Ino80 require extra-nucleosomal linker DNA to re-position nucleosomes (Whitehouse et al., 2003; Kagalwala et al., 2004; Zofall et al., 2004; Gangaraju and Bartholomew, 2007a; Hota and Bartholomew, 2011; Udugama et al.). These chromatin remodelers may pull linker DNA onto nucleosomes, thereby causing nucleosomes to move towards the linker until the linker is too short to promote binding. In vitro, the ISWI group moves nucleosomes from the end of a DNA fragment to the middle, which is an activity that may help equally space nucleosomes in an array (Tsukiyama et al., 1999; Stockdale et al., 2006; Zofall et al., 2006; Udugama et al.).
SWI/SNF and RSC not only transfer histone octamers to exogenous DNA in trans, but they also slide nucleosomes without the requirement of extra-nucleosomal DNA (Whitehouse et al., 1999; Kassabov et al., 2003). Furthermore, if sliding encounters another nucleosome, packing or nucleosome eviction may occur RSC (Boeger et al., 2004; Montel et al., 2011).
The genome-wide contribution of chromatin remodelers to nucleosome positioning in yeast has been examined through remodeler mutants (Whitehouse et al., 2007; Hartley and Madhani, 2009; Tirosh et al., 2010; Gkikopoulos et al., 2011). Isw2 reportedly binds to regions flanking NFRs and repositions nucleosomes towards the NFR to suppress cryptic transcription initiation events (Whitehouse et al., 2007). In contrast, RSC moves nucleosomes away from the NFR (Hartley and Madhani, 2009). Isw1 functions more towards the middle of genes, where it promotes the repositioning of nucleosomes towards the 3′ direction (Tirosh et al., 2010). The combined action of ISWI-type remodelers may help define nucleosome positioning in gene bodies (Gkikopoulos et al., 2011).
An important open question that we address here is whether each remodeler has selectivity for specific nucleosome positions in and around genes. In addition, we expand on existing genome-wide studies to examine a comprehensive set of remodeler deletions to address whether such nucleosome selectivity of binding imparts directionality. We further investigate the notion of remodeler directionality by focusing on Isw2, where we employ a novel ChIP-exo technique to reveal the directional sub-nucleosomal and linker contacts made by Isw2 on a genomic scale. This analysis suggests a mechanism by which Isw2 might pack nucleosomes against Reb1 in the NFR to repress transcription. Our study suggests that remodelers are nucleosome position- and orientation-specific and move nucleosomes with predetermined net directionality relative to NFRs.
RESULTS
Enrichment of Remodelers at Specific Nucleosome Positions
Saccharomyces contains approximately 60,000 nucleosomes. To examine which consensus positions are potentially bound by specific remodelers, we conducted genome-wide remodeler-nucleosome interaction assays, based on an experimental design to detect the interaction of transcription factors with nucleosomes (Koerber et al., 2009). These assays involve standard formaldehyde-crosslinking chromatin immunoprecipitation (ChIP) assays in which the chromatin is fragmented to mononucleosomes using MNase. DNA fragments that immunoprecipitate with a specific chromatin remodeler subunit are then detected by deep sequencing.
Chromatin remodelers are notoriously difficult to ChIP, perhaps owing to inefficient crosslinking and/or a transient presence at any one place during the course of their catalytic cycle of nucleosome remodeling (Ng et al., 2002; Whitehouse et al., 2007). To maximize our “hit” rate, we conducted MNase-ChIP on 20 remodeler subunits that may be in close association with the nucleosome. These 20 subunits are components of the remodeler complexes SWI/SNF, RSC, ISW1a, ISW1b, ISW2, INO80, and CHD1. Consistent with the difficulties associated with immunoprecipitating remodelers in combination with the additional requirement that our MNase-ChIP required the generation of ~150 bp MNase-resistant DNA, some of the tested remodeler proteins did not show detectable interactions (Figure S1A). However, eight remodeler subunits (Ioc3, Isw2, Arp5, Ino80, Rsc8, Snf2, Ioc4, Isw1) were successfully assayed and carried forward for genome-wide analysis (Table S1). These subunits represent a mix of catalytic and regulatory subunits. The catalytic subunits are the definitive core of the complexes, and thus their detection should be definitive of remodeler presence.
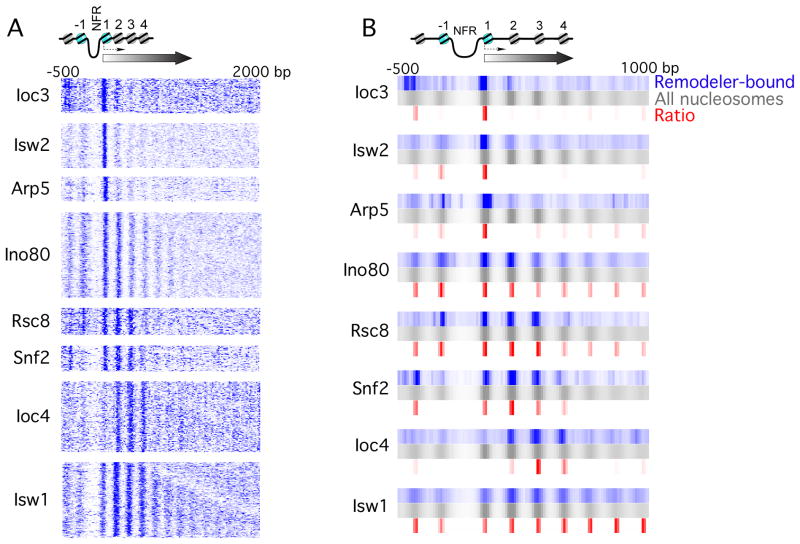
As a control, histone H3 was immunoprecipitated in parallel to identify all genomic nucleosomes. Untagged BY4741 and Sua7-TAP strains served as negative controls. Sua7 is the TFIIB component of the transcription pre-initiation complex. Because it resides in the NFR, its interaction with nucleosomes is expected to be negligible (Koerber et al., 2009). For each sequence tag associated with a remodeler-bound nucleosome, a putative nucleosomal dyad was derived. Dyad distributions were then collapsed to single tracks, one track per gene, aligned by their TSS (David et al., 2006), and k-means clustered (Figures 1A and S1A). Results from multiple k values indicated a single predominant gene cluster within each dataset (not shown). These clusters constituted the set of genes that were enriched with remodeler-bound nucleosomes. We further collapsed individual clusters into a single track (blue tracks in Figure 1B), along with the distribution of all nucleosomes (i.e. H3 MNase ChIP) for the same subset of genes (gray tracks).
Figure 1. ATP-dependent chromatin remodelers bind to specific nucleosome positions at the beginning of genes.
(A) Remodeler-bound nucleosomal tags were aligned by the TSS of the underlying gene, binned (5 bp) and smoothed (3 bin moving average). Color intensity represents tag counts. Shown are raw tags, with no background subtraction or normalization. At least one remodeler was detected at about half of all 5866 yeast genes having an annotated TSS.
(B) Values from panel A were averaged and plotted as in panel A for remodeler-bound nucleosomes (blue) and all (H3-containing) nucleosomes (gray). Red indicates density, where values represented by blue were divided by values represented by gray. See also Figure S1.
We observed that each tested remodeler subunit crosslinked to specific nucleosome positions relative to the TSS (summarized in Table 1). These observations, however, are based on raw tag distributions of remodeler-bound nucleosomes, which did not take into account the underlying intrinsic nucleosome occupancy levels. We therefore made peak calls to derive consensus nucleosome positions and their occupancy levels (tag counts within the peak region, Table S1). These remodeler-bound nucleosome levels were then normalized to the corresponding histone H3 occupancy levels. A composite of position-specific densities is shown in Figure 1B (red). With this normalization, the selectivity of individual remodelers for specific nucleosome positions became even more evident.
