The pioneering experiments of Emerson and Arnold (1) gave rise to the concept of a photosynthetic unit in which many antenna pigments could efficiently pass excited state energy on to a reaction center (RC) where the first photochemistry occurred. The experimental data indicated that the antenna must contain a large set of chlorophyll molecules relative to the reaction center and implied a close geometric relationship between the antenna molecules and the RC that would allow for efficient delivery of excited state energy resulting from a quantum of light absorbed anywhere in the antenna chlorophyll. With the development of membrane biochemistry came the realization that all chlorophyll and bacteriochlorophyll molecules were specifically bound to proteins that held them in a fixed relationship to each other (2–4). In photosynthetic bacteria, the antenna pigments were found to be specifically bound in protein complexes referred to as light-harvesting (LH) complexes whereas the RC cofactors were bound to a separate protein. The isolation and crystallization of these complexes has led to the determination of the structure of the bacterial RC at high resolution (5–7), of a bacterial light-harvesting complex (LH2) at high resolution (8, 9), and of Photosystem I of Synechococcus elongatus (which contains the RC and core LH) at intermediate resolution (10). In addition, electron cryomicroscopy has provided lower resolution structures for a major light-harvesting complex (LHCII) of oxygenic organisms (11), the core light-harvesting complex of Rhodospirillum rubrum (12), and Photosystem II RC of spinach, which also contained part of the core LH (13). As a result of these studies, the early concept of a photosynthetic unit consisting of a kind of “funnel” of antenna pigments that could pass excitation energy to an RC has been transformed into the framework of specific integral membrane protein complexes that form a two-dimensional mosaic in which the RC is at the center and is surrounded by LH complexes. A “core” complex contains the RC and a set of light-harvesting pigments (LH1) that are intimately and specifically associated with each other and the RC. Among the challenges now being addressed in photosynthetic bacteria research is to describe the structure of this supramolecular photosynthetic core complex and its relationship to the bc1 complex, and to determine the structure-function relationships that are critical for the capture of light energy.
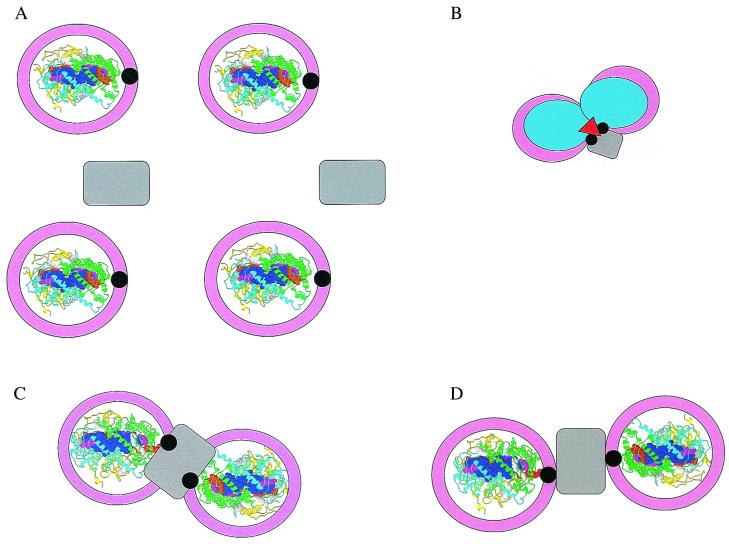
A variety of geometries have been proposed for core complexes. These models are mostly based on the concept of an RC surrounded by LH1, and, if present, more peripheral LH complexes are placed outside the core complex. In addition to satisfying low resolution electron microscopic images (12, 14), such cyclic arrangement of LH1 would also explain the circular degeneracy observed in fluorescence polarization experiments and other spectral properties (15). However, a complication arises in models that propose a ring of LH1 around RC (Fig. 1A). Since it has been well established that reducing equivalents that accumulate at the QB site of the RC must reach the bc1 complex to complete cyclic electron transport and to complete the generation of an electrochemical gradient of protons (16), a ring of LH1 encircling the RC would block the transfer of reducing equivalents to the bc1 complex assumed to be elsewhere in the membrane (Fig. 1A). At this juncture, a protein found in Rhodobacter species, referred to as PufX (encoded by the pufx gene), has been shown to facilitate the necessary transport of reducing equivalents from QBH2 of the RC to the bc1 complex (17–19). It has been speculated that the PufX protein interacts with LH1 and/or the RC to provide a pathway for QBH2 to diffuse from the RC and to be replaced by an oxidized ubiquinone from the Q-pool in the membrane (20, 21). However, experimental evidence to show that PufX has an effect on LH1 and/or RC structure has been lacking. In new experimental work, Frese et al. in this issue of PNAS (22) report measurements of polarized absorption spectra of oriented photosynthetic membranes obtained from two mutants of Rhodobacter sphaeroides, one containing a functional PufX gene and one not. A striking difference in LD spectra was found in the region of 800 nm. In membranes of the pufx- mutant, the orientation of the RC about an axis normal to the membrane appeared to be random while a high degree of orientation was observed in membranes containing Puf X. These data provide the first evidence that PufX induces the formation of a long-range regular array of RC. When the electron microscopic data of Jungas et al. (23) and the biochemical studies of Recchia et al. (24) and Francia et al. (25) are also taken into account, a direct involvement of PufX in the LH1/RC structure is implied that results in a specific orientation of the RC in an LH1 ring. Thus, PufX is thought to play a key structural role not only in the organization of LH1 about the RC, but also in the long-distance arrangement of these core complexes in the membrane, at least in the case of this LH2− mutant of Rb. sphaeroides.
