Background: Conjugation of sugars to chemicals by (UDP-glycosyltransferases) UGTs is a critical detoxification mechanism.
Results: A single amino acid defines the differential sugar specificities of two related UGTs.
Conclusion: The change of a single amino acid during primate evolution has generated a new capacity for small molecule glycosidation.
Significance: Determinants of UGT sugar selectivity are currently poorly understood; novel glycosidation pathways may have important metabolic roles.
Keywords: Drug Metabolism, Glycosyltransferases, Homology Modeling, Protein Evolution, Site-directed Mutagenesis, UDP-glucuronosyltransferase
Abstract
Recent studies in this laboratory characterized the UGT3A family enzymes, UGT3A1 and UGT3A2, and showed that neither uses the traditional UDP-glycosyltransferase UGT co-substrate UDP-glucuronic acid. Rather, UGT3A1 uses GlcNAc as preferred sugar donor and UGT3A2 uses UDP-Glc. The enzymatic characterization of UGT3A mutants, structural modeling, and multispecies gene analysis have now been employed to identify a residue within the active site of these enzymes that confers their unique sugar preferences. An asparagine (Asn-391) in the UGT signature sequence of UGT3A1 is necessary for utilization of UDP-GlcNAc. Conversely, a phenylalanine (Phe-391) in UGT3A2 favors UDP-Glc use. Mutation of Asn-391 to Phe in UGT3A1 enhances its ability to utilize UDP-Glc and completely inhibits its ability to use UDP-GlcNAc. An analysis of homology models docked with UDP-sugar donors indicates that Asn-391 in UGT3A1 is able to accommodate the N-acetyl group on C2 of UDP-GlcNAc so that the anomeric carbon atom (C1) is optimally situated for catalysis involving His-35. Replacement of Asn with Phe at position 391 disrupts this catalytically productive orientation of UDP-GlcNAc but allows a more optimal alignment of UDP-Glc for sugar donation. Multispecies sequence analysis reveals that only primates possess UGT3A sequences containing Asn-391, suggesting that other mammals may not have the capacity to N-acetylglucosaminidate small molecules. In support of this hypothesis, Asn-391-containing UGT3A forms from two non-human primates were found to use UDP-GlcNAc, whereas UGT3A isoforms from non-primates could not use this sugar donor. This work gives new insight into the residues that confer sugar specificity to UGT family members and suggests a primate-specific innovation in glycosidation of small molecules.
Introduction
Glycosidation of lipophilic chemicals by the UDP-glycosyltransferase (UGT)3 superfamily is an important detoxification pathway in all vertebrates (1). Glycosidation increases the water solubility of the acceptor substrate, facilitating its excretion and/or otherwise altering its biological reactivity (2–5). Acceptors are structurally diverse and include steroid hormones, bile acids, biogenic amines, plant and bacterial metabolites, carcinogens, and many therapeutic drugs (6). Thus, glycosidation protects cells against exogenous toxins and accumulations of potentially toxic by-products of metabolism. It can also play a role in modulating signaling pathways such as those mediated by steroid hormones. The glycosyl donor (co-substrate) is usually a UDP-hexose, typically UDP-glucuronic acid (UDP-GlcUA), UDP-Glc, UDP-xylose, UDP-Gal, or UDP-GlcNAc. During the conjugation reaction, the UDP–α-bond between UDP and the hexose moiety is converted into a β-bond between the acceptor and the sugar via an SN2 mechanism to form a β-d-glycoside.
In humans, four UGT families have been identified; UGT1, UGT2 (divided into subfamilies 2A and 2B), UGT3, and UGT8 (6). The UGT1 enzymes are encoded by a single genomic locus on chromosome 2q37, which contains multiple enzyme-specific exons 1A and a shared set of exons 2–5 (7). Differential promoter usage and splicing produce mRNAs for nine separate UGT1A enzymes, each having a unique N-terminal domain encoded by exon 1A and an identical C-terminal domain encoded by exons 2–5. The UGT2 family is divided into the UGT2A and UGT2B subfamilies. UGT2A1 and UGT2A2 are encoded in a similar fashion to the UGT1A family and have identical C-terminal domains encoded by a shared set of five exons. The remaining members of the UGT2 family are encoded by separate genes of six exons each that are arrayed along chromosome 4q13 (6). The UGT3A family has two members denoted UGT3A1 (8) and UGT3A2 (9) that are arranged as a direct repeat on chromosome 5p13. The human UGT8 family contains only one gene (UGT8A1) located on chromosome 4q26.