Table 1.
Summary of chromatin remodeler-nucleosome interactions
| Protein | Complex | ATPase Family | 5′-enda | 3′-endb |
|---|---|---|---|---|
| Arp5 | INO80 | INO80 | +1 | None |
| Ino80 | INO80 | INO80 | All | TN |
| Ioc3 | ISW1a | ISWI | +1 | TN |
| Ioc4 | ISW1b | ISWI | +2, +3, +4 | TN-1 |
| Isw1 | ISW1a/ISW1b | ISWI | All | All |
| Isw2 | ISW2 | ISWI | +1 | TN |
| Rsc8 | RSC | SWI/SNF | −1, +1, +2, +3 | None |
| Snf2 | SWI/SNF | SWI/SNF | −2, +1, +2, +3 | None |
Chromatin remodeler interacting nucleosomes located near the 5′-end of the genes
Chromatin remodeler interacting nucleosomes located near the 3′-end of the genes, in which TN = terminal nucleosomes and None means no interaction is seen in this region
We found that Rsc8 (RSC) and Snf2 (SWI/SNF) crosslinked predominantly to the first three genic nucleosomes, and to selected positions upstream of the TSS, which is consistent with an earlier lower resolution report (Ng et al., 2002). SWI/SNF was largely depleted at the −1 position, but enriched at −2. Based on current models of SWI/SNF action (Boeger et al., 2004), this would place SWI/SNF in position to remove the adjacent −1 nucleosome. Isw2 crosslinked predominantly to the +1 position, as reported previously (Whitehouse et al., 2007). The catalytic Isw1 subunit, found in at least two ISWI-type complexes ISW1a and ISW1b (Vary et al., 2003), was spread across all genic nucleosome positions. However, the regulatory subunit Ioc3 (ISW1a) was particularly enriched at the +1 position, whereas Ioc4 (ISW1b) was enriched at positions +2, +3, and +4, which is consistent with their distinct functional roles (Morillon et al., 2003). Additional Ioc3 interactions were observed at the −2 nucleosome position, which is in line with a previous suggestion (Yamada et al., 2011). Thus, the Ioc subunits may confer distinct genome-wide positional (and functional) specificities on Isw1. Similarly, Ino80 was spread across many positions, although the Arp5 subunit of the INO80 complex was particularly enriched at the +1 position. This suggests that Ino80 may also exist apart from at least one of its subunits.
Several additional experiments and analyses address the robustness of these results. First, similar conclusions were evident when the patterns were normalized to nucleosome occupancy level (Figure 1B). Second, patterns were reproducible across multiple biological replicates. Third, positional selectivity patterns were unique to the remodeler subunit being tested, which is an outcome that would be unlikely if the patterns were arising from noise in the data. Fourth, an untagged BY4741 negative control failed to generate a pattern (Figure S1B). Fifth, the general transcription factor TFIIB (Sua7), which binds to promoter NFRs, generated a distinct pattern (consistent with NFR binding) (Figure S1B). Sixth, the remodeler-bound nucleosome positions did not have intrinsically high or low H3 occupancy (data not shown), indicating that they were not outliers. Taken together, these findings suggest that chromatin remodeling complexes preferentially occupy specific nucleosome positions in and around a subset of all genes across the yeast genome. These positions are defined by their proximity to the TSS.
The analysis of position selectivity within coding regions suggested two major themes, one in which certain remodeler subunits crosslinked predominantly with the +1 nucleosome, and another where remodelers crosslinked at multiple positions within genes. Thus, the +1 nucleosome appears to be handled differently by remodelers compared to the other genic nucleosomes.
Enrichment of ISWI, but not the SWI/SNF Family, at Terminal Nucleosomes
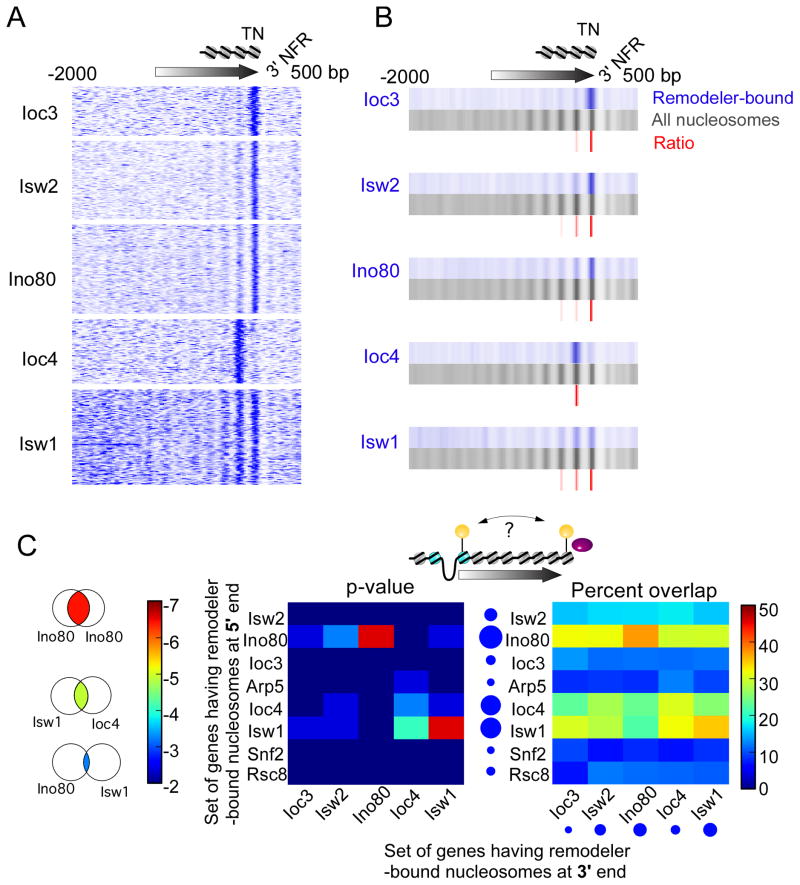
NFRs, positioned nucleosomes, and antisense noncoding transcription exist at the 3′ ends of genes, but little is known about the presence or function of remodelers in organizing these regions. We examined the distribution of remodeler-bound nucleosomes relative to the terminal nucleosome of each gene (as defined by the wild-type dataset). We found that Ioc3 (ISW1a), Isw2, and Ino80 crosslinked directly with the terminal nucleosome (Figure 2A). Isw1 crosslinked minimally with the last three nucleosome positions, consistent with results indicative of association across gene bodies. Ioc4 (ISW1b) crosslinked specifically with the penultimate terminal nucleosome, which is consistent with its role in transcription termination (Alen et al., 2002; Morillon et al., 2003). A composite of position-specific densities is shown in Figure 2B (red), highlighting the selectivity of individual remodelers for specific nucleosome positions at the end of genes.
Figure 2. ISWI remodelers interact with terminal nucleosomes.
(A,B) Same as Figure 1, except genes having remodeler-bound nucleosomes near their 3′ ends were aligned by the terminal nucleosome dyad of the underlying gene.
(C) Heat map representing the venn overlap (illustrated to the left) of those genes containing remodeler-bound nucleosomes near the 5′ end versus 3′ end (see illustration). The overlap is presented as a chi-square distribution (middle) and percentage of overlap (right). The size of the blue circles reflects the number of bound genes.
