SUMMARY
The mitochondrial protein MAVS (also known as IPS-1, VISA, CARDIF) interacts with RLR (RIG-I-like receptors) to induce type 1 interferon (IFN-I) during viral infection. NLRX1 is a mitochondrial NLR (nucleotide-binding, leucine-rich repeats containing) protein that attenuates MAVS-RLR signaling. Using Nlrx1−/− cells we confirmed NLRX1 attenuated IFN-I production, but additionally promoted autophagy during viral infection. This dual function of NLRX1 paralleled the previously described functions of the autophagy-related proteins Atg5-Atg12, but NLRX1 did not associate with Atg5-Atg12. High throughput quantitative mass spectrometry and endogenous protein-protein interaction revealed an NLRX1-interacting partner, mitochondrial Tu translation elongation factor (TUFM). TUFM interacted with Atg5-Atg12 and Atg16L1, and has similar functions as NLRX1 by inhibiting RLR-induced IFN-I but promoting autophagy. In the absence of NLRX1, increased IFN-I and decreased autophagy provide an advantage for host defense against vesicular stomatitis virus. This study establishes a link between an NLR protein and the viral-induced autophagic machinery via an intermediary partner, TUFM.
INTRODUCTION
The mitochondrion is a coordinating site for several key host responses against viral infection including apoptosis and type I IFN (IFN-I) signaling (Brenner and Mak, 2009; Lei et al., 2009; Rehwinkel and Reis e Sousa, 2010). Upon infection with a RNA virus, virus-related RNA species are recognized by RIG-I (retinoic-acid inducible gene I)-like receptors (RLRs), including RIG-I and MDA5 (melanoma differentiation associated gene 5) (Rehwinkel and Reis e Sousa, 2010). The 5’-ppp moiety of cytosolic dsRNA is the molecular signature that activates RIG-I (Hornung et al., 2006; Myong et al., 2009), and RIG-I deficiency leads to compromised IFN-I in response to vesicular stomatitis virus (VSV), Sendai virus (SeV) and Newcastle disease virus (NDV) (Kato et al., 2006). The helicase domain drives the unwinding of dsRNA with 3’-terminal overhang via its ATPase activity (Takahasi et al., 2008). Upon activation, RIG-I binds to its adaptor, the mitochondrial antiviral signaling protein (MAVS, also referred to as IPS-1, VISA or Cardif) via homotypic CARD interaction to activate IFN-I by facilitating the nuclear translocation of the transcription factors NF-κB and IRF3 (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). Similar to MAVS, another adaptor protein STING (also referred to as MITA, ERIS and MPYS) is ubiquitously expressed and is indispensable for inducing IFN-I upon RIG-I activation (Ishikawa and Barber, 2008; Ishikawa et al., 2009; Sun et al., 2009; Zhong et al., 2008; Zhong et al., 2009). Overexpression of STING is less potent than MAVS in the activation of IRF3 (Zhong et al., 2008), and it may employ a separate mechanism via its association with components of the endoplasmic reticulum translocon-associated protein (TRAP) complex to modulate RLR signaling (Ishikawa and Barber, 2008).
Due to the potential detrimental effects of IFN-I, several molecular machineries keep the RLR pathway in-check. A unique member of the nucleotide-binding domain (NBD) and leucine-rich-repeats (LRR)-containing proteins (NLRs), NLRX1, resides in the mitochondria and attenuates the activation of MAVS by binding to its CARD domain, possibly precluding its engagement with RIG-I (Moore et al., 2008). The NLR family was first discovered by bioinformatic data mining based on the sequence of CIITA (Harton et al., 2002). NLRs have diverse biological functions including formation of the caspase-1 activating inflammasome protein complex, modulation of host anti-pathogen response, regulation of gene transcription and cytokine, chemokine and defensin production (Ting et al., 2010). Emerging evidence suggests that NLRs actively participate in antiviral signaling pathways. Among these, NLRX1 is the first NLR found to negatively impact IFN-I production (Allen et al., 2011; Moore et al., 2008). In addition to NLR-based complexes, the autophagy-related protein conjugate, Atg5-Atg12, has also been shown to regulate the RLR signaling pathway. The absence of autophagy, which causes the degradation of misfolded proteins and cytoplasmic organelles to recycle nutrients (Sanjuan and Green, 2008; Virgin and Levine, 2009), results in significantly higher RLR signaling, leading to more IFN-I production in mouse embryonic fibroblasts (MEFs) (Jounai et al., 2007; Ke and Chen, 2011; Tal et al., 2009). One study showed that Atg5-deficient cells and Atg7-deficient cells, in which the Atg5-Atg12 conjugation is impeded, exhibit enhanced IFN-I response to virus (Jounai et al., 2007). Another report showed that the absence of autophagy leads to amplification of the RLR pathway by causing the elevation of MAVS, and by elevating reactive oxygen species (ROS) which increases RLR (Tal et al., 2009). However, the role of autophagy in RLR signaling appears to be cell type specific since such inhibitory effects were not seen in plasmacytoid dendritic cells (Lee et al., 2007). Nonetheless, the relationship between the two regulatory functions of RLR in IFN-I and autophagic signaling remains to be studied and how NLRs might integrate these two pathways has not been addressed.
This study employs quantitative proteomics approaches and uncovered a regulatory mitochondrial protein complex comprised of NLRX1 and mitochondrial Tu translation elongation factor (TUFM). This complex regulates virus-induced IFN-I activation and autophagy by engaging RIG-I and the autophagy-related proteins, Atg5-Atg12 conjugate and Atg16L1. This shows that a complex comprised of NLRX1, TUFM, RIG-I, Atg5-Atg12 and Atg16L1 controls two crucial anti-viral functions: IFN-I induction and autophagy.
RESULTS
NLRX1 inhibits IFN-I signaling in MEFs
Previously we showed that NLRX1 attenuates IFN-I production (Allen et al., 2011). However, the MEFs were generated from different litters. To confirm the role of NLRX1 in modulating IFN-I signaling, we generated littermate-controlled MEFs with or without the Nlrx1 gene by intercrossing heterozygous Nlrx1+/− mice to minimize genetic differences between the MEFs. WT and Nlrx1−/− MEFs were cultured from 13.5 day embryos; and the lack of Nlrx1 transcripts in Nlrx1−/− MEFs was confirmed by RT-PCR (Figure S1A). When MEFs from both genotypes were challenged with VSV, a deficiency in NLRX1 resulted in enhanced cytokines production including IL-6, TNF-α and IFN-I both in transcript levels (Figure S1B) and protein levels (Figure S1C). These findings confirmed recent studies from our group (Allen et al., 2011; Moore et al., 2008). Another group also showed that NLRX1 negatively regulates inflammatory cytokine production in response to viral infection or toll-like receptor activation (Xia et al., 2011).
TUFM associates with NLRX1
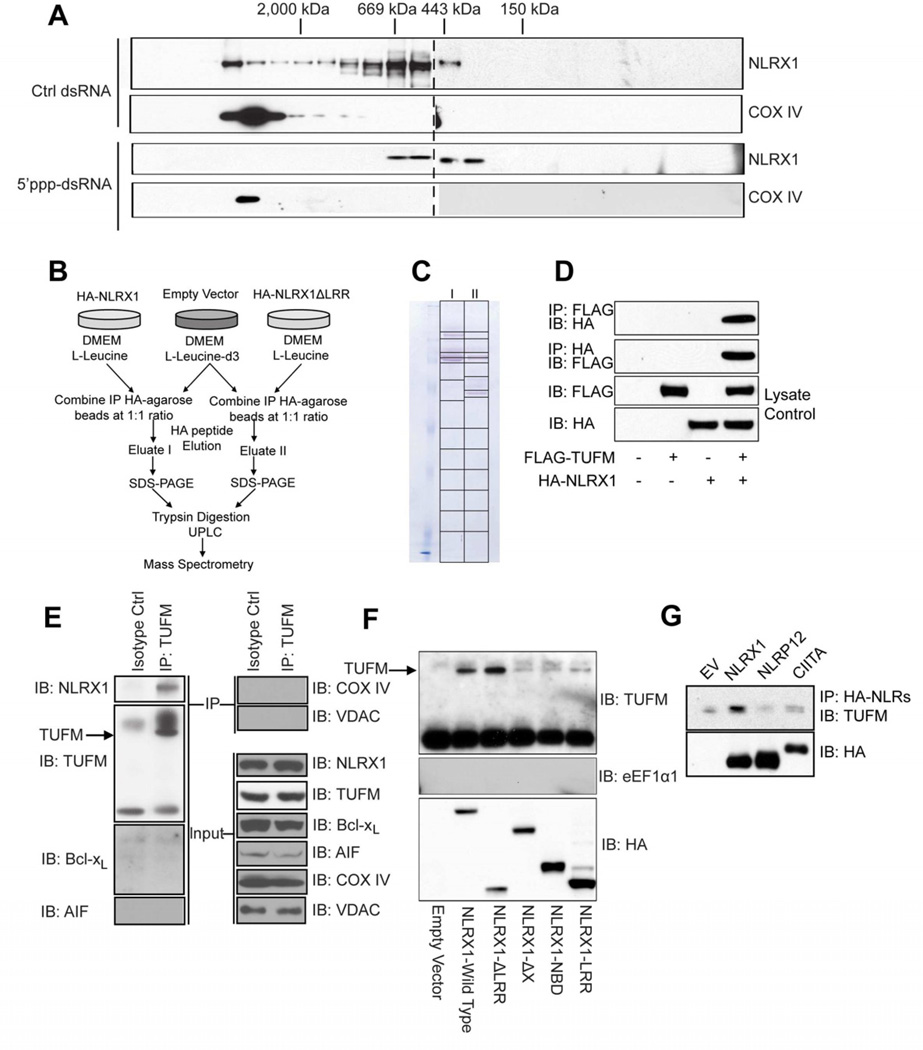
We previously found that NLRX1 interacts with MAVS to impede the latter’s interaction with RIG-I at a basal state, thus suggesting that NLRX1 might form an active inhibitory complex at the unstimulated state (Allen et al., 2011; Moore et al., 2008). To further explore this possibility, we employed size exclusion chromatography and found that NLRX1 is most abundant in a higher molecular mass during the resting state, while it resides in smaller molecular complexes upon RLR activation by the RLR ligand, 5’ppp-dsRNA (Figure 1A, upper panels). As an internal control, the mitochondrial protein COX IV-containing complex did not display an altered size profile in response to 5’-ppp dsRNA challenge (Figure 1A, lower panels).