The UGT1 and UGT2 enzymes use UDP-GlcUA as their preferred sugar donor, although there are examples where other UDP-sugars are used. For example, UGT2B7 and UGT1A1 can utilize UDP-Glc, and UGT1A1 can use UDP-xylose (10–12). However, the activities of UGT1 and UGT2 family enzymes with these alternative UDP-sugars are much lower than those with UDP-GlcUA, and are typically restricted to specific aglycone substrates. There is no evidence that UGT1 and UGT2 forms can use UDP-Gal or UDP-GlcNAc. UGT8 catalyzes the transfer of Gal from UDP-Gal to ceramide in the production of glycosphingolipids; no other sugar donors or acceptors have been identified for this enzyme. Currently, it is believed that UGT8 has an exclusively biosynthetic role and is not involved in detoxification (13).
We recently characterized the substrate specificities of the UGT3A enzymes (8, 9). In striking contrast to the UGT1 and UGT2 families, we found that neither UGT3A enzyme can utilize the prototypic UGT co-substrate UDP-GlcUA. UGT3A1 uses UDP-GlcNAc to conjugate several substrates, including bile acids, steroids, and bioflavones (8). This sugar selectivity is unique among vertebrate UGTs. UGT3A2 preferentially utilizes UDP-Glc with a range of substrates, including bioflavones and estrogens (9). The significance of these unusual sugar specificities remains unclear, although it is possible that the GlcNAc and Glc conjugates have important biological roles in vivo. UGT3A2, in particular, is expressed at very low levels in the human liver, kidney, and gastrointestinal tract, suggesting that its major roles may be not in elimination of exogenous chemicals but in endogenous metabolism.
UGTs can be divided into two domains and typically show much greater sequence conservation in their C-terminal halves than in their N-terminal halves. This is consistent with recognition of diverse aglycone substrates by the N-terminal domain and binding of the UDP-sugar donor by the C-terminal domain. Although sequence determinants of aglycone specificity have been identified by domain swapping and mutagenesis experiments, there has been limited progress in identifying residues involved in sugar specificity, in part because all UGT1 and UGT2 enzymes preferentially use UDP-GlcUA and have only minor activities with other sugars. The recent discovery of differential sugar usage in the UGT3A family now provides a unique opportunity to address this issue. Using cross-species sequence comparisons and site-directed mutagenesis, we have identified a key residue in the putative sugar-binding domain that confers differential sugar specificities to the UGT3A proteins. We also provide evidence that the use of UDP-GlcNAc as a sugar donor is unique to primate UGT3A proteins.
EXPERIMENTAL PROCEDURES
Materials
Radioactive and nonradioactive UDP-sugars were obtained as indicated: [14C]UDP-Glc and [14C]UDP-GlcNAc from GE Healthcare and UDP-Glc and UDP-GlcNAc from Sigma-Aldrich. All other reagents and solvents were of analytical reagent grade.
Production of UGT3A1 and UGT3A2 Mutants
Site-directed mutagenesis was performed using human UGT3A1 and UGT3A2 cDNAs encoded in the pCR2.1 vector (Invitrogen) and the QuikChange mutagenesis method. Asn-391 in UGT3A1 was mutated to Phe-391 using primers 5′-CCCATGGTGGGATTACCAGTCTTTGGAGACCAGCATGGAAACATGG-3′ (forward) and 5′-CCATGTTTCCATGCTGGTCTCCAAAGACTGGTAATCCCACCATGGG-3′ (reverse). Phe-391 in UGT3A2 was mutated to Asn-391 using primers 5′-CCCATGGTGGGGATCCCTCTCAATGGAGACCAGCCTGAAAACATGG-3′ (forward) and 5′-CCATGTTTTCAGGCTGGTCTCCATTGAGAGGGATCCCCACCATGGG-3′ (reverse). The mutated products were verified by resequencing of the complete coding region and then subcloned into the pEF-IRESpuro6 expression vector, which contains a puromycin resistance gene (14). Expression vectors were transfected into human embryonic kidney HEK293T cells, and mixed cell populations stably expressing the mutant proteins were selected with puromycin (2 μg/ml). Lysates were prepared for assays in buffer containing 10 mm Tris and 1 mm EDTA (pH 7.6).
