Background: ADP-ribose transferases ARTD1–3/PARP1–3 have an α-helical domain that closes over the NAD+-binding site.
Results: Human ARTD15/PARP16 is a mono(ADP-ribose) transferase with a novel α-helical domain that interacts with a catalytic domain loop.
Conclusion: The ARTD15 transferase domain is likely regulated by effector binding to the adjacent helical domain.
Significance: This provides a basis for understanding the enzymatic mechanism of this previously uncharacterized enzyme.
Keywords: ADP-ribosylation, Covalent Regulation, NAD, Protein Domains, X-ray Crystallography
Abstract
ADP-ribosylation is involved in the regulation of DNA repair, transcription, and other processes. The 18 human ADP-ribose transferases with diphtheria toxin homology include ARTD1/PARP1, a cancer drug target. Knowledge of other family members may guide therapeutics development and help evaluate potential drug side effects. Here, we present the crystal structure of human ARTD15/PARP16, a previously uncharacterized enzyme. ARTD15 features an α-helical domain that packs against its transferase domain without making direct contact with the NAD+-binding crevice or the donor loop. Thus, this novel domain does not resemble the regulatory domain of ARTD1. ARTD15 displays auto-mono(ADP-ribosylation) activity and is affected by canonical poly(ADP-ribose) polymerase inhibitors. These results add to a framework that will facilitate research on a medically important family of enzymes.
Introduction
Protein ADP-ribosylation, in which ADP-ribose units derived from NAD+ are covalently linked to protein side chains (1), was first reported a half-century ago (2). Among the enzymes that modulate their substrate protein activities by mono(ADP-ribosylation) are toxins from various pathogenic bacteria (3) and the eukaryotic ADP-ribose transferase ectoenzymes (4). Poly(ADP-ribosylation), the formation of branched or unbranched chains of ADP-ribose units on protein side chains, is catalyzed by ADP-ribose transferases with diphtheria toxin homology (ARTD),3 also referred to as poly(ADP-ribose) polymerases (PARPs) (5, 6). ARTD1/PARP1 (7, 8), a multidomain protein with functions in DNA repair, is a target for development of cancer therapeutics (9).
The human genome encodes at least 18 different ARTD proteins. ARTD1 and ARTD2, as well as ARTD5 and ARTD6 (the tankyrases), have proven poly(ADP-ribose) transferase activity. Recently, ARTD10/PARP10 was identified as a mono(ADP-ribose) transferase in vitro, and it was concluded that most of the remaining family members may also catalyze mono(ADP-ribosylation) rather than ADP-ribose chain synthesis (10). It is not known whether the catalytic activities are an intrinsic property of the transferase domains or a consequence of the presence and identity of other functional domains, which vary widely within the family (5, 11).
On the N-terminal side of their transferase domains, ARTD1–3 (and, by prediction, ARTD4) contain an α-helical domain that likely has regulatory functions (11, 12). This domain interacts with an elongated loop of the transferase domain (the donor or D-loop) and partially blocks the NAD+ co-substrate-binding crevice (13–16). We present the crystal structure of ARTD15, a previously uncharacterized animal-specific member of the family (11, 17). ARTD15 has a novel α-helical domain that makes interactions with the transferase domain but without sharing sequence or structural homology with the helical domain of ARTD1–4. ARTD15 has auto-mono(ADP-ribosylation) activity and binds to canonical PARP inhibitors.
EXPERIMENTAL PROCEDURES
Recombinant Protein Production
Human ARTD15 cDNA was obtained from the Mammalian Gene Collection (accession number BC031074), provided by the National Institutes of Health. Expression vectors were constructed by inserting cDNA fragments into pNIC28-Bsa4 by ligation-independent cloning (18). Protein expression in Escherichia coli BL21-Gold(DE3) cells (Stratagene) transformed with the pRARE2 plasmid (Novagen) and protein purification by immobilized metal ion chromatography, size exclusion chromatography, and affinity tag cleavage were performed using standard methods as described under supplemental “Experimental Procedures.” Proteins solved in 20 mm HEPES (pH 7.5), 300 mm NaCl, 10% glycerol, and 2 mm tris(2-carboxyethyl)phosphine were stored at −80 °C. For crystallization experiments, the buffer was exchanged to 10 mm sodium acetate (pH 5.5), and the protein was concentrated to 37 mg/ml by ultrafiltration.