Figure 1. TUFM interacts with NLRX1.
(A) Five million HEK293T cells were transfected with 5’ppp-dsRNA or control dsRNA, cells were harvested and homogenized in hypotonic lysis buffer 16 hr post-transfection. Total cell protein extractions were then separated by size exclusion chromatography, and collected fractions were subjected to western blotting against NLRX1 and COX IV. (B) Scheme of AACT/SILAC mass spectrometry identification of NLRX1-interacting partners. (C) Immunoprecipitated proteins were eluted by HA-peptide and separated by SDS-PAGE, the gel was continuously sliced as indicated for further ultra-high performance liquid chromatography and mass spectrometry analyses. (D) HA-tagged NLRX1 was co-transfected with either empty vector or FLAG-tagged TUFM. Cells were lysed and immunoprecipitated proteins were blotted for either FLAG-TUFM or HA-NLRX1 as indicated. (E) HEK293T cells were lysed and immunoprecipitated with either isotype control antibody or anti-TUFM and blotted for NLRX1, TUFM, Bcl-xL, AIF, COX IV and VDAC. (F) Full-length NLRX1 or its truncation mutants were expressed in HEK293T cells, and protein samples were blotted for endogenous TUFM and eEF1α1. (G) Over-expressed NLRX1, NLRP12 and CIITA were immunoprecipitated from HEK293T cells, and blotted for endogenous TUFM. In A, one of three independent experiments is shown. In each of D-G, the results are representative of two to four independent experiments.
To identify additional molecules which might interact with NLRX1 and participate in NLRX1-mediated immune effects, we employed the amino acid-coded mass tagging (AACT, also known as SILAC)-assisted quantitative mass spectrometry to identify NLRX1-interacting proteins (Chen et al., 2000; Zhu et al., 2002) (Figure 1A). The N-terminal domains of NLRs are considered to mediate protein-protein interaction while LRRs are considered inhibitory (Moore et al., 2008; Ye and Ting, 2008). Hence, we constructed vectors encoding full-length NLRX1 and a mutant lacking the LRR domain, and used these as baits to identify interacting partners. Eluted proteins from both groups (Eluates I and II) (Figure 1B) were separately fractionated by SDS-PAGE (Figure 1C) followed by high-performance liquid chromatography and mass spectrometry. A subcellular compartment localization analysis of the NLRX1 interactome revealed that a high percentage of interacting partners are mitochondrial proteins (Figure S1D). Among these, peptide profiles that matched the mitochondrial Tu translation elongation factor (TUFM) with ion scores of >39 were identified in both eluates (Figure S1E–1H). This indicates with >95% certainty that these peptides represent TUFM (Table S1).
To further validate these findings, reciprocal co-immunoprecipitation experiments were performed to show interaction between ectopically expressed NLRX1 and TUFM (Figure 1D). Most importantly, endogenous TUFM co-immunoprecipitated with endogenous NLRX1 (Figure 1E). This interaction appears to be specific, since TUFM did not co-immunoprecipitate with multiple mitochondrial proteins with various sub-mitochondrial locations, including Bcl-xL (mitochondrial outer membrane), apoptosis-inducing factor (AIF, mitochondrial intermembrane space), cytochrome c oxidase (COX) IV (mitochondrial inner membrane) and voltage-dependent anion channel (VDAC, mitochondrial outer membrane) (Figure 1E). NLRX1-TUFM interaction requires the N-terminus of NLRX1 as its deletion (NLRX1-ΔX) abolished this interaction (Figure 1F). Deletion of the LRR domain (NLRX1-ΔLRR) slightly increased this interaction while constructs expressing NBD or LRR alone (NLRX1-NBD, NLRX1-LRR) did not interact with TUFM. As negative controls, NLRX1 did not bind to the cytoplasmic homolog of TUFM, eEF1α1 (Figure 1F). Furthermore, TUFM did not co-precipitate with other NLRs such as NLRP12 and CIITA when compared to an empty vector (EV) control (Figure 1G).
TUFM potently inhibits RIG-I signaling
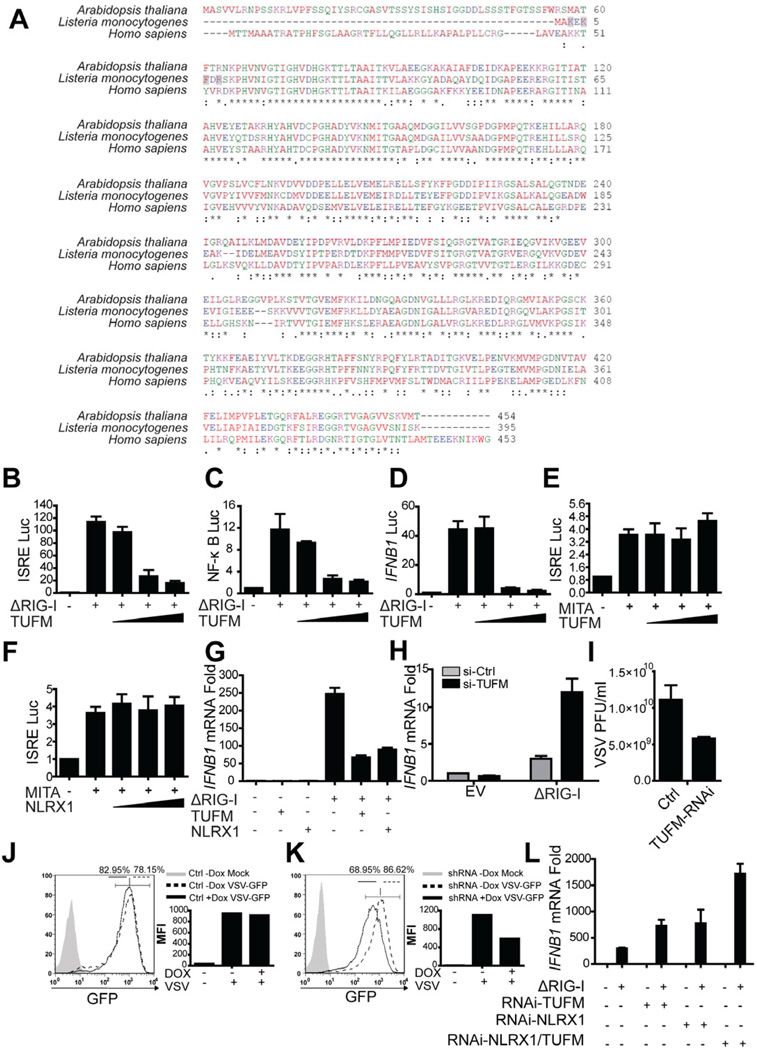
Similar to NLRX1 (Moore et al., 2008), TUFM is ubiquitously expressed in multiple tissue and cell types (Figure S2A–B). NLRX1 is a highly conserved protein, thus we also evaluated the evolutionary conservation of TUFM. TUFM is conserved from humans to its bacterial counterpart. The bacterial elongation factor Tu (EF-Tu) serves as a pathogen-associated molecular pattern (PAMP) in plant cells to elicit immune activation (Kunze et al., 2004; Zipfel et al., 2006). However, despite this general sequence homology, the key activating sequence of bacterial EF-Tu KxKFxR no longer exists in the human or A. thaliana counterparts; instead the N-terminus of TUFM has undergone drastic changes (Figure 2A).
Figure 2. TUFM inhibits RIG-I mediated type I IFN production.