This specific targeting of remodelers to the 3′ end of genes was similar but inverted to the pattern observed at the 5′ end. For example, ISW1a, INO80, and ISW2 targeted both the first and last nucleosomes, whereas Ioc4 (ISW1b) was particularly enriched at penultimate nucleosomes. Our clustering analysis did not detect significant interactions between RSC or SWI/SNF with terminal nucleosomes, indicating that they may be selective for sense-driven mRNA gene expression, at least with respect to measurable binding in this assay.
We next asked whether a remodeler binding at one end of a gene might work coordinately, independently, or mutually exclusive with any remodeler at the other end of the gene. We calculated the correlation between remodeler-nucleosome interactions at the 5′ and 3′ ends of the same gene (Figure 2C). As expected, nucleosome association with Isw1, which is found across gene bodies, was highly correlated between the 5′ and 3′ ends. The same was observed for Ioc4, which was more position-selective than Isw1. We interpret these findings to indicate that Isw1 works across the body of genes from end to end, but as part of ISW1b (as defined by Ioc4) it may be more restricted to penultimate nucleosome positions at both ends of coding genes. Whether the continuity of Isw1 throughout gene bodies contributes to the coordinated presence of Ioc4 at both ends is unclear. INO80-nucleosome interactions were also highly linked between the 5′ and 3′ ends (Figure 2C), which further supports our suggestion of gene-specific 5′-3′ coordination of certain remodelers.
Chromatin Remodelers Target Similar Sets of Genes
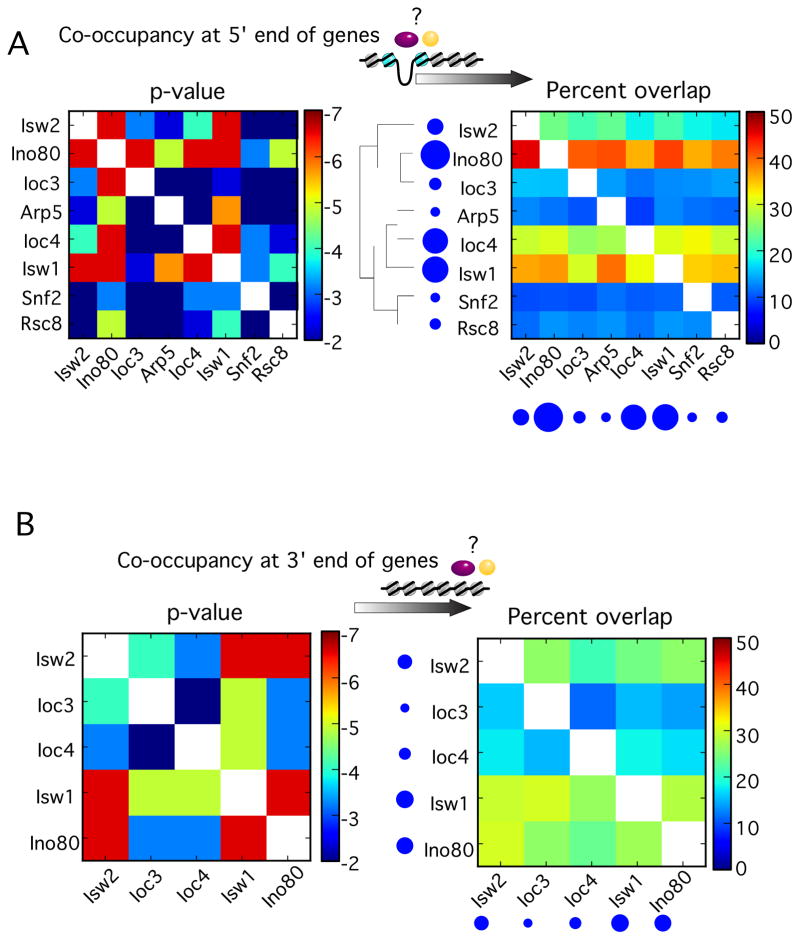
The notion that chromatin remodelers target specific nucleosome positions raises the question as to whether there is coordination among the remodelers so as to regulate entire genic nucleosomal arrays. We examined all pair-wise co-occurrence of genes (as opposed to nucleosomes) enriched with any two remodelers by calculating both the p-value and the percentage of co-occurrence (Figure 3).
Figure 3. Chromatin remodelers target similar sets of genes.
(A) Heat map representing the venn overlap of genes containing each type of remodeler-bound nucleosome near their 5′-ends, either by chi-square distribution (left) or percentage overlapping (right). Circles reflect population sizes.
(B) Same as panel A, but for the 3′ ends of genes.
Nucleosomes crosslinked to Ino80, Isw1, and Isw2 had the largest number of genes in common (Figure 3A, right panel), suggesting that these remodelers work together. As expected, both Ioc3 and Ioc4 overlapped substantially with Isw1. However, Ioc3 and Ioc4 overlapped very little with each other, indicating that ISW1a and ISW1b may target distinct sets of genes, as previously reported (Vary et al., 2003).
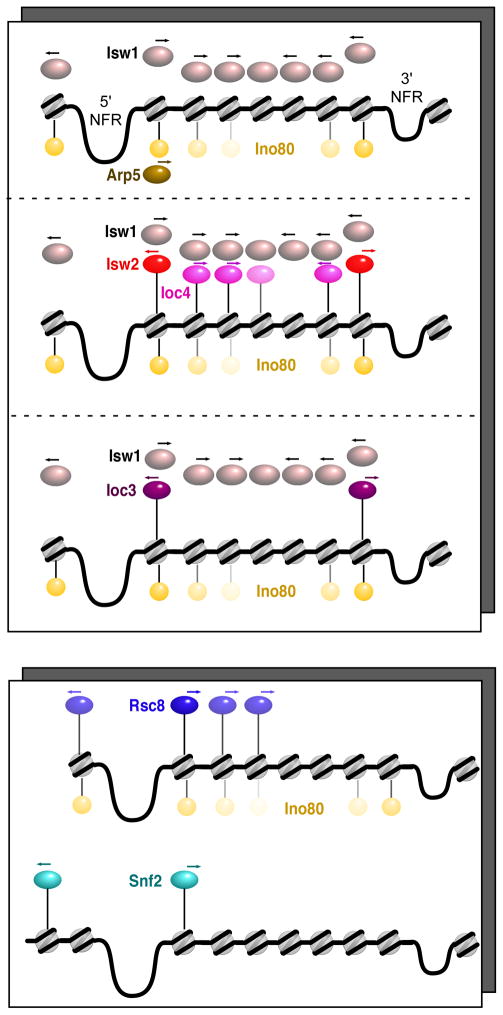
RSC and SWI/SNF tended to be bound to different genes than the ISWI and INO80 remodelers (although some overlap between INO80 and RSC was observed). RSC and SWI/SNF overlapped very little with each other, all of which may reflect fundamental differences in the function of ISWI-type vs SWI/SNF-type remodelers. An illustration of the distinct groups of genes bound by the various remodelers, and their occupied nucleosome positions is illustrated in Figure 4. These findings suggest that the INO80 and ISWI complexes act collectively over many nucleosome positions within genic arrays, whereas the RSC and SWI/SNF complexes act separately and are more restricted to nucleosomes at the 5′ ends of genes.
Figure 4. Model of chromatin remodeler targeting and directionality.
Each illustration represents an approximate grouping of genes similarly enriched with remodelers, separated into ISWI and RSC classes (upper and lower panels). Spheres represent the predominant locations of remodelers relative to 5′ and 3′ NFRs. Arrows depict the direction to which the indicated chromatin remodeler moves nucleosomes.