Crystallization and Structure Determination
Crystallization of ARTD15 Gly-5–Ala-279; derivatization using heavy atoms; native and multiple anomalous diffraction data collection; phasing by multiple isomorphous replacement with anomalous scattering; and model building and refinement using autoSHARP (19), Refmac5 (20), BUSTER (21), and Buccaneer (22) are described in detail under supplemental “Experimental Procedures.” ARTD15 crystallized in space group C2221, and the structure was refined to a resolution of 2.7 Å. A complete overview of data collection and refinement statistics is given in supplemental Table S1. The final model contained the complete α-helical domain, whereas a number of loop residues in the transferase domain were not resolved (supplemental Table S2).
ADP-ribosylation Assay
ARTD15 automodification was measured essentially as described by Langelier et al. (23), involving capture of hexahistidine-tagged ARTD15 Gly-5–Ala-279 on Ni2+-chelating plates (5 PRIME), automodification using 2% biotinylated NAD+ (Trevigen), and chemiluminescence detection of modified reaction products. Further details are given under supplemental “Experimental Procedures.” Km values were estimated by nonlinear regression analysis using Michaelis-Menten kinetics, and curve fitting was done with GraphPad Prism (GraphPad Software).
RESULTS AND DISCUSSION
Bacterial Expression of Catalytically Active ARTD15
We set out to express a number of ARTD15 protein fragments in E. coli. We observed that expression constructs spanning only the predicted ADP-ribose transferase domain were poorly soluble and that extension of expression constructs toward the N terminus strongly increased the yields of soluble protein (supplemental Fig. S1). No protein was recovered when the N-terminal segment was expressed on its own. The TMHMM server (24) predicted a transmembrane helix including Trp-290–Ala-312 near the C terminus of ARTD15. Little protein was recovered when the putative transmembrane segment was included in the expression constructs (supplemental Fig. S1).
Overall Structure of ARTD15
We conducted crystallization experiments with the ARTD15 protein and obtained well diffracting crystals of ARTD15(5–279) in the presence of the small canonical PARP inhibitor 3-aminobenzamide. As no suitable molecular replacement model could be identified and as the wild-type protein lacks methionine residues necessary for selenomethionine single-wavelength anomalous dispersion approaches, we performed heavy atom soaks. Anomalous scattering experiments were performed using gold- and platinum-soaked crystals, and the structure was solved using three complete three-wavelength data sets. The crystal structure of ARTD15 shows a canonical transferase domain with one molecule of 3-aminobenzamide bound in the nicotinamide pocket paired with a novel α-helical domain on its N-terminal side (Fig. 1A). The N-terminal domain consists of six α-helices that form an irregular bundle with a large hydrophobic core. Three-dimensional structure searches using different algorithms (25, 26) did not identify any domains with significant structural similarity, and BLAST searches did not identify proteins with significant sequence similarity. We conclude that ARTD15 is a two-lobed enzyme with a novel N-terminal α-helical domain and a canonical diphtheria toxin homology transferase domain (Fig. 1, B and C).
FIGURE 1.
Crystal structure of ARTD15/PARP16. A, crystal structure of ARTD15(5–279) with the canonical PARP inhibitor 3-aminobenzamide (3-AB), a nicotinamide mimic, bound in the active site. The α-helical domain is shown in blue, and the transferase domain is shown in green and orange. The positions of the N and C termini are indicated. B, domain architecture of ARTD15 in comparison with ARTD1/PARP1 and ARTD8/PARP14. α-hel., α-helical domain; cat., catalytic; TM, predicted transmembrane helix. C, structure-based sequence alignment of the transferase domains of ARTD15, ARTD8, and ARTD1. Sequences were aligned using ClustalW and adjusted manually based on the crystal structures of the three proteins (Protein Data Bank codes 4F0D, 3SMJ, and 3L3M, respectively). ARTD15 and ARTD1 sequences of the α-helical domains are shown on a blue background; the ARTD8 sequence is shown truncated at the transferase domain. Secondary structural elements were annotated following previous nomenclature for ARTD1 (13) and ARTD10 (10) with an additional prime to mark the α-helices of the novel domain of ARTD15. Red stars mark the positions of the HYE motif residues (6, 11), and the blue star highlights the conserved nicotinamide-stacking aromate (6).