(A) The protein sequences of human TUFM (Accession: NP_003312), mitochondrial EF-Tu in A. thaliana (Accession: NP_192202) and EF-Tu in L. monocytogenes (Accession: YP_002759310). (B-F) HEK293T cells were transfected with ΔRIG-I or MITA/STING expression plasmid plus luciferase reporter plasmids for ISRE, NF-κB or IFNB1 activation. FLAG-TUFM plasmid was titrated into the system and cells were lysed 24 hr post-transfection for luciferase assays. (G) ΔRIG-I was co-transfected with either TUFM or NLRX1 into HEK293T cells; and cells from all groups were harvested for real time PCR analysis 24 hr post-transfection. (H) Endogenous TUFM was partially reduced by transfecting a pool of 4 targeting siRNA into the cells; ΔRIG-I was transfected to induce IFNB1 mRNA as measured by RT-PCR. (I) HEK293T cells with reduced TUFM expression and control cells were challenged with VSV, and plaque assay was performed to assess the viral titers. (J) HEK293T cells were transduced with control lentiviruses and infected with VSV-GFP (MOI of 0.1) for 16 hr and analyzed by flow cytometry for GFP as a marker for VSV. (K) HEK293T cells transduced with sh-TUFM were treated and analyzed as described in (I). (L) HEK293T cells transduced with scrambled control shRNA or sh-NLRX1 were treated with control or siRNA pool targeting TUFM. Cells were then harvested 16 hr post transfection of either empty vector control or ΔRIG-I plamid. IFNB1 mRNA abundance was analyzed by RT-PCR. Results of all panels are representative of three independent experiments.
Given the interaction of NLRX1 with TUFM and previous findings that NLRX1 attenuates MAVS and RIG-I induced IFN-I production and NF-κB signaling (Allen et al., 2011; Moore et al., 2008; Xia et al., 2011), we analyzed the role of TUFM in these same pathways. Similar to NLRX1, TUFM inhibited ISRE-, NF-κB-dependent and IFNB1 promoter activation by ΔRIG-I in a dose-dependent fashion (Figure 2B–D). IFN-I signaling in response to cytosolic viral PAMPs is also regulated by the adaptor MITA (STING) (Ishikawa and Barber, 2008; Ishikawa et al., 2009; Zhong et al., 2008). However, neither TUFM nor NLRX1 affected ISRE activation by MITA, indicating specificity of these two proteins in the RIG-I pathway (Figure 2E–F). In addition to reporter assays, TUFM inhibited ΔRIG-I-induced IFNB1 mRNA production, similar to NLRX1 (Figure 2G). RNA interference targeting of TUFM (si-TUFM) partially reduced TUFM expression (Figure S2C) and increased IFNB1 mRNA induction by ΔRIG-I (Figure 2H). Reduction of TUFM expression also resulted in lower viral plaque forming units (PFU) compared to control cells (Figure 2I). As an additional approach to assess the IFN-I-regulatory function of TUFM, GFP-expressing VSV was used to infect HEK293T cells stably transduced with a doxycycline-inducible shRNA lentivirus (Figure S2D). In the control group, doxycycline did not affect the percentage of infected cells and viral infection as assayed by the GFP-positive population and mean fluorescence intensity (MFI) (Figure 2J). By contrast, induction of TUFM shRNA reduced both signals (Figure 2K). These results indicate that TUFM is similar to NLRX1 in attenuating RLR signaling. Reducing the expression of both NLRX1 and TUFM further resulted in enhanced IFNB1 mRNA (Figure 2L).
NLRX1 is essential for VSV-induced autophagy in MEFs
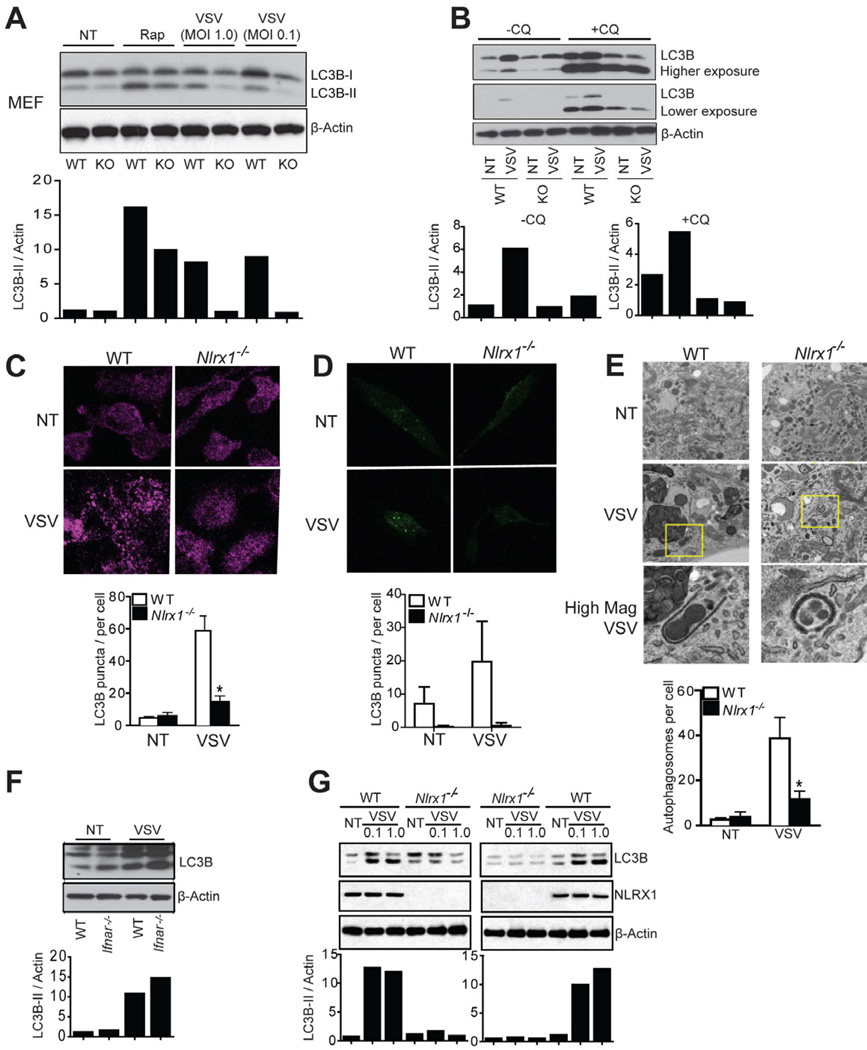
Another key host response that is elicited by virus is autophagy. In addition to IFN-I induction, VSV is known to induce autophagy (Jounai et al., 2007; Shelly et al., 2009) during which the light chain 3 (LC3) protein is converted from a cytosolic form (LC3B-I) to a LC3B-lipidated, membrane-associated form (LC3B-II) and displays a punctate appearance (Kabeya et al., 2000; Mizushima et al., 2010). To study the role of NLRX1 in autophagy, WT and Nlrx1−/− cells were inoculated with VSV. Autophagosome formation was measured by LC3B conversion by immunoblot, punctate appearance by immunofluorescence staining, and direct observation of autophagosomes by transmission electron microscopy (TEM) (Klionsky et al., 2008). In WT MEFs, VSV infection increased LC3B-II level relative to β-actin (Figure 3A). Two widely used lysosomal protease inhibitors, chloroquine (Figure 3B) and bafilomycin A1 (Figure S3), did not reverse the reduced amount of LC3B-II in VSV-infected Nlrx1−/− MEFs compared to control MEFs. This suggests that decreased autophagic activity in Nlrx1−/− MEFs was due to decreased autophagosome synthesis, and not an increased degradation of autophagic substrates. The inclusion of a lower exposure in Figure 3B is necessary since chloroquine treatment caused increased and over-exposed LC3B signals. VSV infection promoted the punctated staining of endogenous LC3B (Figure 3C) and exogenously expressed GFP-LC3B (Figure 3D) and increased number of double-membrane bound autophagosomes (Figure 3E), as noted by others (Jounai et al., 2007; Shelly et al., 2009). All of these were dramatically decreased in Nlrx1−/− MEFs (Figure 3A–E). Rapamycin, an inhibitor of the mTOR signaling and an inducer of autophagy, resulted in a modest reduction in LC3B-II in Nlrx1−/− compared to WT cells (Figure 3A).
Figure 3. NLRX1 is essential for VSV-induced autophagy in MEFs.