Western Blotting
Proteins in lysates from HEK293T cells stably expressing wild-type and mutant UGT3A cDNAs were separated on SDS-polyacrylamide gels and transferred to nitrocellulose membranes as described previously (8, 15). Wild-type and mutant UGT3A proteins were detected with anti-UGT3A1 and anti-UGT3A2 antibodies generated previously (8, 9) and a peroxidase-conjugated goat anti-rabbit secondary antibody (NeoMarkers). Immunocomplexes were visualized with the SuperSignal West Pico chemiluminescent kit (Thermo Fisher Scientific) and quantified with a GE Healthcare LAS4000 scanner.
Enzyme Assays
For assays to assess co-substrate preference, glycosidation reactions were performed as described previously (8). Briefly, incubations were performed at 37 °C for 1 h in 100 mm phosphate buffer (pH 7.5), 4 mm magnesium chloride, 100 μg of HEK293T cell lysate, 200 μm aglycone, and 2 mm [14C]UDP-sugar (0.1 μCi/mmol). Radioactive products were separated by thin layer chromatography (8) and quantified by exposure to a Molecular Dynamics phosphor screen, which was scanned with a GE Healthcare Typhoon 9400 scanner. Standard curves with known amounts of [14C]UDP-sugar were constructed to quantify product formation. For comparisons of activities between lysates expressing UGT3A proteins, activities relative to the amount of UGT protein present in the lysate (as determined by Western blotting) were used.
In Silico Modeling and Docking
A human UGT3A1 homology model was constructed using FUGUE and ORCHESTRAR (SYBYL-X 1.3, Tripos). Homologs identified by FUGUE were refined against profile hidden Markov model (16) data constructed from the UGT3A1 coding sequence. Crystal templates of grape UDP-Glc:flavonoid 3-O glucosyltransferase (codes 2C1X, 2C1Z, and 2C9Z), glycosyltransferase UGT78G1 from Medicago truncatula (codes 3HBF and 3HBJ), and the cofactor-binding domain of UGT2B7 (code 2O6L) (17) were obtained from the Brookhaven Protein Data Bank, and sequence alignments were refined using Cn3D relative to secondary structure. The highest acceptable inter-Cα distance between equivalent residues within a sequence conserved region was 1.5 Å, with a root mean square difference of 0.00001Å considered significant. Loops not modeled were found by loop threading all crystal structures of the HOMSTRAD Database (SYBYL-X 1.3). Refinement of the model was achieved by undertaking independent minimizations of biopolymer hydrogens, side chains, the biopolymer omitting Cα carbons, and finally the biopolymer as a whole. Each independent minimization allowed a maximum of 10,000 iterations using the method reported by Powell (18) with a termination gradient of <0.05 kcal/mol/Å. Automated docking of UDP-GlcNAc and UDP-Glc was achieved using the Surflex-Dock (SFXC) suite (SYBYL-X 1.3). Co-substrate docking was consensus-scored (CScore) relative to protein interactions with hydrogen flexibility allowed. The relative strengths of co-substrate and protein covalent force fields were set to 1.00 and 0.10, respectively. Ring flexibility was additionally allowed when generating the molecular co-substrate fragments and minimized using a Broyden-Fletcher-Goldfarb-Shanno quasi-Newton method and an internal DREIDING force field (19). In silico mutants were generated by substitution of the desired amino acid. Substituted residues were energy-minimized as a subset of the entire protein molecule using the Powell conjugate gradient method with an energy cutoff set to 0.05 kcal/mol/Å. A “hot region” of 6 Å surrounding the substituted residue was established where the side chains of all residues were minimized. A further “intermediate region” of 12 Å was generated to set the minimization environment without side chain movement. Minimization by this method allowed changes in the energetic forces experienced by residues that either adjoin or neighbor the substituted amino acid. Distance measurements were collected to characterize the orientation of docked co-substrate poses. Distances were measured between the anomeric carbon (C1) of the co-substrate sugar moiety (i.e. the site of SN2 attack) and the N2 of the catalytic base, His-35.