Figure 1.
(A) Model of the core complex in which each RC (Rasmol structure of the Rb. sphaeroides RC, Protein Data Bank (RCSB) accession no. 1aij) is surrounded by a closed ring of LH1 (pink) interrupted by PufX (black) and interacting with the RC at the QB site (orange space-fill tail of QB). The bc1 complex (gray) is elsewhere in the membrane. In the Rasmol presentation, the RC is viewed from the extracellular side of the membrane, the H-polypeptide is yellow, the L-polypeptide is cyan, the M-polypeptide is green, bacteriochlorophyll are blue, bacteriopheophytin are magenta, and QA and QB are orange. (B) Model of an intimate RC dimer (light blue) in which PufX and another possible component (red) link LH1 with the RC and cytochrome b (28, 29). Other color assignments are as for A. (C) Model of an RC-cytochrome b-RC complex in which two RC are intimately associated with a bc1 complex linked by PufX (23). Colors are as in A. (D) Model of a core complex-cytochrome b-core complex in which the linkage to cytochrome b is through PufX (22). Colors are as in A.
A criticism might be raised that the mutants studied by Jungas et al. (23) and Frese et al. (22) are unique in that they exhibit an unusual membranous tube showing long-range organized structures, that they were grown under dark, partial aerobic conditions rather than photosynthetically, and that they do not contain LH2. Even so, the experimental results, along with those of Francia et al. (25), implicate PufX as intimately involved in the organization of LH1 and the RC. And since structure-function relationships of core complexes of photosynthetic bacteria seem highly conserved across species, it is likely that these results will be of general importance. It would be of great interest if similar linear absorption measurements were extended to other photosynthetic bacteria such as Rs. rubrum and Rhodopseudomonas viridis since a puf x gene has not been found in these species.
In the experiments of Recchia et al. (24), the PufX proteins from Rhodobacter capsulatus and Rb. sphaeroides were isolated and examined for their effect on in vitro reconstitution of LH1. Each protein inhibited LH1 formation in proportion to its ratio to the LH1 α-polypeptide. To explain these results, it was proposed that PufX binds to the α-polypeptide, perhaps in the presence of bacteriochlorophyll (Bchl), and prevents its participation in further oligomerization. In the association of LH1 with the RC, it is likely that the α-polypeptide is on the inside of the ring as in LH2 (8, 9). Thus, PufX may interact with both an α-polypeptide of LH1 and with the RC presumably at a location near the QB site, thus playing the dual role of a linker and causing interruption of a homo-oligomeric ring of LH1.
One might think that ring formation requires specificity in the α- and β-polypeptides of the B820 subunit of LH1 and LH2. However, it should be noted that B850- and B870-type complexes can be readily formed with a single α- or β-polypeptide that has been substantially shortened at its N terminus (26, 27). These complexes exhibited both the absorption and CD properties similar to those of LH2/LH1 complexes. Thus, from these observations, it can be concluded that it is the specific geometry of the two Bchl bound in a dimeric structure that drives circular oligomerization to achieve the overlapping ring of Bchl. Small changes in the relative orientation of some of the Bchl pairs in a ring might be expected to give rise to somewhat ellipsoidal, rather than strictly circular geometries without sacrificing the energy transfer function of the complex. This would perhaps allow a more intimate fit of LH1 around the RC than is indicated in the models of Fig. 1 A, C, and D.