(A) WT and Nlrx1−/− MEFs were infected with VSV for 12 hr, or treated with rapamycin for 2 hr. Protein samples were immunoblotted for LC3B. Densitometry analysis to quantify ratio of LC3B-II to β-actin is shown at the bottom. (B) WT and Nlrx1−/− MEFs were infected with VSV (MOI of 0.1) for 12 hr in the absence or presence chloroquine (20 µM). Immunoblotting for LC3B was performed and densitometry analysis as described in (A) is shown at the bottom. (C) WT and Nlrx1−/− MEFs were infected with VSV (MOI of 0.1), fixed and stained for endogenous LC3B 12 hr post-infection and examined by confocal imaging. Quantitation of autophagosomes was performed by counting LC3B puncta in 50 cells (C, bottom). (D) WT and Nlrx1−/− MEFs were transduced with LC3B-GFP expression vector for 24 hr, and then infected with VSV (MOI of 0.1) for 12 hr. Cells were fixed and examined by confocal imaging. Quantitation of autophagosomes was performed by counting LC3B puncta in 20 cells (D, bottom). (E) WT and Nlrx1−/− MEFs were infected with VSV (MOI of 0.1) for 12 hr and examined by transmission electron microscopy for autophagosomes. Quantitation is based on counting the number of autophagosomes in 10 cells per treatment (E, bottom). (F) WT and Ifnar−/− MEFs were infected with VSV (MOI of 0.1) for 12 hr. Protein samples were immunoblotted for LC3B. Densitometry analysis as described in (A) is shown at the bottom. (G) WT and Nlrx1−/− MEFs were respectively placed in the upper and lower chamber (left panel) or vice versa (right panel) in the transwell culture system. Cells were infected with VSV for 12 hr. Protein samples were immunoblotted for LC3B. Densitometry analysis as described in (A) is shown at the bottom. The results are representative of three independent experiments. Values in Figure C–E are expressed as mean ± s.d. * P < 0.05, versus controls.
Since NLRX1 is an attenuator of several inflammatory cytokines, Nlrx1−/− MEFs is known to produce increased proinflammatory cytokines such as IFN-I, TNF-α and IL-6 (Figure S1B–C) (Allen et al., 2011; Xia et al., 2011). To determine if the defective VSV-induced autophagy observed in Nlrx1−/− MEFs is caused by a reduced level of IFN-I, MEFs with a deficiency in IFN-α receptor (Ifnar−/−), a common receptor for IFN-I, was tested. WT and Ifnar1−/− MEFs showed a similarly increase in LC3B-II amounts upon VSV infection, indicating that VSV-induced autophagy is independent of IFN-I signaling (Figure 3F). We also tested the possibility that difference in the amounts of multiple cytokines controlled by NLRX1 may cause defective autophagy in Nlrx1−/− MEFs. To address this, WT and Nlrx1−/− MEFs were respectively cultured in the upper and lower chamber (Figure 3G, left panel), or vice versa (Figure 3G, right panel), in a transwell system where the culture milieu is shared. VSV-infected Nlrx1−/− MEFs still exhibited a greatly decreased LC3B-II level relative to β-actin compared to WT MEFs in both setups (Figure 3G). Therefore, defective autophagy in Nlrx1−/− MEFs is not due to extrinsic alterations in soluble mediators, but rather due to a cell-intrinsic mechanism.
NLRX1 affects virus-induced autophagy in peritoneal macrophages
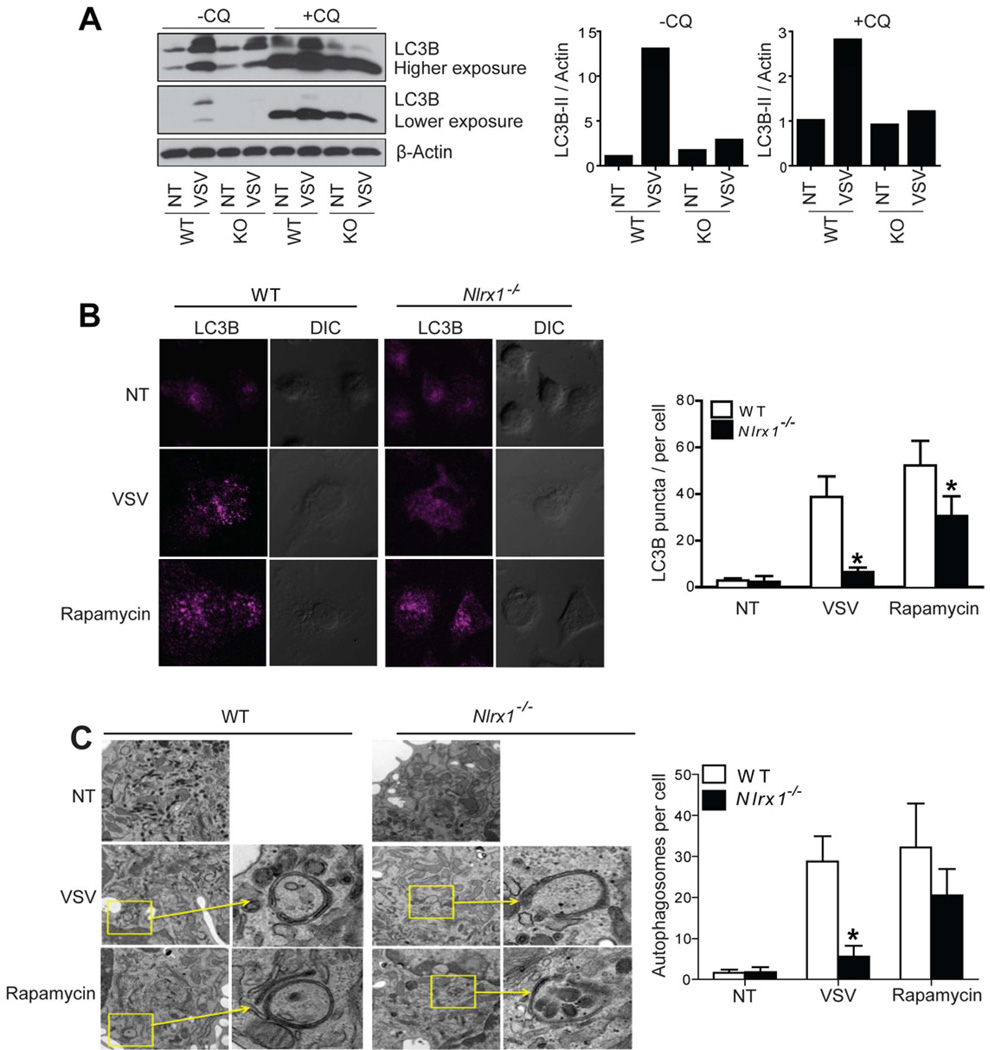
We next explored the role of NLRX1 in autophagy in a primary immune cell type, peritoneal macrophages. Similar to MEFs, VSV-infected Nlrx1−/− peritoneal macrophages showed decreased autophagy markers when compared to similarly treated WT controls. These observations include decreased LC3B-II : β-actin ratio in the presence or absence of chroloquine (Figure 4A), decreased endogenous LC3B punctate staining (Figure 4B), and less autophagosomes (Figure 4C).
Figure 4. NLRX1 is essential for VSV-induced autophagy in peritoneal macrophages.
(A) Naïve peritoneal macrophages were isolated from WT or Nlrx1−/− (KO, knockout) mice and infected with VSV for 12 hr in the presence or absence of chloroquine (20 µM). Protein samples were immunoblotted for LC3B. Densitometry analysis to quantify ratio of LC3B-II to β-actin was shown at the right. (B and C) Naïve peritoneal macrophages isolated from WT or Nlrx1−/− mice were infected with VSV (MOI of 0.1) for 12 hr. Cells were fixed and stained for endogenous LC3B (B, left). Quantitation of autophagosomes was performed by counting LC3B puncta in 50 cells (B, right). Cells were fixed and examined by transmission electron microscopy for autophagosomes (C left). Quantitation is based on counting the number of autophagosomes in 10 cells per treatment (C, right). The results are representative of two independent experiments. Values in Figure B and C are expressed as mean ± s.d. * P < 0.05, versus controls.