Cloning and Expression of Non-human UGT3A cDNAs
Mouse Ugt3a1 and Ugt3a2 cDNAs were obtained by reverse transcription of neonatal (up to 1-week old) mouse liver or kidney RNA. Briefly, first-strand cDNA was synthesized with the SuperScriptTM first-strand synthesis system (Invitrogen). The Ugt3a1 coding region was amplified from the cDNA using primers 5′-CGGAATTCATGGCTGCACATCGGAGTTGGC-3′ (forward) and 5′-CGGAATTCTTATGCCTGCTTGACCTTCCTTG-3′ (reverse). The mouse Ugt3a2 coding region was amplified using primer 5′-CGGAATTCATGGCAGCACATCGGCGTTGG-3′ (forward) and 5′-CGGAATTCTTATGCCTCCTTGACCTTCGT-3′ (reverse). The initiation and stop codons in the forward and reverse primers, respectively, are in italic type. The gene nomenclature follows that of the NCBI Gene Database. PCR was performed in a volume of 50 μl with 200 ng of cDNA, 0.5 μm forward and reverse primers, and PfuTurbo DNA polymerase (Stratagene). The cycling parameters consisted of one cycle at 95 °C for 2 min; 35 cycles at 95 °C for 0.5 min, 60 °C for 0.5 min, and 72 °C for 2 min; followed by a single cycle at 72 °C for 5 min. PCR products were excised from a 1% agarose gel, purified using the QIAquick gel extraction kit (Qiagen), and cloned into the pCR2.1 shuttle vector for sequencing. Both cDNAs were subcloned into the pEF-IRESpuro6 expression vector.
Full-length cDNAs for chimp (Pan troglodytes) UGT3A1 (XM_526949), rhesus (Macaca mulatta) UGT3A1 (XM_001093373) and UGT3A2 (XM_001093598), Cow (Bos taurus) UGT3A2 (NM_001076087), and rabbit (Oryctolagus cuniculus) UGT3A (XM_002714108) were obtained from GenScript in the pUC57 shuttle vector. The cDNAs were excised and subcloned into the pEF-IRESpuro6 expression vector. Sequencing confirmed the identities of each clone.
The human and chimp UGT3A2 proteins differ by only one amino acid (Asn-309 in human is Tyr-309 in chimp). To obtain a cDNA with the equivalent sequence to chimp UGT3A1, we performed site-directed mutagenesis of the human UGT3A2 cDNA to introduce the chimp-specific amino acid (Tyr-309). The mutagenesis primer sequences were as follows: 5-gtcagtatccggaaatcttcaaggag-3′ (forward) and 5′-cggatactgacaggtgttcaccatg-3′ (reverse).
All non-human UGT cDNAs were transfected into HEK293T cells. Either transiently expressing lysates or lysates from puromycin-selected cell populations stably expressing the UGT of interest were used in assays.
Analysis of Endogenous Ugt3a Gene Expression in Mouse Tissue
To measure the levels of endogenous Ugt3a1 and Ugt3a2 mRNAs in neonatal mouse livers and kidneys, we used quantitative RT-PCR with the following primers: Ugt3a1, 5′-ACCGTGTGTCGCAAATTCTG-3′ (forward) and 5′-ACCTGGTATGATGAGTTTTCC-3′ (reverse); and Ugt3a2, 5′-TTCTCATGAGCTTCCTTTTCC-3′ (forward) and 5′-GCGACACACGGCTTATCAC-3′ (reverse). The raw data were converted to copy number using standard curves constructed from the cloned cDNAs. Data were then normalized to the mRNA abundance of the ribosomal protein S26 (Rps26) housekeeping gene. Reactions were carried out using SYBR Green reagent (Qiagen) or GoTaq (Promega) and the Rotor-Gene 300 thermal cycler (Corbett Life Sciences).
Preparation of Mouse Liver and Kidney Microsomes
Livers and kidneys from 2–3-week-old mice were snap-frozen on dry ice. Microsomes were prepared as described previously (20), and assays were performed as described above.