ARTD15 Catalytic Site Structure
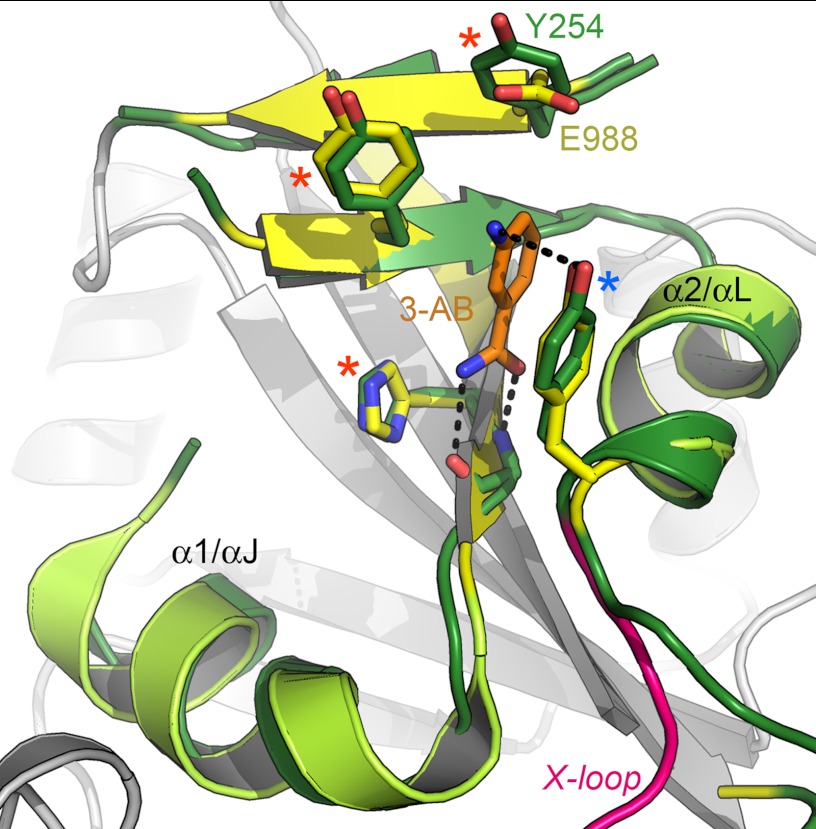
The NAD+-binding crevice of ARTD15 is lined by the conserved side chains His-152 and Tyr-182 of the HYE motif, as well as the nicotinamide-stacking Tyr-193. In diphtheria toxin, mutagenesis of the side chains corresponding to His-152 and Tyr-182 resulted in a strongly reduced affinity for the NAD+ co-substrate (27, 28). All three ARTD15 side chains align well with the corresponding ones in ARTD1 (Fig. 2) and other family members. By contrast, the third residue of the motif, the catalytic glutamate (Glu-988) of ARTD1, is replaced by a tyrosine (Tyr-254) in ARTD15. Although this was anticipated from sequence alignments, the crystal structure shows that the tyrosine side chain has a similar orientation as Glu-988 of ARTD1. This is suggestive of a role of Tyr-254 in catalysis, and we speculate that the side chain might stabilize a reaction intermediate. Notably, ARTD15 is the only family member that contains neither a glutamate (ARTD1–4 and the tankyrases ARTD5 and ARTD6) nor an aliphatic residue (the remainder of the family) in this position.
FIGURE 2.
Detail of the active site of ARTD15. The structures of ARTD15 (green and gray) and ARTD1 (yellow) were superimposed as in Fig. 2. The side chains of the canonical HYE motif (6) and the nicotinamide-stacking tyrosine are indicated by red and blue asterisks, respectively, as in Fig. 1C. Note that the position of the catalytic glutamate of ARTD1 is occupied by a tyrosine that maintains a similar orientation with respect to the active site. The bound ligand of ARTD1 was omitted for clarity. 3-AB, 3-aminobenzamide.
Structure and Potential Role of the Novel Helical Domain
The N-terminal α-helical domain of ARTD15 packs against the catalytic domain, burying a mainly hydrophobic interface of 1260 Å2 as determined using the PISA server (29). As in ARTD1, this likely explains why the domains cannot be produced in isolation (Ref. 13 and supplemental Fig. S1). The helical domain of ARTD15 does not share any obvious sequence similarity with the regulatory domains of ARTD1–4. A structure-based sequence alignment illustrates that these domains are quite dissimilar (Fig. 1C).