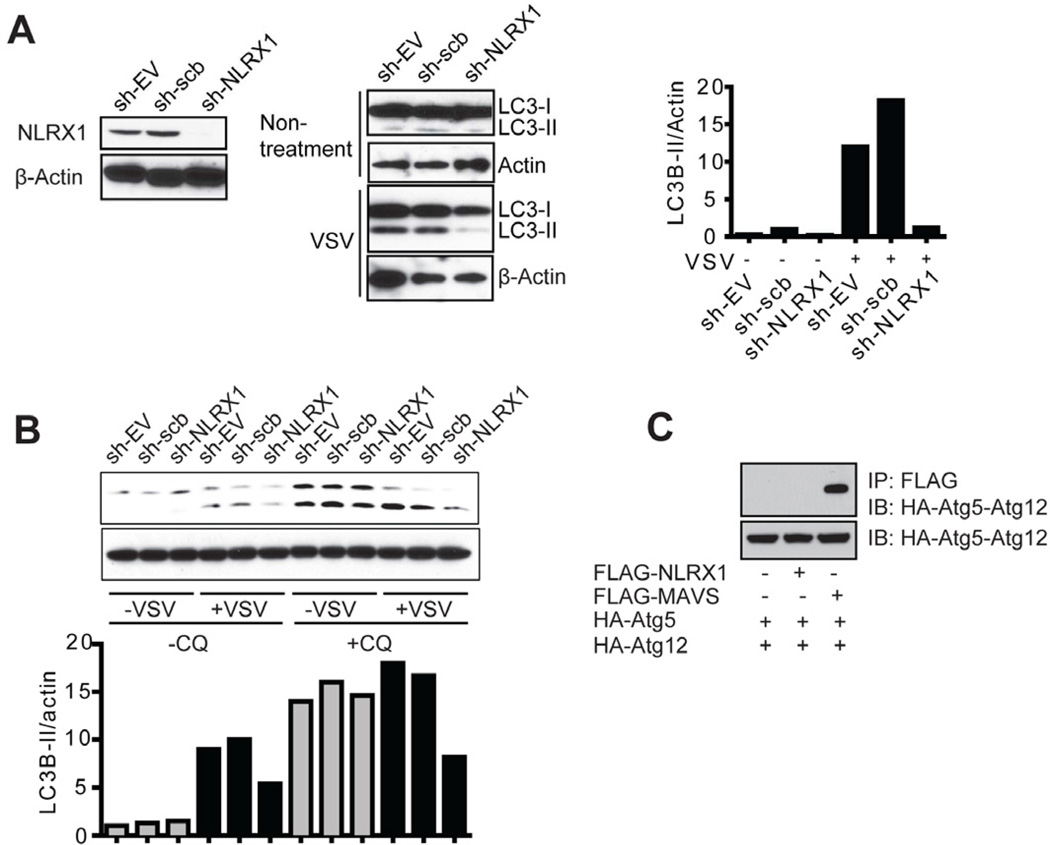
To extend these findings to human cells, lentiviral vector harboring shRNA targeting human NLRX1 and its controls were produced. HEK293T cells with sh-NLRX1 construct showed reduced endogenous NLRX1 expression and reduced LC3B-II : β-actin ratio compared to controls in the presence or absence or chloroquine (Figure 5A and 5B). The association of the Atg5-Atg12 conjugate with MAVS has been shown to inhibit MAVS-mediated IFN-I production (Jounai et al., 2007). Thus we explored the interaction of Atg5-Atg12 with NLRX1, but did to detect an association under stringent lysis and wash conditions (Figure 5C). This led us to hypothesize that an additional protein exists to engage both IFN-I and autophagy pathways and we explored if TUFM is such an intermediary.
Figure 5. NLRX1 is essential for VSV-induced autophagy in human cells.
(A) HEK293T cells were transfected with control lentiviral sh-EV, sh-scrambled or sh-NLRX1 (left). Cells were infected with VSV (MOI of 0.1) and harvested 12 hr post-infection for LC3B immunoblot (middle). Densitometry analysis to quantify ratio of LC3B-II to β-actin is shown at the right. (B) HEK293T cells transfected with sh-EV, sh-scrambled or sh-NLRX1 were infected with VSV (MOI of 0.1) in the absence or presence of chloroquine (20 µM). Cells were harvested 12 hr post-infection for LC3B immunoblot. Densitometry analysis as described in (A) is shown at the bottom. (C) FLAG-tagged NLRX1 or MAVS and HA-tagged Atg5, Atg12 expression plasmids were transfected into HEK293T cells. Cell extracts were harvested 24 hr post-transfection and subjected to co-immunoprecipitation as indicated. Results of all experiments are representative of at least 2–3 repeats.
TUFM associates with Atg5-Atg12 conjugate
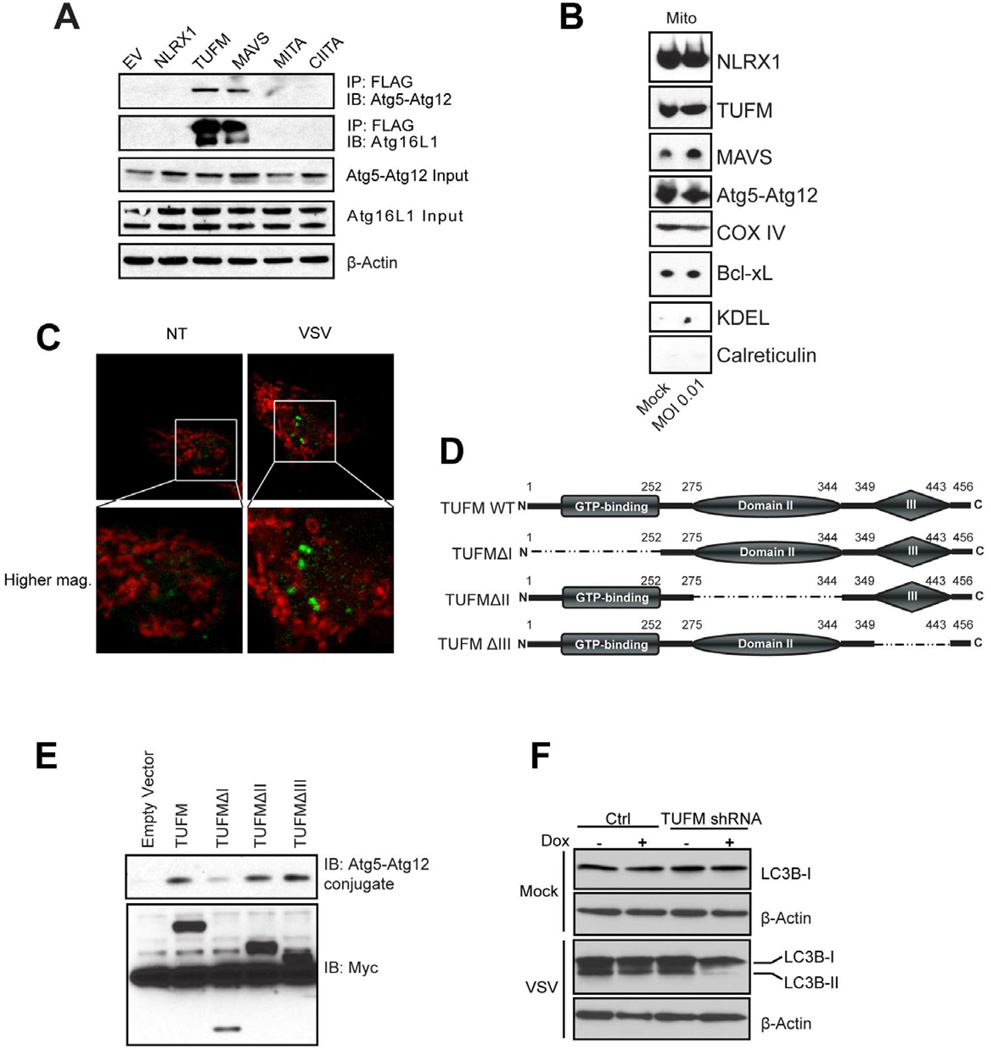
To test the hypothesis that TUFM engages with both NLRX1 and Atg5-Atg12 conjugate, we performed a hemi-endogenous co-immunoprecipitation analysis. This revealed strong interaction of Atg5-Atg12 with TUFM but not with NLRX1, MITA or CIITA (Figure 6A). As a positive control, MAVS also interacted with Atg5-Atg12 as reported (Jounai et al., 2007). TUFM and MAVS also associate with Atg16L1, an essential autophagy partner of Atg5-Atg12 (Levine et al., 2011).
Figure 6. TUFM associates with the Atg5-Atg12 conjugate.
(A) HEK293T cells were transfected with empty vector or FLAG-tagged NLRX1, TUFM, MAVS, MITA/STING or CIITA. Cell lysates were immunoprecipitated with anti-FLAG M2 affinity gel and then blotted for Atg5 or Atg16L1. The immunoblot of Atg5 represents the Atg5-Atg12 conjugate form. (B) Crude mitochondria were isolated from cells untreated or infected with VSV using the protocol described in Figure S4. All samples were subjected to SDS-PAGE and immunoblotting. (C) HEK293T cells were transfected with LC3B-GFP expression vector. After VSV infection (MOI of 0.1) for 12 hr, cells were fixed and stained with MitoTracker® Deep Red. (D) Domain structures of TUFM and truncation mutants. (E) FLAG-tagged TUFM wildtype and domain truncation mutants were co-transfected with HA-Atg5 and HA-Atg12 into HEK293T cells. Cell extracts were immunoprecipitated with anti-FLAG and immuno-blotted for HA-Atg5-Atg12 conjugate. (F) HEK293T cells transduced with tetracycline-inducible shRNA for TUFM and or control shRNA were infected with VSV (MOI of 0.1) and harvested for LC3B immunoblot. Results of all experiments are representative of at least 2–3 repeats.
A previous study has shown that Atg5-Atg12 can be transiently localized to the mitochondria (Hailey et al., 2010). If so, a model can be evoked where MAVS, NLRX1, TUFM and Atg5-Atg12 association takes place within the mitochondria. To assess this, we prepared mitochondria-enriched fractions using the described protocol (Figure S4). In these fractions, we detected NLRX1, TUFM, MAVS and Atg5-Atg12 conjugate, but not the ER protein calreticulin or the ER marker detected by anti-KDEL (Figure 6B). As positive controls, these fractions were enriched for mitochondrial COX IV and Bcl-xL. We further tested if LC3B is recruited to the mitochondria after VSV infection. Ectopically overexpressed LC3B shows punctate appearance after VSV infection, but does not colocalize with the mitochondria identified by MitoTracker staining, suggesting that autophagy of mitochondria (mitophagy) was not occurring under these test conditions (Figure 6C) although further analysis is needed to address this issue fully.