In Vivo Directionality of Chromatin Remodelers
Chromatin remodelers position nucleosomes on genomic DNA, perhaps by translocation in cis until some barrier is reached. How a division of labor among remodelers leads to well-defined nucleosome organization across genomes remains unclear. In particular, do remodelers translocate nucleosomes towards or away from promoters? Is the directionality reversed at the 3′ ends of genes? The question of remodeler directionalities has been examined in part for Isw2 (Whitehouse et al., 2007), RSC (Hartley and Madhani, 2009), and Isw1 (Tirosh et al., 2010) at varying degrees of resolution and coverage.
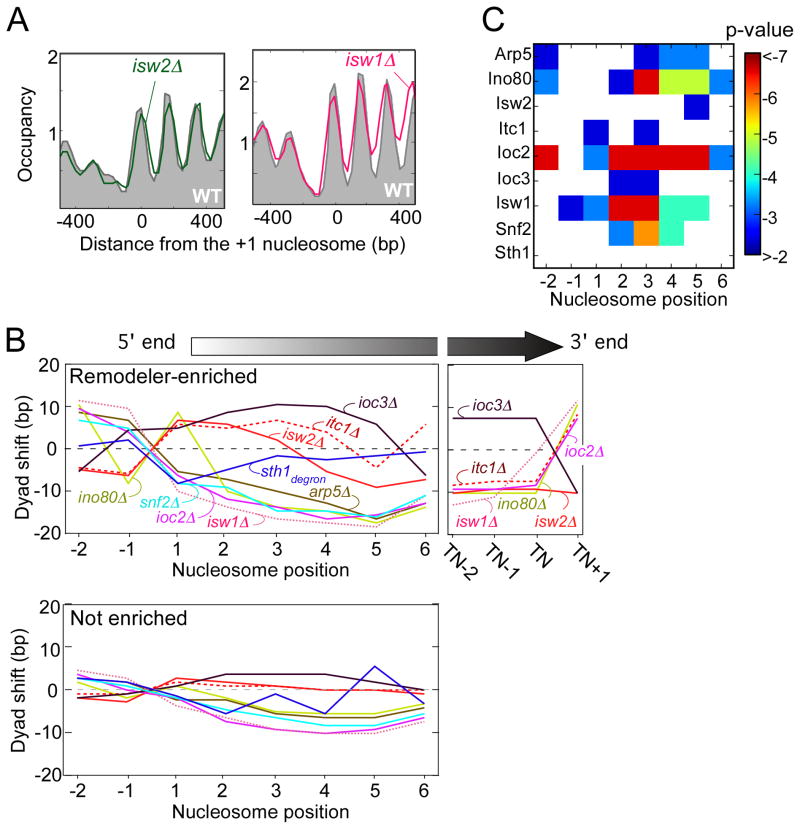
To determine the contribution of all remodelers to nucleosome positioning on a genome-wide scale, we generated a high-resolution map of all nucleosome positions (via MNase H3 ChIP-seq) in strains harboring deletions of various chromatin remodeler subunits. From a population of nucleosomal tags, we calculated a single consensus dyad location for each nucleosome (Figure 5A). A portion of all nucleosomes are inherently delocalized or randomly positioned, and thus repositioning from one random location to another in a population is inherently not meaningful and not applicable to measuring remodeler-dependent positioning. We quantified nucleosome delocalization as the standard deviation of individual tag locations. We then applied a t-test between mutant and wild-type positions so as to further analyze only those nucleosomes that underwent a statistically significant change in position (Table S2) (Tirosh et al., 2010). We examined the nucleosomal shifts occurring at remodeler-enriched genes separately from all other genes (Figure 5B).
Figure 5. Directionality of chromatin remodelers.
(A) Composite plot of nucleosome positions at the 5′ ends of genes of either wild type (WT, gray fill) or mutant (green or pink trace).
(B) Line graphs of nucleosome dyad shifts from chromatin remodeler null mutants. The shift is reported as the median distance between a mutant and wild type dyad position for those genes either having (upper panel) or lacking (lower panel) remodeler-bound nucleosomes, as defined in the Methods section. Nucleosome positions are relative to the 5′ NFR, or the terminal nucleosome (TN) at the 3′ end of genes. RSC (Sth1degron) data is from (Hartley and Madhani, 2009).
(C) P-value of the nucleosomal shifts observed in the mutants. Log10 p-values are reported as a heat-map table. White block indicate p-values > 0.01.
See also Figure S2.
Loss of ISW1b (isw1Δ or ioc2Δ), INO80 (ino80Δ or arp5Δ), CHD1 (chd1Δ), or SWI/SNF (snf2Δ) generally resulted in nucleosomal shifts in the 5′ direction for nucleosomes located immediately downstream of 5′ NFRs (Figures 5B, and S2A, with statistical evaluation in panel 5C), indicating that these complexes normally promote nucleosome shifts in the 3′ direction, as was previously shown for Isw1 (Tirosh et al., 2010). The 5′ shifts were substantially more pronounced at remodeler-bound genes, indicating a direct effect (Figure 5B).
In contrast, loss of ISW2 (isw2Δ or itc1Δ) or ISW1a (ioc3Δ) resulted in a shifting of genic nucleosome positions towards the 3′ direction (Figure 5B). Again, the effect was more pronounced at genes bound by these remodelers. Thus, ISW2 and ISW1a, which crosslinked to the +1 nucleosome, normally slide nucleosomes towards the 5′ NFR, as previously reported for ISW2 (Whitehouse et al., 2007).
The observation that nucleosomes downstream of where a remodeler was bound, were also shifted when the remodeler was deleted, is consistent with a packing mechanism of nucleosome organization (Zhang et al., 2011b). According to this model, remodelers play a role in positioning nucleosomes against a barrier. If that barrier was a positioned +1 nucleosome, then movement of the barrier in a remodeler mutant would indirectly involve repositioning of adjacent nucleosomes as well. Those adjacent nucleosomes may be actively repositioned by other remodelers that are present.
The trends observed downstream of the 5′ NFR were reversed upstream of the 5′ NFR (Figure 5B, left side of left panel). Similar trend reversals were found at 3′ NFRs. Thus, the focal point for directionality of remodelers is the NFR at the start and end of genes. ISW1a and ISW2 tend to move nucleosomes toward NFRs, whereas all others move nucleosomes away from NFRs.
In the ioc3Δ (ISW1a-defective) strain, the magnitude of the shift towards the 5′ ends of genes was low, but it increased towards the middle of genes. The rather small shift at the 5′ end might be due to functional redundancy between ISW2 and ISW1a at some genes. We examined this possibility by further separating the Ioc3-enriched +1 nucleosomes into those that were also occupied by Isw2 and those that were not (Figure S2B). The +1 nucleosomes that normally lacked Isw2 had a more prominent shift in the ioc3Δ mutant, which supports the notion of functional redundancy between ISW2 and ISW1a at a subset of genes.
Transient Positioning of Nucleosomes
Nucleosome positions at the start and end of genes may not be in their intrinsically favored positions (Whitehouse and Tsukiyama, 2006; Zhang et al., 2009; Zhang et al., 2011b). Such nucleosomes may be kinetically trapped, whereby they remain in their metastable positions long after the remodeler has dissociated. Alternatively, they may be dynamic, moving rapidly between intrinsically favorable and unfavorable locations. As such, a continuous and perhaps dynamic association of a remodeler would be required to maintain the nucleosome over unfavorable sequences.