A dimeric structure for the bacterial RC has been suggested to explain several experimental results, first based on electrochemical and quinone extraction experiments (28, 29), then on the basis of cytochrome c interaction with the RC (30) and more recently on the basis of isolation of RC dimers apparently requiring PufX to be formed (25) and the electron microscopic results of Jungas et al. (23). In each of these cases, the interaction between two RC in the dimer was considered to be more intimate (see Fig. B and C) than in the model suggested by Frese et al. (22). In the latter case, an LH1 ring surrounds each RC and contains one PufX that interfaces with a single shared bc1 complex (also represented in Fig. 1D). The results obtained by Francia et al. are of particular interest because they have provided evidence for in vivo dimerization of RC in the presence of PufX and because they determined the mole ratio of PufX/RC to be one (25). In their work, no dimeric RC were observed in preparations from PufX− mutant membranes.
Many questions remain to be addressed regarding the structure of the core complexes of photosynthetic bacteria. For example, why has a pufx gene been found only in the puf operon of Rhodobacter species and not in the puf operon of other bacteria such as Rs. rubrum and Rs. viridis? Since the role of PufX seems crucial for function in Rb. capsulatus and Rb. sphaeroides, is an x-type gene located elsewhere in other bacteria, or does some other protein fulfill this role? Are there other proteins that are part of the core structure but have not yet been identified (e.g., Fig. 1B)? It should be kept in mind that the structure-function relationships for core complexes of all known photosynthetic bacteria are highly homologous. As indicated above, the RC are highly similar, all have a core LH with highly similar biochemical and spectral properties, all exhibit very nearly the same ratio of Bchl of LH1:RC, and all RC are tightly coupled to the bc1 complex to achieve cyclic electron transport and develop an electrochemical gradient of protons. Isn't it compelling to expect that all will also share a similar mechanism for the fundamentally important process of moving reducing equivalents from QBH2 of the RC to the bc1 complex?
Other questions might also be raised. Does the PufX protein play a regulatory as well as a structural role such that it controls the expression of another protein? Does PufX interact with the bc1 complex to establish an even larger supramolecular structure, as is implied by three of the four models shown in Fig. 1? When considering the very high structural similarity between bacterial RC and PSII and PSI RC, why do the core LH complexes appear to be so different (10, 13)?
It may be safely concluded that the growing knowledge of structure and the many experimental tools now in place will enable these supramolecular structures to be carefully probed. Future research in this area should be able to elucidate how these remarkable converters of solar energy work.
Footnotes
See companion article on page 5197.
References
- 1.Emerson R. Annu Rev Plant Physiol. 1958;9:1–24. [Google Scholar]
- 2.Reed D W, Clayton R K. Biochem Biophys Res Commun. 1968;30:471–475. doi: 10.1016/0006-291x(68)90075-2. [DOI] [PubMed] [Google Scholar]
- 3.Okamura M Y, Feher G, Nelson N. In: Photosynthesis. Govindjee, editor. Vol. 1. New York: Academic; 1982. pp. 195–272. [Google Scholar]
- 4.Zuber H, Cogdell R J. Adv Photosynth. 1995;2:316–348. [Google Scholar]
- 5.Deisenhofer J, Epp O, Miki F, Huber R, Michel H. J Mol Biol. 1984;180:380–398. doi: 10.1016/s0022-2836(84)80011-x. [DOI] [PubMed] [Google Scholar]
- 6.Allen J P, Feher G, Yeates T O, Rees D C, Deisenhofer J, Michel H, Huber R. Proc Natl Acad Sci USA. 1986;83:8589–8593. doi: 10.1073/pnas.83.22.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang C H, Tiede D M, Tang J, Smith U, Norris J, Schiffer M. FEBS Lett. 1986;205:82–86. doi: 10.1016/0014-5793(86)80870-5. [DOI] [PubMed] [Google Scholar]