RESULTS
Functional Analysis of Residue 391 in Human UGT3A1 and UGT3A2
We have previously shown that UGT3A1 is unique among human UGTs in using UDP-GlcNAc as its preferred sugar donor (8), whereas human UGT3A2 utilizes UDP-Glc as its preferred co-substrate (9). The UGT signature sequence is a 30-amino acid region that is highly conserved in all UGTs and implicated in the binding of UDP-sugars (see Ref. 6 and references therein). When the signature sequences of all human UGTs are aligned, residue 391 is clearly divergent; this residue is asparagine in UGT3A1 and phenylalanine in all other human UGTs, including UGT3A2 (Fig. 1). We hypothesized that Asn-391 may be important for conferring the unique ability of human UGT3A1 to utilize UDP-GlcNAc.
FIGURE 1.

Alignment of UGT signature sequences of all human UGTs. The residue at position 391 is divergent in human (h) UGT3A1.
Site-directed mutagenesis was used to substitute Asn-391 in UGT3A1 with Phe (N391F), and, conversely, to substitute Phe-391 in UGT3A2 with asparagine (F391N). The stable expression of both mutant proteins in HEK293T cells was confirmed by Western blotting (data not shown). The activities of both mutant and wild-type proteins were examined using either UDP-Glc or UDP-GlcNAc and the aglycone substrate genistein. Genistein was selected because it was previously found to be a good substrate for both UGT3A1 and UGT3A2 with their preferred sugar donors (8, 9, 21).
Using sensitive assays for glycosidation, we observed that wild-type UGT3A1 in fact has a weak inherent capacity to conjugate glucose to genistein (supplemental Fig. 1). UGT3A1 mutation N391F dramatically enhanced glucosidation activity and simultaneously abolished activity with UDP-GlcNAc (Table 1). UGT3A2 mutation F391N reduced the enzymatic activity with UDP-Glc by ∼150-fold (Table 1); however, it did not confer activity with UDP-GlcNAc.
TABLE 1.
Activities of wild-type and mutant forms of human UGT3A1 and UGT3A2 with genistein and either UDP-Glc or UDP-GlcNAc as sugar donor