To explore these possibilities, we compared the positioning of remodeler-bound nucleosomes (e.g., Ioc3 MNase-ChIP) to the local positioning of all nucleosomes (i.e., H3 MNase-ChIP) at the same position for the same genes in a wild-type strain. The differences between the two are mechanistically informative. For example, Ioc3-bound nucleosomes were shifted more 5′ compared to all nucleosomes at the same position for the same genes (Figure 1B, compare blue/gray alignments). This demonstrates that ISW1a (Ioc3) may be transiently shifting nucleosomes, which then return relatively quickly to their pre-shifted state when ISW1a is not engaged. The same was seen for Isw2, although as expected from the isw2Δ analysis in Figure 3, the 5′ shift was smaller. Arp5 (INO80)-bound nucleosomes were shifted 3′, as expected of the arp5Δ analysis, which suggests that it too acts transiently.
Genome-wide +1 Packing Implicated by the Subnucleosomal Organization of Isw2
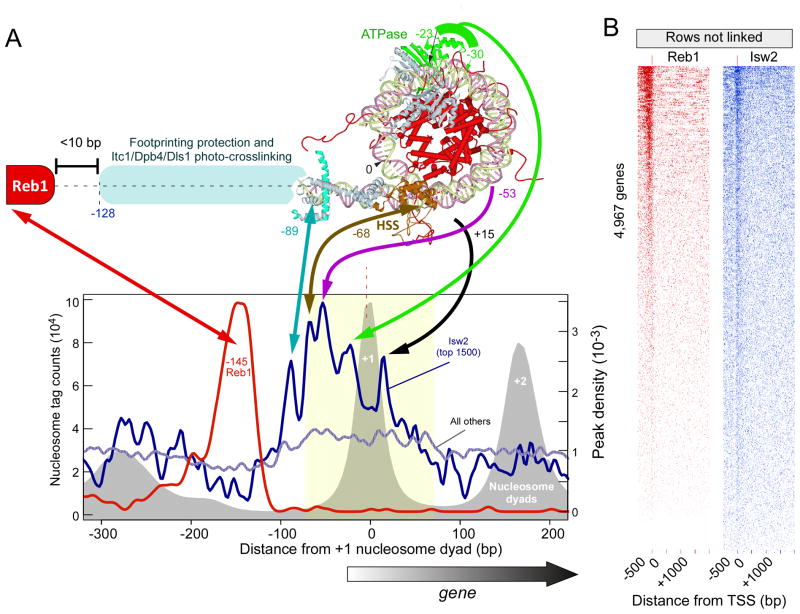
The binding of ISW2 to the +1 nucleosome provides a clear example of position-specific binding and directional re-positioning of nucleosomes, with the functional consequence of occluding the promoter region from transcription (Whitehouse et al., 2007). Still unknown is how ISW2 specifies the +1 nucleosome (and to some extent -1), and how it engages this nucleosome in a directional manner that mechanistically reflects how it biochemically positions nucleosomes. We addressed this problem on a genomic scale by applying a novel ultra-high resolution mapping strategy termed ChIP-exo to map Isw2 locations (Rhee and Pugh, 2011, 2012).
In brief, ChIP-exo applies a 5′-3′ strand-specific exonuclease to a ChIP sample. After deep sequencing to detect the exonuclease stop sites, the location of a precisely-positioned protein across a genome can be determined to within a few bp as a pair of peaks separated by a fixed distance. Fuzzier complexes, as might be expected of ISW2 bound to fuzzy nucleosomes, may produce broader peaks. If the protein is denatured prior to exonuclease treatment, then peak pairs are generated around each crosslinking point, which may be interpretable in light of structural information about the bound complex. Importantly, mapping by ChIP-exo is not affected by the presence of underlying nucleosomal histone-DNA contacts (Rhee and Pugh, 2011, 2012).
As expected, ChIP-exo tags were concentrated around the +1 nucleosome (and to a lesser extent −1). The signal was concentrated within ~100 bp upstream of the +1 nucleosome dyad, part of which corresponds to the core promoter region (graph in Figure 6A). Importantly, five prominent sub-nucleosomal peak-pair midpoints (points of crosslinking) were evident within the averaged Isw2-bound locations. All other detected peak-pairs across the genome failed to display such patterns. We mapped these peaks onto a previously reported crystallographic-based model of the ISW2/nucleosome complex, which was guided by in vitro footprinting and photo-crosslinking experiments with purified ISW2 and homogenous nucleosomes (shown in Figure 6A) (Dang and Bartholomew, 2007). One caveat to the ISW2 structure is that it was computationally assembled using homologous Sulfolobus Rad54 ATPase and Drosophila ISWI-C domain structures, and thus reflects a hypothetical organization that remains to be validated in vivo.
Figure 6. Directional binding of Isw2 to the +1 nucleosome.
(A) Matching of a crystallographic-based model of an Isw2/nucleosome complex (Dang and Bartholomew, 2007) to Isw2 ChIP-exo data. The graph plots the distribution of ChIP-exo crosslinking points (peak-pair midpoints corresponding to exonuclease stop sites) relative to the dyad position of the +1 nucleosome, and orientated such that the nearest TSS is directed to the right. The top 1500 occupied Isw2 peaks were selected, and compared to all others. Also shown is the distribution of Reb1-bound locations (Rhee and Pugh, 2011) for the same collection of genes. The distribution of nucleosome dyads is shown as a gray filled plot.
(B) Most genes contain detectable levels of Reb1 and Isw2. All 4,967 genes having an annotated TSS were aligned by their TSS, and the intensity level and positions of ChIP-exo Reb1 and Isw2 peak-pairs plotted. Genes were sorted by intensity level. The order of genes in the two panels are not the same.
Based on biochemical mapping (and supported by the modeled crystal structure), Isw2 makes three major contacts and one minor contact across approximately 90 bp of nucleosomal and adjoining linker DNA (Kagalwala et al., 2004; Dang et al., 2006). The Isw2 ATPase domain makes contact on one side of the nucleosome dyad, and its HAND/SANT/SLIDE (HSS) domain contacts the opposite side of the nucleosome near the DNA entry/exit point and continues along the same DNA to make contacts as far as 30 bp into the linker region. When the ISW2 Itc1, Dpb4, and Dls1 subunits are considered, biochemical contacts as far as ~130 bp from the nucleosome dyad are made (~70 bp of linker).
Remarkably, ChIP-exo detected each of the biochemically-defined Isw2 contacts (peaks at −23/−30, −53, −68, and −89 bp from the dyad in Figure 6A). Importantly, this not only provided supporting in vivo evidence for the in vitro model of Isw2 specifically bound to a nucleosome, but it also uniquely defines its orientation and organization at the +1 nucleosome on a genomic scale. A fifth and novel contact was detected on the second DNA gyre (at +15 bp ) just below the region of the first gyre to which crosslinking to the presumed HSS domain was detected. Similar patterns were evident around the −1 and genic terminal nucleosomes (Figure 6A, and data not shown), but at lower levels and in an inverted orientation.
The orientation of Isw2 at +1 (and other positions) is entirely consistent with the direction by which ISW2 moves nucleosomes, both in vitro and in vivo, further supporting the interpretation of the ChIP-exo peak locations. ISW2 uses ATP hydrolysis to pull the linker DNA towards the nucleosome core, which propagates the DNA across the histone surface (Langst and Becker, 2001). ISW2 continues this process until a barrier is reached. This model therefore predicts that a barrier should reside immediately upstream to where ISW2 makes linker contacts.