The helical domains of ARTD15 and ARTD1–3 occupy partially overlapping overall positions relative to the transferase domains. The only similarity in architecture is, however, between ARTD15 helix α2′ and ARTD1 α-helix A, which align roughly with one another (Fig. 3A). Nonetheless, the domain arrangement in both ARTD15 and ARTD1–3 is suggestive of a regulatory role for their α-helical domains, albeit by different mechanisms.
FIGURE 3.
Positioning of the novel α-helical domain of ARTD15. A, the bound ligands and central β-sheet of the transferase domains of ARTD15 and ARTD1 (Protein Data Bank code 3L3M) were superimposed to illustrate the partial overlap and relative positioning of the N-terminal helical domains of both proteins. Transferase domains are shown in dark green (ARTD15) and light green (ARTD1). The helical domains are shown in dark blue (ARTD15) and light blue (ARTD1). The ARTD15 X-loop (which makes contacts with helix α2′) is shown in pink, and the ARTD1 D-loop (not resolved in our ARTD15 structure) is shown in black. Secondary structural elements mentioned under “Results and Discussion” are annotated as described for Fig. 1C. The area magnified in B is boxed. B, details of the interaction between the ARTD15 helical domain and X-loop. The position of helix α2 is shown to indicate the linkage between the X-loop and co-substrate-binding site. The surface of the helical domain is shown rendered according to surface charge, where blue indicates positive charges, red indicates negative charges, and colorless indicates hydrophobicity. Key side chains referred to under “Results and Discussion” are shown as sticks.
The helical domain of ARTD1–3 closes over the NAD+ site like a lid, with αF restricting access to the co-substrate-binding pocket (Fig. 3A) (13–16). In ARTD1, the D-loop (Pro-881–Gly-888) makes extensive hydrophobic contacts with α-helix D of the helical domain (Fig. 3A). This interaction has been interpreted to keep the catalytic domain in a meta-stable state in which it can accept the adenine base of NAD+ (12). A similar arrangement is observed in the structures of ARTD2 and ARTD3 (14–16), whereas the structure of ARTD4 has not been determined. In our ARTD15 structure, the donor loop is partially disordered, but its position is not in the vicinity of the helical domain. Also, there is no secondary structural element that corresponds to ARTD1 helix D. Thus, if the ARTD15 helical domain is involved in the regulation of cofactor binding or activity of the catalytic domain, this must be accomplished by a mechanism that is distinct from that in ARTD1–3.
The crystal structure of ARTD15 suggests two ways by which the helical domain might relay a putative effector-binding event to the NAD+-binding site. (i) The extended or X-loop between α2 and β3 (Ser-193–Ile-208) makes both polar and hydrophobic contacts with α2′ (Fig. 3B). The most extensive contact is formed by the Trp-200 side chain, which is inserted into a hydrophobic pocket created between α2′ and the loop connecting α4′ and α5′. Also, the His-202 and Leu-204 side chains pack against hydrophobic patches in that region (Fig. 3B). Effector binding to the helical domain might therefore affect the positioning of side chains at the C-terminal end of α2 that interact with the cofactor, including Tyr-193, which stacks on the nicotinamide moiety. (ii) Helix α2′ and the loop-helix-loop motif that follows in sequence also pack onto α1 of the catalytic domain. Modulation of this interaction might have direct consequences on the positioning of side chains at the C-terminal end of β3, including His-152, the histidine that is part of the ARTD signal motif HYE (6).
Catalytic Activity of ARTD15
To investigate whether ARTD15 has ADP-ribose transferase activity, we conducted activity assays in the presence of 2% biotinylated NAD+ followed by either Western blotting or capture of hexahistidine-tagged ARTD15 on nickel-chelating multiplates and detection of modified products using anti-biotin antibodies and bioluminescence. ARTD15 displayed automodification activity (Fig. 4) but was unable to modify histone proteins (data not shown). We observed no apparent signs of poly(ADP-ribose) polymer formation, although we did not study this aspect in detail. Next, we used the protein capture strategy in multiplate format to determine an apparent Km of 267 ± 52 μm with respect to NAD+ and automodification. This is ∼2–3-fold higher than the Km determined for DNA-independent ARTD1 automodification using the same method (23). We recently showed that several compounds, including the widely used unselective PARP inhibitor EB-47, can stabilize ARTD15 in thermal unfolding assays (30). Along with our crystal structure in complex with 3-aminobenzamide, this implies that ARTD15 can be targeted by canonical PARP inhibitors. We conclude that ARTD15 has ADP-ribose transferase activity, is capable of automodification, and can be inhibited by canonical PARP inhibitors.