To map the essential domains for the Atg5-Atg12 conjugate recruitment, TUFM truncation mutants were generated (Figure 6D). The N-terminal domain I of TUFM was required for optimal interaction with Atg5-Atg12, while domains II and III were not as critical (Figure 6E). To explore the physiological significance of such interaction, we utilized the TUFM stable knockdown cells described above to investigate if TUFM also affects autophagy. Lentiviral vector transduced HEK293T cells were challenged with VSV, which induced LC3B-II formation. However, cells with sh-TUFM displayed significantly reduced LC3B-II (Figure 6F). This indicates that TUFM, similar to NLRX1, also regulates viral-induced autophagy.
Autophagy and IFN-I cooperatively regulate VSV replication
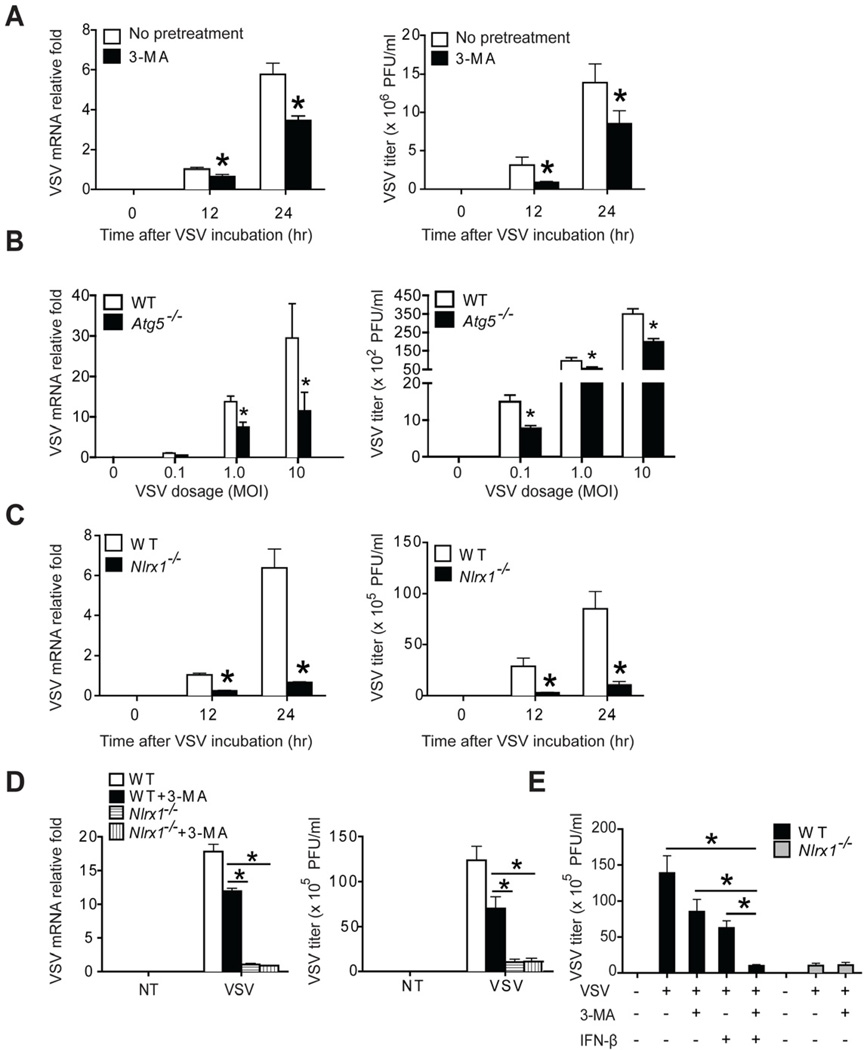
Thus far, we have demonstrated that NLRX1 and TUFM regulated autophagy and IFN-I production during VSV infection. An important question is the biological effects of IFN-I and autophagy on virus replication in mammalian cells. Although the anti-viral effect of IFN-I has been well established, the role of autophagy in viral replication is less established. Both pro- and anti-viral effects of autophagy have been proposed depending on the virus and host cells. For example, dengue virus use autophagy to obtain essential nutrients and macromolecules to support its replication (Heaton and Randall, 2010), while coxsackievirus B3 requires autophagy for optimal infection and pathogenesis (Alirezaei et al., 2012). Other studies show that autophagy enhances viral clearance (Orvedahl et al., 2010; Shelly et al., 2009). Therefore, we sought to determine whether autophagy promotes or inhibits VSV replication. 3-Methyladenine (3-MA), a widely used autophagy inhibitor of class III phosphoinositide 3-kinase (PI3K), moderately but significantly decreased VSV mRNA expression (Figure 7A, left panel) and VSV particles in the supernatant (Figure 7A, right panel), suggesting that VSV might utilize the host autophagy pathway to facilitate its replication in mammalian cells. Furthermore, macrophages with deficiency in autophagy molecule Atg5 (Atg5−/−) show similarly decreased VSV mRNA expression (Figure 7B, left panel) and VSV particles in the supernatant (Figure 7B, right panel) that are statistically significant. Reduced virus replication observed in 3-MA-treated cells or Atg5−/− cells are in agreement with the observation that Nlrx1−/− cells, which have defective autophagy, showed a dramatic repression of virus replication (Figure 7C). However, defective autophagy could not fully explain VSV replication defect in Nlrx1−/− cells, since VSV mRNA level and VSV titers in WT MEFs treated with 3-MA were still significantly higher than those in Nlrx1−/− cells (Figure 7D). Based on the higher production of IFN-I by viral-infected Nlrx1−/− cells [Figure S1B–C; (Allen et al., 2011)], we hypothesized that defective autophagy and enhanced IFN-I might act cooperatively to inhibit VSV replication in Nlrx1−/− cells. To this end, the combination of 3-MA treatment and the addition of recombinant mouse IFN-β (600 pg/ml) to WT MEF at a similar level as that produced by VSV-infected Nlrx1−/− MEF (Figure S1C) decreased VSV titer to the level observed in Nlrx1−/− MEFs (Figure 7E). This experiment suggests that both reduced autophagy and enhanced IFN-I can lead to better control of VSV replication.
Figure 7. NLRX1-mediated IFN-I inhibition and autophagy enhancement cooperatively promotes virus replication.
(A) WT MEFs were infected with VSV (MOI of 0.1) for 12 or 24 hr in the absence or presence of autophagy inhibitor, 3-methyladenine (3-MA, 10 mM). VSV replication was determined by measuring VSV transcripts by RT-PCR (A, left) or virus titer by standard plaque assay (A, right). (B) WT and Atg5−/− bone marrow-derived macrophages were infected with VSV with indicated dosage for 24 hr. VSV replication was determined as described in (A). (C) WT and Nlrx1−/− MEFs were infected with VSV (MOI of 0.1) for 12 or 24 hr. VSV replication was determined as described in (A). (D) WT and Nlrx1−/− MEFs were infected with VSV (MOI of 0.1) for 24 hr in the absence or presence of 3-MA. VSV replication was determined as described in (A). (E) WT and Nlrx1−/− MEFs were treated as indicated. Exogenous IFN-β (600 pg/ml) is determined by the difference in IFN-β produced by WT versus Nlrx1−/− MEFs in Figure S1C. Values are expressed as mean ± s.d. * P < 0.05, versus controls.
DISCUSSION
Our previous analysis showed that NLRX1 is a negative regulator of the MAVS-RIG-I induced IFN-I response (Allen et al., 2011; Moore et al., 2008). In the current study, we showed that NLRX1 dually regulated IFN-I and autophagy through the engagement of a mitochondrial protein, TUFM. TUFM associated with NLRX1 and the Atg5-Atg12 conjugate, serving as an intermediary between NLRX1 and Atg5-Atg12. Similar to NLRX1, TUFM inhibited NF-κB-, ISRE-dependent and IFNB1 promoter activities in a dose-dependent manner and the over-expression of TUFM suppressed IFNB1 mRNA transcription. Reduction of both NLRX1 and TUFM resulted in enhanced IFN-I activation, thus demonstrating a partnership between NLRX1 and TUFM to control host anti-viral responses.