We searched for DNA sequence motifs in and around Isw2-enriched regions. We found strong enrichment of the Reb1 motif to which Reb1 was bound (Figure 6A). The p-value for Reb1 and ISW2 enrichment at the same set of genes, using the enrichment threshold defined in Figure 1, was 10−36. Reb1 is well known to organize nucleosomes (Fedor et al., 1988; Angermayr et al., 2003; Raisner et al., 2005; Hartley and Madhani, 2009), and serves as a polar barrier to transcription and DNA replication (Singh et al., 2010). Remarkably, Reb1 binding was highly focused at a position centered at −145 bp relative to the +1 dyad. Reb1 is expected to cover a region that extends ~6 bp beyond its site (Rhee and Pugh, 2011), which places it <10 bp from the predicted ISW2 edge, taking into account additional contacts that are likely to be made by Itc1 (Kagalwala et al., 2004; Dang et al., 2006). Thus, the combined data presented here suggest a model whereby orientation-specific binding of ISW2 to the +1 nucleosome (and −1 to some extent) moves the +1 nucleosome towards the NFR until it encounters a barrier such as Reb1. With the exception of poly (dA-dT) tracts, we found no other enriched element associated with ISW2 binding.
We took advantage of the high signal-to-noise inherent in ChIP-exo assays to examine the extent to which Isw2 (and Reb1) can be detected at all genes. We were surprised to find that the vast majority of all genes contained detectable levels of these proteins at the precise canonical distance from the TSS (Figure 6B). However, most locations had very low binding levels and thus would not have been detected in other lower sensitivity assays. These binding events do not represent noise since their locations are not random. We suspect that the low binding may be a consequence of transient interactions of Reb1 and ISW2 with their target sites.
DISCUSSION
Coordinated Remodeling through Position-specific Nucleosomal Interactions
A variety of chromatin remodeling complexes are largely responsible for organizing nucleosomes across eukaryotic genomes (Whitehouse et al., 2007; Hartley and Madhani, 2009; Tirosh et al., 2010; Gkikopoulos et al., 2011). Within genes, remodelers organize nucleosomes by using the energy of ATP hydrolysis, possibly packing them into arrays that start at a fixed distance from the TSS (Zhang et al., 2011b), leaving an NFR upstream of the TSS and a 3′ NFR just downstream of the end of genes. Strongly repressed genes may in addition have a nucleosome placed over their promoter.
The genome-wide nucleosomal interactions of remodelers detected here may be separated into three broad groups: 1) those that predominantly interact with +1 nucleosomes (Arp5, Ioc3 and Isw2), 2) those that primarily interact with nucleosomes inside coding regions but are depleted at the +1 nucleosome (Ioc4 and Isw1), and 3) those that interact more broadly with nucleosomes flanking the NFR and into the coding region (Ino80, Rsc8 and Snf2). Thus, remodelers either bind to or are excluded from specific nucleosome positions relative to the start and end of genes.
Our observation that position-specific binding for ISWI-type remodellers is mirrored at the 5′ and 3′ ends of the same genes raises the question of how such binding is coordinated. This would include the maintenance of position specificity relative to nearby NFRs. One intriguing possibility arises from the notion that genes are looped (O’Sullivan et al., 2004; Ansari and Hampsey, 2005). Such looping would place the 5′ and 3′ ends in close proximity, allowing coordinated loading at both ends.
Additional positional specificity may arise through position-specific combinations of histone modifications. It is clear that nucleosome positions within at least the first ~800 bp of gene start sites are distinctly identifiable by combinations of histone marks (Liu et al., 2005; Zhang et al., 2011a), and thus could help remodelers identify specific nucleosome positions. Patterns of H3K36 methylation mirror patterns of Isw1b binding, and thus it is of interest to determine whether the two events are linked. Remodelers are enriched with domains that interact with specific histone modifications. Additionally, sequence-specific factors might contribute toward specificity through the recruitment of remodelers to nucleosome neighborhoods (Hassan et al., 2001; Hassan et al., 2002).
Conceivably, some remodelers might work rather indiscriminately across a genome to enhance nucleosome fluidity. Our observations do not favor that notion since positions-specific interactions were detected. Possible exceptions include Isw1 and Ino80, which show broad distributions. However, these catalytic subunits may be linked to a variety of regulatory subunits that may be nucleosome position specific. We cannot exclude the possibility of nonspecific remodeler interactions that exist transiently across the genome and below the detection threshold, but nonetheless are sufficient to promote widespread nucleosome repositioning and fluidity.
Multiple different remodelers tend to work at the same genes, and so any placement of nucleosomes is likely a net consequence of their coordinated involvement. One consequence of coordinated action is that the transient action of one remodeler may be counteracted by the action of a different remodeler. Our findings support this notion in that only a fraction of the nucleosomes at a given location may be bound by a remodeler, and these nucleosomes often are at positions that are shifted from the bulk population at the same location. Thus, for some remodelers to have a lasting effect on nucleosome positioning, it may be necessary to keep other remodelers away.
The presence of multiple remodelers at the same genes raises questions as to whether they are present at the same time or have a temporal order of binding that is predicated on prior events. Such temporal ordering might occur during the transcription cycle, since transcription elongation-coupled nucleosome eviction is a directional ordered process. However, when transcription is not occurring, then there may exist an ebb and flow of nucleosome repositioning and occupancy that is lubricated by remodelers (Dion et al., 2007). This may not involve a prescribed order of events.
NFRs are Focal Points for Directional Packing of Nucleosomes
Our findings suggest that NFRs are focal points or organizing centers upon which nucleosomes are moved either towards or away from, depending on the remodeler. These focal points create an inversion of directionality on either side of the NFR. Thus, ISW1a and ISW2 remodelers, which are generally repressive towards transcription, move nucleosomes towards NFRs, whereas all other moved nucleosomes away. What comprises a focal point will likely be a subject of deeper investigation, although at least Reb1 and poly (dA:dT) tracts appear to be important contributors (Raisner et al., 2005). We find Reb1 positioned <10 bp from where ISW2 is predicted to contact NFR DNA. Loss of Reb1 or its binding sites results in at least partial collapse of NFRs and a failure to position adjacent nucleosomes (Fedor et al., 1988; Angermayr et al., 2003; Hartley and Madhani, 2009). The placement of a barrier at the 5′ end of genes may help set the register of the +1 nucleosome. Any activity that positions the +1 nucleosome may indirectly cause positioning of adjacent or nearby nucleosomes in the array. This effect would arise from the action of remodelers such as Isw1b and other potential Isw1 remodelers that may adjust nucleosome positioning and spacing relative to the position of the +1 nucleosome.
How Remodelers Might Position and Space Entire Arrays
Several important questions arise from this study. 1) How do remodelers orientate themselves on nucleosomes to establish directional movement? 2) How is the register (distance from TSS) of a nucleosomal array established? 3) How is uniform nucleosome spacing maintained? Analysis of the ISWI-type remodelers provides some insight. The presence of a linker-binding HSS domain plus associated regulatory subunits (Itc1, Dbp4, and Dls1) may constrain the ISW2 complex to only those nucleosomes having an adjacent NFR of ~60 bp or more (Kagalwala et al., 2004). In the case of ISW1a, that limit may be set at approximately 25 bp (Stockdale et al., 2006; Gangaraju and Bartholomew, 2007a; Yamada et al., 2011). In both cases, an NFR on one side of a nucleosome and a short linker on the other side may suffice to also determine the orientation of these remodelers, and thus set the directionality of nucleosome repositioning towards the NFR. Such directionality is entirely consistent with the established biochemical mechanism of ISW2 movement of nucleosomes (Dang et al., 2006).