FIGURE 4.
Catalytic activity of ARTD15. A nonlinear regression plot of the automodification rates of ARTD15(5–279) yielded an estimate for Km of 267 ± 52 μm with respect to NAD+. Measurements were carried out in duplicate, and the error bars represent the S.E. of the fitted parameter. RLU, relative luminescence units.
While this manuscript was under review, Di Paola et al. (31) published a cell biological and biochemical characterization of ARTD15. In agreement with our results, ARTD15 was shown to possess mono(ADP-ribose) transferase activity. Furthermore, the protein was shown to localize to the cytosolic face of endoplasmic reticulum membranes and the nuclear envelope, a localization that depended on the presence of the predicted transmembrane helix (Ser-288–Ile-308). Finally, Di Paola et al. (31) identified the nuclear transport factor karyopherin-β1 as a substrate protein for ARTD15.
In summary, we have shown that ARTD15 contains a novel α-helical domain that is in contact with the active site of the transferase domain. Important functions of the helical domain are supported by its conservation from insects to humans (11), and we anticipate that this domain functions in ARTD15 regulation. The crystal structure of ARTD15 reported here will guide future research on (i) ADP-ribose transferase mechanisms, (ii) regulation of ARTD15 and the role of the helical domain therein, (iii) evaluation of the specificity of preclinical and clinical PARP inhibitors, and (iv) development of ARTD15-selective inhibitors as research tools.
Supplementary Material
Acknowledgments
We thank the beamline staff at Synchrotron Radiation Source BESSY II (Berlin, Germany) for excellent support and personnel at the Structural Genomics Consortium for technical assistance.
This work was supported by the Structural Genomics Consortium, a registered charity (1097737) that received funds from the Canadian Institutes of Health Research, the Canada Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, the Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust. This work was also supported by Swedish Foundation for Strategic Research Grant RBc08-0014.

This article contains supplemental “Experimental Procedures,” Fig. S1, Tables S1 and S2, and additional references.
The atomic coordinates and structure factors (code 4F0D) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ARTD
- ADP-ribose transferase with diphtheria toxin homology
- PARP
- poly(ADP-ribose) polymerase.
REFERENCES
- 1. Bürkle A. (2005) Poly(ADP-ribose). The most elaborate metabolite of NAD+. FEBS J. 272, 4576–4589 [DOI] [PubMed] [Google Scholar]
- 2. Chambon P., Weill J. D., Mandel P. (1963) Nicotinamide mononucleotide activation of a new DNA-dependent polyadenylic acid-synthesizing nuclear enzyme. Biochem. Biophys. Res. Comm. 11, 39–43 [DOI] [PubMed] [Google Scholar]
- 3. Holbourn K. P., Shone C. C., Acharya K. R. (2006) A family of killer toxins–exploring the mechanism of ADP-ribosylating toxins. FEBS J. 273, 4579–4593 [DOI] [PubMed] [Google Scholar]
- 4. Corda D., Di Girolamo M. (2003) Functional aspects of protein mono(ADP-ribosylation). EMBO J. 22, 1953–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hakmé A., Wong H. K., Dantzer F., Schreiber V. (2008) The expanding field of poly(ADP-ribosyl)ation reactions. EMBO Rep. 9, 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hottiger M. O., Hassa P. O., Lüscher B., Schüler H., Koch-Nolte F. (2010) Toward a unified nomenclature for mammalian ADP-ribosyltransferase. Trends Biochem. Sci. 35, 208–219 [DOI] [PubMed] [Google Scholar]
- 7. Cherney B. W., McBride O. W., Chen D. F., Alkhatib H., Bhatia K., Hensley P., Smulson M. E. (1987) cDNA sequence, protein structure, and chromosomal location of the human gene for poly(ADP-ribose) polymerase. Proc. Natl. Acad. Sci. U.S.A. 84, 8370–8374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uchida K., Morita T., Sato T., Ogura T., Yamashita R., Noguchi S., Suzuki H., Nyunoya H., Miwa M., Sugimura T. (1987) Nucleotide sequence of a full-length cDNA for human fibroblast poly(ADP-ribose) polymerase. Biochem. Biophys. Res. Comm. 148, 617–622 [DOI] [PubMed] [Google Scholar]