- 8.McDermott G, Prince S M, Freer A A, Hawthornthwaite-Lawless A M, Papiz M Z, Cogdell R J, Isaacs N W. Nature (London) 1995;374:517–521. doi: 10.1016/s0969-2126(96)00050-0. [DOI] [PubMed] [Google Scholar]
- 9.Koepke J, Hu X, Muenke C, Schulten K, Michel H. Structure (London) 1996;4:581–597. doi: 10.1016/s0969-2126(96)00063-9. [DOI] [PubMed] [Google Scholar]
- 10.Schubert W-D, Klukas O, Krauss N, Saenger W, Fromme P, Witt H T. J Mol Biol. 1997;272:741–769. doi: 10.1006/jmbi.1997.1269. [DOI] [PubMed] [Google Scholar]
- 11.Kühlbrandt W, Wang D N, Fujiyoshi Y. Nature (London) 1994;367:614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- 12.Karrasch S, Bullough P A, Ghosh R. EMBO J. 1995;14:631–638. doi: 10.1002/j.1460-2075.1995.tb07041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee K-H, Morris E P, Barber J, Kühlbrandt W. Nature (London) 1998;396:283–286. doi: 10.1038/24421. [DOI] [PubMed] [Google Scholar]
- 14.Jay F, Lambillotte M, Stark W, Mühlethaler K. EMBO J. 1984;3:773–776. doi: 10.1002/j.1460-2075.1984.tb01883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Grondelle R, Dekker J P, Gillbro T, Sundstrom V. Biochim Biophys Acta. 1994;1187:1–65. [Google Scholar]
- 16.Gennis R B, Barquera B, Hacker B, Van Doren S R, Arnaud S, Crofts A R, Davidson E, Gray K A, Daldal F. J Bioenerg Biomembr. 1993;25:195–209. doi: 10.1007/BF00762582. [DOI] [PubMed] [Google Scholar]
- 17.Farchaus J W, Gruenberg H, Oesterhelt D. J Bacteriol. 1990;172:977–985. doi: 10.1128/jb.172.2.977-985.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farchaus J W, Gruenberg H, Gray K A, Wachveitl J, DeHoff B, Kaplan S, Oesterhelt D. In: Molecular Biology of Membrane-Bound Complexes in Phototropic Bacteria. Drews G, Dawes E A, editors. New York: Plenum; 1990. pp. 65–76. [Google Scholar]
- 19.Lilburn T G, Haith C E, Prince R C, Beatty J T. Biochim Biophys Acta. 1992;1100:160–170. doi: 10.1016/0005-2728(92)90077-f. [DOI] [PubMed] [Google Scholar]
- 20.Barz W P, Verméglio A, Francia F, Venturoli G, Melandri B A, Oesterhelt D. Biochemistry. 1995;34:15248–15258. doi: 10.1021/bi00046a033. [DOI] [PubMed] [Google Scholar]
- 21.Lilburn T G, Prince R C, Beatty J T. J Bacteriol. 1995;177:4593–4600. doi: 10.1128/jb.177.16.4593-4600.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frese R N, Olsen J D, Branvall R, Westerhuis W H J, Hunter C N, van Grondelle R. Proc Natl Acad Sci USA. 2000;97:5197–5202. doi: 10.1073/pnas.090083797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jungas C, Ranck J-L, Rigaud J-L, Joliot P, Verméglio A. EMBO J. 1999;18:534–542. doi: 10.1093/emboj/18.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Recchia P A, Davis C M, Lilburn T G, Beatty J T, Parkes-Loach P S, Hunter C N, Loach P A. Biochemistry. 1998;37:11055–11063. doi: 10.1021/bi980657l. [DOI] [PubMed] [Google Scholar]
- 25.Francia F, Wang J, Venturoli G, Melandri B, A, Barz W P, Oesterhelt D. Biochemistry. 1999;38:6834–6845. doi: 10.1021/bi982891h. [DOI] [PubMed] [Google Scholar]
- 26.Meadows K A, Iida K, Kazuichi T, Recchia P A, Heller B A, Antonio B, Nango M, Loach P. Biochemistry. 1995;34:1559–1574. doi: 10.1021/bi00005a012. [DOI] [PubMed] [Google Scholar]
- 27.Meadows K A, Parkes-Loach P S, Kehoe J W, Loach P A. Biochemistry. 1998;37:3411–3417. doi: 10.1021/bi972269+. [DOI] [PubMed] [Google Scholar]
- 28.Loach P A. In: Progress in Bioorganic Chemistry. Kaiser E T, Kezdy F J, editors. Vol. 4. New York: Wiley Interscience; 1976. pp. 89–192. [Google Scholar]
- 29.Morrison L, Runquist J, Loach P A. Photochem Photobiol. 1977;25:73–84. doi: 10.1111/j.1751-1097.1977.tb07426.x. [DOI] [PubMed] [Google Scholar]
- 30.Verméglio A, Joliot P, Joliot A. Biochim Biophys Acta. 1993;1183:352–360. [Google Scholar]