In the case of ISW1b, its linker length requirement is less defined. Its enrichment at the tightly spaced nucleosome positions +2, +3, and +4 suggests that it has a rather small lower limit, perhaps as low as 15–20 bp (Stockdale et al., 2006; Gangaraju and Bartholomew, 2007a), which represents the canonical linker length in this region and the distance covered by the HSS domain. Since ISW1b moves nucleosomes away from NFRs, one expectation is that ISW1b is orientated in the opposite direction of ISW2 and ISW1a. This raises the question as to what orientates ISW1b. In principle, ISW1b could approach both sides of a nucleosome and elicit bidirectional movement if sufficient linker length exists (Racki et al., 2009; Narlikar, 2010; McKnight et al., 2011). However, if packing of nucleosomes against the 5′ end of genes results in too short of a proximal linker to allow ISW1b entry, then the remodeler might be limited to only the distal accessible side, thereby producing nucleosome movement away from the NFR until a steady-state of bidirectionality is achieved.
Further into the body of the gene where other ISW1-type complexes potentially exist (because neither Ioc4 or Ioc3 are highly enriched there), the linker length is more variable. Without the influence of directional packing, nucleosome placement in the middle of genes may be directed by a combination of remodeler-facilitated stochastic positioning (Kornberg and Stryer, 1988), and underlying sequence-directed preferences (Ioshikhes et al., 2006; Segal et al., 2006).
The +1 Nucleosome as a Gateway to Transcription
How does positioning of nucleosomal arrays regulate transcription? The +1 nucleosome is strikingly unique in character. Compared to all other nucleosomes it is highly enriched with the histone variant H2A.Z (Albert et al., 2007), is highly acetylated, and its position may influence the positions of downstream nucleosomes. The bromodomain factor Bdf1 is highly enriched at the +1 nucleosome due to interactions with resident histone acetylation marks (Koerber et al., 2009). Bdf1 is linked to H2A.Z deposition via interactions with the SWR1 complex (Krogan et al., 2003), and may be involved in TFIID recruitment (Matangkasombut et al., 2000; Sanders et al., 2002).
The +1 nucleosome can be repositioned such that the TSS resides closer to the midpoint of the nucleosome, which is a more repressive location (Schmid et al., 1992; Morillon et al., 2003; Whitehouse and Tsukiyama, 2006; Kelly et al., 2010; Zhang et al., 2011a). Genes that become activated tend to move the +1 nucleosome downstream so that the TSS resides in a more favorable position. Recent studies find that TFIID/Bdf1 engages the +1 nucleosome of most yeast genes of the “housekeeping” class (Rhee and Pugh, 2012). It was also suggested that the +1 nucleosome positions at least the front end of the transcription pre-initiation complex (PIC) at the TSS, with the TATA element positioning the back end. Thus, positioning of a +1 nucleosome upstream of its canonical location may inhibit PIC assembly, whereas canonical positioning may promote PIC assembly.
Remarkably, mutations that decrease nucleosome assembly in the body of genes in the wake of transcription, have dramatically lower effects on the occupancy and positioning of the +1 nucleosome (Batta et al., 2011; Celona et al., 2011). Depletion of nucleosomes downstream of the +1 position negatively impacts transcription, including inhibiting the translocation of RNA polymerase (Pol) II into the body of genes (Batta et al., 2011). In this sense, the +1 nucleosome may serve not only to regulate PIC assembly and the positioning of the TSS, but it may also be a gatekeeper of elongation whereby proper nucleosome arrays in gene bodies may be required to open the gate. Similarly, at the ends of genes, proper nucleosome positioning may be important for transcription termination, disassembly of the elongation complex, and/or anti-sense transcription (Alen et al., 2002; Morillon et al., 2003).
In multicellular eukaryotes, the +1 nucleosome is further downstream of the TSS compared to fungi, and Pol II pauses its elongation at the +1 nucleosome doorstep. An important question to address is whether remodelers “see” such an arrangement as it is seen in yeast. If so, what constitutes the barrier and what measures the distance from the barrier to the +1 nucleosome? These questions may be addressable in vivo using a combination of ChIP-exo and shRNA knockdown to eliminate candidate organizing factors, and examining their effect on nucleosome positions.
The results presented here suggest that remodelers work in concert at subsets of genes through remodeler-specific interactions at select nucleosome positions relative to the start and end of genes. Genomic chIP-exo mapping of Isw2 suggests that at least the ISWI-type remodelers bind nucleosomes in an orientation-specific manner that may be dictated largely by constraints imposed by linker/NFR lengths as well as corresponding linker-binding domain length. This then sets the direction of nucleosome movement.
EXPERIMENTAL PROCEDURES
Strains and Sample Preparation
C-terminally TAP tagged and deletion strains were obtained from Open Biosystems, and were grown in YPD media at 25 C to OD600 ~0.8. MNase ChIP-seq experiments were performed as described (Koerber et al., 2009). Briefly, cells were crosslinked with 1% formaldehyde at 25 C for 15 min. Cells were harvested, disrupted by bead-beating, and chromatin pellets washed extensively with FA lysis buffer (50 mM Hepes pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, and 0.1% Sodium Deoxycholate). Mononucleosomes were solubilized via digestion with MNase to ~80% completion by gel analysis (see Figure S1). Mononucleosomes crosslinked to TAP-tagged factors were immunoprecipitated with IgG sepharose, washed with FA lysis buffer and TEV eluted. Stringent washes were used so that nucleosome isolation depended upon the use of formaldehyde and TAP tags. Mononucleosomes bound to TAP-tagged factors were further purified via calmodulin sepharose. Eluate DNA was subjected to ligation-mediated PCR (LM-PCR) and gel purification of approximately 250 bp nucleosomal libraries. The gel images are representative of at least three biological replicates, and sequencing is representative of at least two biological replicates. Replica data sets were analyzes separately until reproducibility was confirmed, then were combined.
SOLiD Sequence Analysis
SOLiD sequencing tags (35 bp) were mapped to the reference yeast genome (S. cerevisiae 2007-Jan-19 version from SGD) using SHRiMP(v2.1.0) software by allowing 6 mismatches. Gene cluster graphs represent the tag count per 5 bp bin, smoothed with a 3 bin moving average. Reference coordinates for alignments were either TSSs or the dyad location of the terminal nucleosome for each gene. K-means and hierarchical clustering was performed with Cluster and visualization with Treeview (Eisen et al., 1998).
Statistical Analysis
P-values reported in the heat-map were calculated via a chi-squared test assuming a Gaussian distribution of the population. The null hypothesis posits that the genes containing chromatin remodeler-bound nucleosomes are distributed randomly among the 6,226 total genes.
Nucleosome shift Analysis
Nucleosomes whose positions were called as previously described (Jiang and Pugh, 2009a) and differed between mutants and wild type having a t-test p-value lower than 0.05 were regarded as a valid nucleosome shift, as previously described (Tirosh et al., 2010), although we did not impose the previously-described 15 bp minimum shift threshold. Briefly, the t-test was performed by comparing the distribution of read positions of the mutants and wild type around the dyad positions of the respective nucleosome, taking all reads that map to within 73 bp of the dyad position of the respective nucleosomes. Statistically valid (p-value < 0.05) shifted nucleosomes were then aligned back to the TSSs (Figure 5A), parsed into whether those genes did or did not have the remodeler-bound nucleosomes, as defined by the clustering in Figure 1. This was done separately for each remodeler. We then calculated the median shift for those two classes (Figure 5A), and plotted those shifts according to nucleosome position relative to the TSS (Figure 5B). The statistical significance of the median shifts was determined using the Wilcoxon-Mann-Whitney test (Figure 5C).
ChIP-exo mapping of Isw2
ChIP-exo mapping was performed essentially as described (Rhee and Pugh, 2011). Briefly, cells were grown, crosslinked, and disrupted as described above. Chromatin pellets were then sonicated in FA-lysis buffer containing 0.1% SDS, and the supernate subjected to chromatin immunoprecipitation in 0.05% SDS using IgG-dynabeads. After washing the beads, partial library construction and exonuclease digestion was performed on the beads, followed by elution and completion of library construction. Libraries were then amplified by PCR, gel purified, then subjected to deep sequencing.