| Protein variant | Activity with UDP-Glc | Activity with UDP-GlcNAc |
|---|---|---|
| pmol/min/mg | pmol/min/mg | |
| WT UGT3A1 | 1.75 ± 0.05 | 65.74 ± 13.13 |
| UGT3A1(N391F) | 15.03 ± 1.33 | NDa |
| WT UGT3A2 | 137.65 ± 6.59 | ND |
| UGT3A2(F391N) | 0.82 ± 0.08 | ND |
a ND, not detected.
Structure Modeling and UDP-sugar Docking
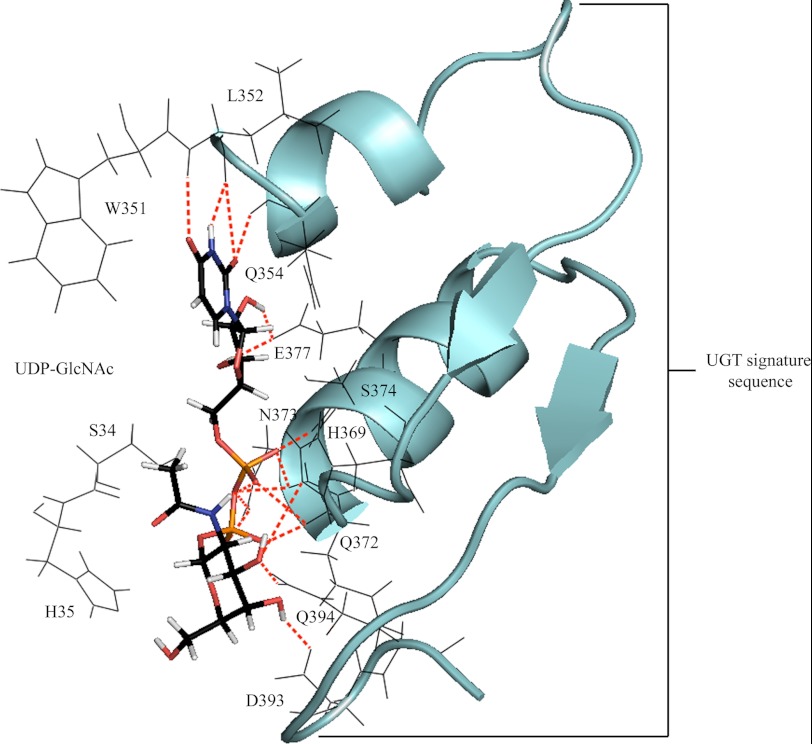
To examine the structural basis underpinning the importance of residue 391, homology models of UGT3A1 and mutant N391F were generated and docked with UDP-GlcNAc or UDP-Glc (Figs. 2 and 3). The docking of UDP-GlcNAc to UGT3A1 revealed direct side chain H-bonding interactions from Glu-377 to both hydroxyl hydrogens of the ribose, Asn-373 to two oxygens of the diphosphate, His-369 and Gln-394 to the 3′-hydroxyl oxygen of the ribose, and Asp-393 to the 4′-hydroxyl hydrogen of the sugar moiety. Numerous main chain (both nitrogen and oxygen) H-bonding interactions provided further stabilization of the UDP moiety; these included Leu-352 and Gln-354 to uracil and Gln-372, Asn-373, and Ser-374 to the diphosphate. Trp-351 was in a position to form π-π and π-σ orbital interactions with the uracil. The residues utilized in orienting UDP-GlcNAc for nucleophilic attack by the aglycone are located within the UGT signature sequence (Fig. 2).
FIGURE 2.
Docked UDP-GlcNAc (capped sticks) positioned for productive SN2 nucleophilic attack. Stabilization of the uridine, diphosphate, and sugar substituents of the cofactor involves H-bonding interactions with highly conserved residues of the UGT signature sequence (schematic). Bonding interactions are observed from Leu-352/Gln-354/Glu-377 to the uridine, Gln-372/Asn-373/Ser-374 to the diphosphate, and His-369/Gln-394 to the sugar moiety.
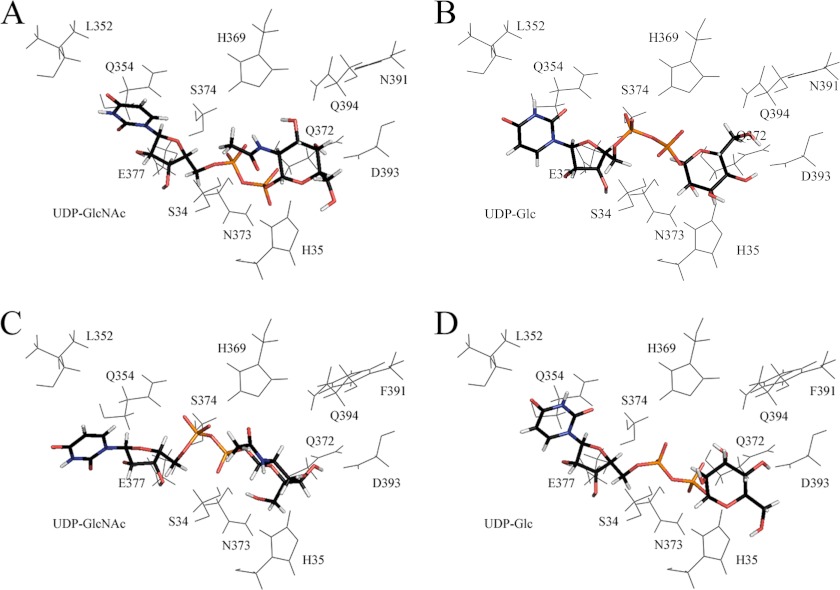
FIGURE 3.
Energetically favored poses of cofactor (capped sticks) after automated in silico docking. A, UDP-GlcNAc with wild-type UGT3A1. B, UDP-Glc with wild-type UGT3A1. C, UDP-GlcNAc with the N391F mutant. D, UDP-Glc with the N391F mutant.