Previous work has shown the association of multiple checkpoint proteins with MAVS, since MAVS is found to be in a supramolecular complex in the quiescent state, migrates into a lower molecular weight complex when cells are activated (Yasukawa et al., 2009). This suggests that during viral activation, MAVS is released from these molecular brake, presumably so that it can engage the RLR protein that are themselves activated by the binding of viral RNAs. This report confirms that NLRX1 resides in a larger molecular complex in the quiescent state but is in a smaller molecular mass in response to cytosolic 5’ppp-dsRNA treatment. This led us to identify TUFM as an NLRX1-interacting partner. Functional analysis of NLRX1 and TUFM indicates that they phenocopy each other. Thus TUFM also serves to attenuate RIG-I dependent IFN-I production and promotes viral-induced autophagy. Autophagy has been increasingly appreciated as a central mechanism for modulating host innate antiviral responses. Several proteins are found to dually control IFN-I and autophagy. For example, the Atg5-Atg12 conjugate are found to regulate the RLR signaling pathway and negatively impact IFN-I activation (Jounai et al., 2007; Tal et al., 2009). In MEFs, the absence of Atg5 results in enhanced IFN-I partially because of the accumulation of intracellular ROS due to a deficiency of autophagy (Tal et al., 2009). Additionally, the Atg5-Atg12 conjugate has been shown to intercalate between RIG-I and MAVS, abolishing RLR-induced IFN-I gene activation (Jounai et al., 2007). Another protein that has been found to dually regulate autophagy and IFN-I is Mfn2. Mfn2 is a mitochondrial outer membrane GTPase and regulates mitochondrial fusion together with its close homologue Mfn1 (Chan, 2006). Recent evidence suggests that Mfn2 is needed for autophagosome formation (Hailey et al., 2010), whereas Mfn2−/− cells exhibit enhanced IFN-I production (Yasukawa et al., 2009). Interestingly, Mfn2 also interacts with MAVS to preclude its association with RLR, providing a plausible mechanism for how it interferes with the RLR-MAVS signaling complex (Yasukawa et al., 2009). Thus the downregulation of IFN-I and induction of autophagy appear to be mediated by a group of proteins with dual functions, including NLRX1 and TUFM described here. It is noteworthy that while a previous study of VSV in Drosophila showed that autophagy is an anti-viral host response (Shelly et al., 2009), this study shows that autophagy enhances viral transcript and titer. This is likely attributed to species or cell type differences. Their paper used the Drosophila melanogaster Schneider 2 cell line (which has features of a macrophage-like lineage) and hemocytes for their in vitro work, while our functional studies were performed in fibroblasts. In fact autophagy has been implicated in facilitating the replication of several viruses, such as Encephalomyocarditis virus and Chikungunya virus, in mammalian fibroblastic cell lines (Krejbich-Trotot et al., 2011; Zhang et al., 2011).
Although the immune function of TUFM has not demonstrated before, its bacterial homologue EF-Tu is found across the bacterial kingdom and is known to function as a PAMP in plants (Kunze et al., 2004; Zipfel et al., 2006). The immune-eliciting activity of EF-Tu resides in its N-terminal 12 amino acids (Kunze et al., 2004). It is possible that bacterial EF-Tu is incorporated into host cells and gradually evolves to become TUFM, while its function evolved from immune-activation to immune-suppression. A comparison of EF-Tu and TUFM shows that the N-terminal immune-activating sequence of EF-Tu is lacking in TUFM, and a mitochondria-targeting sequence is found in the mammalian homologue. In addition to immune modulation, TUFM has also been implicated in protein translation elongation and biosynthesis, oncogenesis, oxidative phosphorylation and protein quality control (Bhargava et al., 2004; Suzuki et al., 2007; Valente et al., 2007; Wells et al., 1995). More investigations are necessary to elucidate if the diverse functions of TUFM are inter-related.
In summary, previous studies have shown a central role for Atg5-Atg12 in autophagy induction and IFN-I inhibition. This work identifies a mitochondrial protein interaction between NLRX1 and TUFM that coordinately controls IFN-I and autophagy through association of the latter with Atg5-Atg12 and Atg16L1. By recruiting Atg5-Atg12 and NLRX1, TUFM serves as a nodal checkpoint of the RIG-I-MAVS axis. These data expand our understanding of the players involved in controlling diverse mitochondria-based anti-viral responses.
EXPERIMENTAL PROCEDURES
Cell culture
Nlrx1−/− MEFs and their littermate WT controls were made from 13.5-day embryos and maintained in complete DMEM medium. Cells transduced with tetracycline-inducible shRNA delivery lenti-viruses were cultured in DMEM complete medium containing 10% tet system approved FBS (Cat. 631105, Clontech, Mountain View, CA). All cells were grown in 37°C incubator supplied with 5% CO2.
Experimental animals
Nlrx1−/− C57BL/6 mice have been described previously (Allen et al., 2011). Mice with the floxed Atg5 locus (Hara et al., 2006) were crossed with Cre-recombinase transgenic mice under the control of the Lysosomal-M promoter. All experimental mice were bred in a pathogen-free facility at the University of North Carolina at Chapel Hill in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and the Institutional Animal Care.
Plasmids and molecular cloning
FLAG-tagged TUFM expression plasmid was a kind gift from Dr. Nono Takeuchi at the University of Tokyo, Japan. FLAG-tagged MITA expression plasmid and ISRE reporter plasmid were generously provided by Dr. Hongbing Shu at Wuhan University, China. The domain truncation mutants of TUFM were generated according to standard molecular cloning protocols. LC3B-GFP expression vector was purchased from Addgene.
RT-PCR and gene tissue expression
RT-PCR was performed as previously described (Allen et al., 2011). The expression data of TUFM in multiple cell and tissue types was made available by the BioGPS project (http://biogps.gnf.org/), funded by the Genomics Institute of the Novartis Research Foundation.
RNAi-based protein expression knockdown
NLRX1 mRNA was targeted using a lentiviral delivery system to stably introduce shRNA expressed from a pol III promoter. The promoter, targeting shRNA or scrambled shRNA hairpin sequence, and polyT termination signal were amplified by PCR from a U6 promoter template and cloned into the lentiviral vector FG12. To generate the tetracycline-inducible lentiviral shRNA delivery system targeting TUFM, we obtained Expression Arrest™ non-silencing TripZ control bacteria stock (Cat. RHS4743, Thermo Scientific Open Biosystems, Huntsville, AL) and TUFM-targeting shRNA-containing construct bacteria stock (Oligo ID V2THS_222080, Cat. RHS4696-99362407, Thermo Scientific Open Biosystems, Huntsville, AL). The ON-TARGETplus SMARTpool of 4 siRNA targeting TUFM (Cat. L-016741-01-0005, Thermo Scientific, Huntsville, AL) and the control pool of 4 siRNA (Cat. D-001810-10-05, Thermo Scientific, Huntsville, AL) were transfected into the cells according to the manual.
Gel filtration
2×107 HEK293T cells were transfected with either control dsRNA or 5’-ppp dsRNA (Invivogen, San Diego, CA). Cells where harvested 16 hr post-transfection and lysed in hypotonic lysis buffer with a Dounce homogenizer. The homogenate was centrifuged at 500g for 15 min to separate nuclei. The supernatants were filtered and loaded onto Superose 6 HR-10/30 columns (GE Healthcare). 500 µl of each fraction was collected and separated by SDS-PAGE.
AACT-based mass spectrometry
AACT-based mass spectrometry was performed as previously described (Chen et al., 2000). Briefly, HEK293T cells were transduced with retroviruses expressing empty vector, HA-tagged NLRX1 full-length or NLRX1ΔLRR truncation. Cells with empty vector were grown in DMEM L-Leucine-deficient medium (Cat. D9816, US Biological, Swampscott, MA) supplemented with L-Leucine-5,5,5-d3 (Cat. 486825, Sigma-Aldrich, St.Louis, MO). NLRX1-expressing or NLRX1ΔLRR-expressing cells were cultured in regular medium. Cells were lysed and immunoprecipitated with anti-HA affinity matrix (Cat. 11815016001, Roche, Indianapolis, IN). The eluted fractions were subjected to SDS-PAGE and Coomassie staining. The Coomassie-stained gel lanes were continuously excised and the peptides were lyophilized and then resuspended in buffer A (2% acetonitrile, 98% water and 0.1% formic acid). LC-MS/MS analysis was performed on a nanoAcquity UPLC - QTOF API US system (Waters, UK). All the raw files acquired on QTOF were processed using Mascot Distiller software (version 2.3, Matrix Science, UK) and then searched against human International Protein Index (IPI) protein database (version 3.26, 67665 sequences). Only proteins that had at least two unique peptides achieving an ion score above 39 and contained at least five consecutive amino acids matching the data base peptides sequences were considered to be positive identifications. All the raw spectrum and SILAC ratios were manually evaluated. Gene Ontology annotation analysis was performed using DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov/).
Transfection and viral infection
HEK293T cells were plated so that they could reach about 80% confluence the next day for transfections. FuGENE® 6 (Cat. 11814443001, Roche, Indianapolis, IN) was used to transfect cells. VSV-GFP virus was propagated in VERO cells (ATCC, Manassas, VA). Cell lines or primary cells were plated in 24-well plate 24 hr prior to infection to reach 80% confluence the next day. Virus inoculum was prepared in pre-warmed serum-free DMEM and incubated with cells for 1 hr at 37°C. Then the virus inoculum was removed and cells were replenished with complete media.