The 5′ end of mapped tags, representing exonuclease stop sites, were then consolidated into peak calls (sigma = 5, exclusion = 20) using GeneTrack (Albert et al., 2008), then peak pairs were matched when found on opposite strands and 0–100 bp apart in the 3′ direction. Peaks were thresholded to have at least 3 tags. The top 1500 peaks were selected to have their peak pair midpoints plotted (SupplementalDataFile1.xls). Consequently, peak pairs having both peaks in the top 1500 were represented twice. The patterns shown in Fig. 6 were reproducible across multiple replicates.
Supplementary Material
Acknowledgments
We thank Charles V. Meade, David Goffman, Anna Chan, Kasthuri Kannan, and Yunfei Li for assistance in nucleosome preparation, DNA sequencing, and data analysis. This work was funded by NIH grant 5R01HG4160 and The Pennsylvania Department of Health using Tobacco Settlement Funds.
Footnotes
ACCESSION NUMBERS
Sequencing data is available at NCBI Sequence Read Archive under accession number SRA051347.
Supplemental Information includes four figures and three tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Albert I, Wachi S, Jiang C, Pugh BF. GeneTrack - a genomic data processing and visualization framework. Bioinformatics. 2008 doi: 10.1093/bioinformatics/btn119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alen C, Kent NA, Jones HS, O’Sullivan J, Aranda A, Proudfoot NJ. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol Cell. 2002;10:1441–1452. doi: 10.1016/s1097-2765(02)00778-5. [DOI] [PubMed] [Google Scholar]
- Angermayr M, Oechsner U, Bandlow W. Reb1p-dependent DNA bending effects nucleosome positioning and constitutive transcription at the yeast profilin promoter. J Biol Chem. 2003;278:17918–17926. doi: 10.1074/jbc.M301806200. [DOI] [PubMed] [Google Scholar]
- Ansari A, Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 2011;25:2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Celona B, Weiner A, Di Felice F, Mancuso FM, Cesarini E, Rossi RL, Gregory L, Baban D, Rossetti G, Grianti P, et al. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol. 2011;9:e1001086. doi: 10.1371/journal.pbio.1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Dang W, Bartholomew B. Domain architecture of the catalytic subunit in the ISW2-nucleosome complex. Mol Cell Biol. 2007;27:8306–8317. doi: 10.1128/MCB.01351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Kagalwala MN, Bartholomew B. Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol Cell Biol. 2006;26:7388–7396. doi: 10.1128/MCB.01159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor MJ, Lue NF, Kornberg RD. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988;204:109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Bartholomew B. Dependency of ISW1a chromatin remodeling on extranucleosomal DNA. Mol Cell Biol. 2007a;27:3217–3225. doi: 10.1128/MCB.01731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju VK, Bartholomew B. Mechanisms of ATP dependent chromatin remodeling. Mutat Res. 2007b;618:3–17. doi: 10.1016/j.mrfmmm.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen-Hughes T. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science. 2011;333:1758–1760. doi: 10.1126/science.1206097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- Hota SK, Bartholomew B. Diversity of operation in ATP-dependent chromatin remodelers. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshikhes IP, Albert I, Zanton SJ, Pugh BF. Nucleosome positions predicted through comparative genomics. Nat Genet. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009a;10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009b;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 2004;23:2092–2104. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassabov SR, Zhang B, Persinger J, Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- Kelly TK, Miranda TB, Liang G, Berman BP, Lin JC, Tanay A, Jones PA. H2A.Z maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes. Mol Cell. 2010;39:901–911. doi: 10.1016/j.molcel.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber RT, Rhee HS, Jiang C, Pugh BF. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol Cell. 2009;35:889–902. doi: 10.1016/j.molcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, Stryer L. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 1988;16:6677–6690. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Langst G, Becker PB. ISWI induces nucleosome sliding on nicked DNA. Mol Cell. 2001;8:1085–1092. doi: 10.1016/s1097-2765(01)00397-5. [DOI] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-Nucleosome Mapping of Histone Modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matangkasombut O, Buratowski RM, Swilling NW, Buratowski S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- McKnight JN, Jenkins KR, Nodelman IM, Escobar T, Bowman GD. Extranucleosomal DNA Binding Directs Nucleosome Sliding By Chd1. Mol Cell Biol. 2011 doi: 10.1128/MCB.05735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J, Morillon A. ISWI complexes in Saccharomyces cerevisiae. Biochim Biophys Acta. 2004;1677:100–112. doi: 10.1016/j.bbaexp.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Montel F, Castelnovo M, Menoni H, Angelov D, Dimitrov S, Faivre-Moskalenko C. RSC remodeling of oligo-nucleosomes: an atomic force microscopy study. Nucleic Acids Res. 2011;39:2571–2579. doi: 10.1093/nar/gkq1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, O’Sullivan J, Kent N, Proudfoot N, Mellor J. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell. 2003;115:425–435. doi: 10.1016/s0092-8674(03)00880-8. [DOI] [PubMed] [Google Scholar]
- Narlikar GJ. A proposal for kinetic proof reading by ISWI family chromatin remodeling motors. Curr Opin Chem Biol. 2010;14:660–665. doi: 10.1016/j.cbpa.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Racki LR, Yang JG, Naber N, Partensky PD, Acevedo A, Purcell TJ, Cooke R, Cheng Y, Narlikar GJ. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature. 2009;462:1016–1021. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive Genome-wide Protein-DNA Interactions Detected at Single-Nucleotide Resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012 doi: 10.1038/nature10799. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol. 2002;22:4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A, Fascher KD, Horz W. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell. 1992;71:853–864. doi: 10.1016/0092-8674(92)90560-y. [DOI] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Sabatinos S, Forsburg S, Bastia D. Regulation of replication termination by Reb1 protein-mediated action at a distance. Cell. 2010;142:868–878. doi: 10.1016/j.cell.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem. 2006;281:16279–16288. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Sigal N, Barkai N. Widespread remodeling of mid-coding sequence nucleosomes by Isw1. Genome Biol. 2010;11:R49. doi: 10.1186/gb-2010-11-5-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama M, Sabri A, Bartholomew B. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol Cell Biol. 2011;31:662–673. doi: 10.1128/MCB.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary JC, Jr, Gangaraju VK, Qin J, Landel CC, Kooperberg C, Bartholomew B, Tsukiyama T. Yeast Isw1p forms two separable complexes in vivo. Mol Cell Biol. 2003;23:80–91. doi: 10.1128/MCB.23.1.80-91.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Stockdale C, Flaus A, Szczelkun MD, Owen-Hughes T. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol Cell Biol. 2003;23:1935–1945. doi: 10.1128/MCB.23.6.1935-1945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Tsukiyama T. Antagonistic forces that position nucleosomes in vivo. Nat Struct Mol Biol. 2006;13:633–640. doi: 10.1038/nsmb1111. [DOI] [PubMed] [Google Scholar]
- Yamada K, Frouws TD, Angst B, Fitzgerald DJ, DeLuca C, Schimmele K, Sargent DF, Richmond TJ. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature. 2011;472:448–453. doi: 10.1038/nature09947. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ma H, Pugh BF. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res. 2011a doi: 10.1101/gr.117465.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science. 2011b;332:977–980. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Persinger J, Bartholomew B. Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2. Mol Cell Biol. 2004;24:10047–10057. doi: 10.1128/MCB.24.22.10047-10057.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.