- 9. Rouleau M., Patel A., Hendzel M. J., Kaufmann S. H., Poirier G. G. (2010) PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer 10, 293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kleine H., Poreba E., Lesniewicz K., Hassa P. O., Hottiger M. O., Litchfield D. W., Shilton B. H., Lüscher B. (2008) Substrate-assisted catalysis by PARP10 limits its activity to mono(ADP-ribosylation). Mol. Cell 32, 57–69 [DOI] [PubMed] [Google Scholar]
- 11. Otto H., Reche P. A., Bazan F., Dittmar K., Haag F., Koch-Nolte F. (2005) In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genomics 6, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ruf A., de Murcia G., Schulz G. E. (1998) Inhibitor and NAD+ binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling. Biochemistry 37, 3893–3900 [DOI] [PubMed] [Google Scholar]
- 13. Ruf A., Mennissier de Murcia J., de Murcia G., Schulz G. E. (1996) Structure of the catalytic fragment of poly(ADP-ribose) polymerase from chicken. Proc. Natl. Acad. Sci. U.S.A. 93, 7481–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oliver A. W., Amé J. C., Roe S. M., Good V., de Murcia G., Pearl L. H. (2004) Crystal structure of the catalytic fragment of murine poly(ADP-ribose) polymerase-2. Nucleic Acids Res. 32, 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lehtiö L., Jemth A. S., Collins R., Loseva O., Johansson A., Markova N., Hammarström M., Flores A., Holmberg-Schiavone L., Weigelt J., Helleday T., Schüler H., Karlberg T. (2009) Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3. J. Med. Chem. 52, 3108–3111 [DOI] [PubMed] [Google Scholar]
- 16. Karlberg T., Hammarström M., Schütz P., Svensson L., Schüler H. (2010) Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888. Biochemistry 49, 1056–1058 [DOI] [PubMed] [Google Scholar]
- 17. Citarelli M., Teotia S., Lamb R. S. (2010) Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes. BMC Evol. Biol. 10, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gileadi O., Burgess-Brown N. A., Colebrook S. M., Berridge G., Savitsky P., Smee C. E., Loppnau P., Johansson C., Salah E., Pantic N. H. (2008) High throughput production of recombinant human proteins for crystallography. Methods Mol. Biol. 426, 221–246 [DOI] [PubMed] [Google Scholar]
- 19. Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 20. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 21. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Smart O. S., Vonrhein C., Womack T. O. (2011) BUSTER, Version 2.11.1, Global Phasing Ltd., Cambridge, United Kingdom [Google Scholar]
- 22. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 23. Langelier M. F., Ruhl D. D., Planck J. L., Kraus W. L., Pascal J. M. (2010) The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 285, 18877–18887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 25. Krissinel E., Henrick K. (2004) Secondary structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 26. Tung C. H., Huang J. W., Yang J. M. (2007) Kappa-alpha plot-derived structural alphabet and BLOSUM-like substitution matrix for rapid search of protein structure database. Genome Biol. 8, R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blanke S. R., Huang K., Wilson B. A., Papini E., Covacci A., Collier R. J. (1994) Active-site mutations of the diphtheria toxin catalytic domain: role of histidine 21 in nicotinamide adenine dinucleotide binding and ADP-ribosylation of elongation factor 2. Biochemistry 33, 5155–5161 [DOI] [PubMed] [Google Scholar]
- 29. Wilson B. A., Blanke S. R., Reich K. A., Collier R. J. (1994) Active-site mutations of diphtheria toxin. Tryptophan 50 is a major determinant of NAD affinity. J. Biol. Chem. 269, 23296–23301 [PubMed] [Google Scholar]
- 30. Wahlberg E., Karlberg T., Kouznetsova E., Markova N., Macchiarulo A., Thorsell A. G., Pol E., Frostell Å., Ekblad T., Öncü D., Kull B., Robertson G. M., Pellicciari R., Schüler H., Weigelt J. (2012) Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat. Biotechnol. 30, 283–288 [DOI] [PubMed] [Google Scholar]
- 31. Di Paola S., Micaroni M., Di Tullio G., Buccione R., Di Girolamo M. (2012) PARP16/ARTD15 is a novel endoplasmic reticulum-associated mono(ADP-ribose) transferase that interacts with and modifies karyopherin-β1. PLoS One 10.1371/journal.pone.0037352 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.