Co-immunoprecipitation and Western blots
Cells were lysed in RIPA buffer (1% Triton X-100, 0.25% DOC, 0.05% SDS, 50mM Tris-HCl pH8.0, 150mM NaCl and 50mM NaF) or NP-40-based lysis buffer (1% NP-40, 50mM Tris-HCl pH8.0, 150mM NaCl) containing complete protease inhibitor cocktail (Cat. 11873580001, Roche, Indianapolis, IN). For the endogenous co-immunoprecipitation experiments, whole cell lysates were pre-cleared with protein A resin (Cat. L00210, GenScript, Piscataway, NJ) and protein G resin (Cat. L00209, GenScript, Piscataway, NJ) for 1 hr at 4°C, and then incubated with primary antibody overnight. The protein A/G UltraLink® resin (Cat. 53132, Thermo Scientific, Huntsville, AL) was added and incubated for 2 hr at 4°C. Beads were washed 5 times before resuspension and boiling in the Laemmli’s sample buffer. Protein samples were separated by NuPAGE Bis-Tris 4–12% precast gels (Cat. NP0322BOX, Invitrogen, Carlsbad, CA) or Tris-Glycine 16% precast gels (Cat. EC64952BOX, Invitrogen, Carlsbad, CA). The antibodies used in this study are as following: High-affinity anti-HA-peroxidase (Cat. 12013819001, Roche, Indianapolis, IN), Anti-FLAG® M2 affinity gel (Cat. A2220, Sigma-Aldrich, St.Louis, MO), anti-LC3B (Cat. 2775, Cell Signaling, Danvers, MA), anti-TUFM (Cat. ab67991, abcam, Cambridge, MA), anti-TUFM (Cat. HPA018991, Sigma-Aldrich, St. Louis, MO), anti-eEF1α1 (Cat. sc-28578, Santa Cruz, Santa Cruz, CA), anti-Bcl-xL (Cat. 2762, Cell Signaling, Danvers, MA), anti-Atg5 (Cat. 2630, Cell Signaling, Danvers, MA), anti-calreticulin (Cat. 2891, Cell Signaling, Danvers, MA), anti-Atg16L1 (Cat. 8089, Cell Signaling, Danvers, MA), anti-Actin-HRP (Cat. sc-1615HRP, Santa Cruz, Santa Cruz, CA).
Luciferase assay
Assays were performed as previously described (Moore et al., 2008).
Laser confocal imaging analysis
WT or Nlrx1−/− MEFs were seeded on 8-well glass slide (Cat. 154534, Thermo Fisher Scientific, Rochester, NY) 24 hr prior to treatments. The next day cells were incubated with VSV-GFP at the MOI of 0.1 for 1h in serum-free DMEM, and then the virus inoculum was removed. Cells were replenished with complete medium and incubated for 4 hr. Cells were fixed and permeabilized with 4% paraformaldehyde containing 0.1% Triton-X 100 for 20 min at room temperature. The cells were washed 3 times with 1× PBS. Samples were incubated with blocking buffer (7% BSA, 1% FBS, 2% goat serum, 1% Fc block) for 1 hr at room temperature. LC3B antibody was diluted at 1:400 in the blocking buffer. The samples were incubated with primary antibody overnight at 4°C followed by the 2 hr incubation with Alexa Fluor® 647 goat anti-rabbit antibody (Cat. A-21244, Invitrogen, Carlsbad, CA) diluted at 1:2000 ratio in the blocking buffer. Images were acquired with an Olympus FV500 confocal microscope.
Supplementary Material
HIGHLIGHTS.
NLRX1 suppresses VSV-mediated type 1 IFN production but enhances autophagy.
TUFM works together with NLRX1 to inhibit RLR-induced type 1 IFN signaling.
TUFM enhances autophagy and interacts with the autophagy mediators, Atg5-Atg12 and Atg16L1.
VSV utilizes NLRX1-mediated autophagy and IFN-I inhibition to enhance its gene expression and replication.
ACKNOWLEDGEMENT
This work was supported by NIH grants U54-AI057157 (SERCEB), U19AI077437, U19AI067798 and CA156330 awarded to J.P.-Y.T., NIH grants R01AI064806, 1U24CA160035 awarded to X.C., NIH grant DE018281 awarded to J.P.-Y.T. and B.D., and NIH grant F32-AI094941 awarded to D.G.W. H.W. is supported by the Postdoctoral Fellowship of the American Heart Association, Mid-Atlantic Affiliate and Postdoctoral Fellowship of the Cancer Research Institute. We thank Dr. Hongbing Shu for providing MITA and ISRE reporter plasmids, Dr. Nono Takeuchi for the TUFM expression plasmid, Dr. Fumihiko Takeshita for the Atg5 and Atg12 expression plasmids, Dr. Noboru Mizushima for mice with the floxed Atg5 locus, and Dr. David Baltimore for the FG12 lentiviral vector plasmid.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alirezaei M, Flynn CT, Wood MR, Whitton JL. Pancreatic acinar cell-specific autophagy disruption reduces coxsackievirus replication and pathogenesis in vivo. Cell Host Microbe. 2012;11:298–305. doi: 10.1016/j.chom.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, Gris D, Roney KE, Zimmermann AG, Bowzard JB, et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity. 2011;34:854–865. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava K, Templeton P, Spremulli LL. Expression and characterization of isoform 1 of human mitochondrial elongation factor G. Protein Expr Purif. 2004;37:368–376. doi: 10.1016/j.pep.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Brenner D, Mak TW. Mitochondrial cell death effectors. Current opinion in cell biology. 2009;21:871–877. doi: 10.1016/j.ceb.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Chan DC. Mitochondrial fusion and fission in mammals. Annual review of cell and developmental biology. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- Chen X, Smith LM, Bradbury EM. Site-specific mass tagging with stable isotopes in proteins for accurate and efficient protein identification. Analytical chemistry. 2000;72:1134–1143. doi: 10.1021/ac9911600. [DOI] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Harton JA, Linhoff MW, Zhang J, Ting JP. Cutting edge: CATERPILLER: a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J Immunol. 2002;169:4088–4093. doi: 10.4049/jimmunol.169.8.4088. [DOI] [PubMed] [Google Scholar]
- Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, Okuda K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO journal. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nature immunology. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejbich-Trotot P, Gay B, Li-Pat-Yuen G, Hoarau JJ, Jaffar-Bandjee MC, Briant L, Gasque P, Denizot M. Chikungunya triggers an autophagic process which promotes viral replication. Virology journal. 2011;8:432. doi: 10.1186/1743-422X-8-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. The Plant cell. 2004;16:3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Lei Y, Moore CB, Liesman RM, O'Connor BP, Bergstralh DT, Chen ZJ, Pickles RJ, Ting JP. MAVS-mediated apoptosis and its inhibition by viral proteins. PloS one. 2009;4:e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5'-triphosphate-dependent translocase on double-stranded RNA. Science (New York, N.Y. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, MacPherson S, Sumpter R, Jr, Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Reis e Sousa C. RIGorous detection: exposing virus through RNA sensing. Science. 2010;327:284–286. doi: 10.1126/science.1185068. [DOI] [PubMed] [Google Scholar]
- Sanjuan MA, Green DR. Eating for good health: linking autophagy and phagocytosis in host defense. Autophagy. 2008;4:607–611. doi: 10.4161/auto.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Ueda T, Taguchi H, Takeuchi N. Chaperone properties of mammalian mitochondrial translation elongation factor Tu. The Journal of biological chemistry. 2007;282:4076–4084. doi: 10.1074/jbc.M608187200. [DOI] [PubMed] [Google Scholar]
- Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr, Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Molecular cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JP, Duncan JA, Lei Y. How the noninflammasome NLRs function in the innate immune system. Science. 2010;327:286–290. doi: 10.1126/science.1184004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente L, Tiranti V, Marsano RM, Malfatti E, Fernandez-Vizarra E, Donnini C, Mereghetti P, De Gioia L, Burlina A, Castellan C, et al. Infantile encephalopathy and defective mitochondrial DNA translation in patients with mutations of mitochondrial elongation factors EFG1 and EFTu. American journal of human genetics. 2007;80:44–58. doi: 10.1086/510559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Levine B. Autophagy genes in immunity. Nature immunology. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J, Henkler F, Leversha M, Koshy R. A mitochondrial elongation factor-like protein is over-expressed in tumours and differentially expressed in normal tissues. FEBS letters. 1995;358:119–125. doi: 10.1016/0014-5793(94)01403-n. [DOI] [PubMed] [Google Scholar]
- Xia X, Cui J, Wang HY, Zhu L, Matsueda S, Wang Q, Yang X, Hong J, Songyang Z, Chen ZJ, Wang RF. NLRX1 negatively regulates TLR-induced NF-kappaB signaling by targeting TRAF6 and IKK. Immunity. 2011;34:843–853. doi: 10.1016/j.immuni.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Molecular cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, Seya T, Kawabata S, Koshiba T. Mitofusin 2 inhibits mitochondrial antiviral signaling. Science signaling. 2009;2:ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- Ye Z, Ting JP. NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Current opinion in immunology. 2008;20:3–9. doi: 10.1016/j.coi.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li Z, Xinna G, Xin G, Yang H. Autophagy promotes the replication of encephalomyocarditis virus in host cells. Autophagy. 2011;7:613–628. doi: 10.4161/auto.7.6.15267. [DOI] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Zhu H, Pan S, Gu S, Bradbury EM, Chen X. Amino acid residue specific stable isotope labeling for quantitative proteomics. Rapid communications in mass spectrometry : RCM. 2002;16:2115–2123. doi: 10.1002/rcm.831. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.