All UGTs appear to have a histidine (His-35 in UGT3A1) that acts as a catalytic base to facilitate activation of the sugar-acceptor group on the aglycone by deprotonation for subsequent nucleophilic attack on the anomeric carbon (C1) of the sugar donor (22). When UDP-GlcNAc is docked into UGT3A1 (Fig. 3A), the distance between the catalytic histidine and C1 is 4.02 Å, which increases to 6.21 Å when UDP-Glc is docked into the protein (Fig. 3B). In contrast, when UDP-GlcNAc or UDP-Glc is docked into UGT3A1 Phe-391 (Fig. 3, C and D), the distances are 6.18 and 3.60 Å, respectively. Hence, the UGT3A1 proteins that are most active with their respective sugar donors (i.e. UGT3A1 with UDP-GlcNAc and UGT3A1 Phe-391 with UDP-Glc) have shorter distances between the catalytic histidine and C1 compared with the UGT3A1 protein/UDP-sugar combinations that have little activity (i.e. UGT3A1 with UDP-Glc and UGT3A1 Phe-391 with UDP-GlcNAc).
Cloning and Functional Analysis of Mouse Ugt3a1 and Ugt3a2
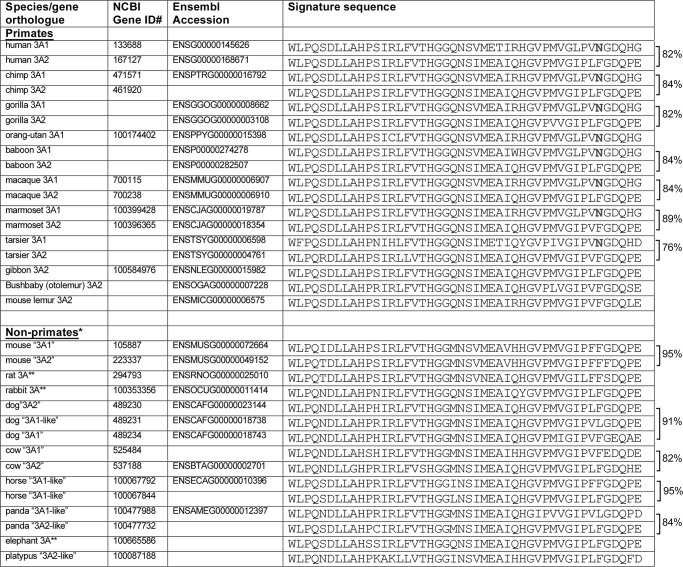
We examined UGT3A sequences in the genomes of multiple species to determine when Asn-391 may have evolved. In all species examined, UGT3A genes are located in regions of shared synteny, including the flanking genes LMBRD2 (LMBR1 domain-containing-2) and CAPSL (calcyphosine-like). These flanking genes were used as markers to guide our identification of UGT3A genes and pseudogenes in each genome. We found that the human, chimp, gorilla, baboon, macaque, marmoset, and tarsier genomes each encode one UGT3A form with Asn-391 and one with Phe-391 (Fig. 4). The orangutan genome appears to encode only one UGT3A form, and it bears Asn-391. The genomes of the gibbon, bushbaby (Otolemur) and mouse lemur and of non-primates (including mouse, rat, rabbit, dog, cow, horse, panda, elephant, and platypus) contain one or more UGT3A genes that each encode Phe-391 (or, in some cases, Leu-391) and no genes that encode Asn-391. In general, the signature sequences of primate UGT3A paralogues show less similarity to one another than those of the non-primate UGT3A paralogues. For example, the human UGT3A1 and UGT3A2 proteins are only 82% identical within their signature sequences, whereas the mouse Ugt3a1 and Ugt3a2 proteins are 95% identical within their signature sequences.
FIGURE 4.
Alignment of UGT3A signature sequences from multiple primate and non-primate species. Genomes were accessed using the NCBI and Ensembl genome browsers. Accession numbers are given for each database where the gene had been annotated. BLAST analysis was also used to identify all possible homologues within each locus. Pairwise comparisons were performed using ClustalW, and the percentage identity between the signature sequences of paralogous genes is given to the right. For dog, the percentage identity is given as the mean of the three pairwise comparisons. *, in these species, the homologues of human UGT3A1 and UGT3A2 cannot generally be determined by sequence analysis alone; thus, the names are placed in quotation marks. **, rat, human, and elephant have only one UGT3A form.
Given the effect of UGT3A1 mutation N391F on sugar preference, we hypothesized that only genes that encode Asn-391 may efficiently utilize UDP-GlcNAc as a sugar donor in conjugation reactions. To test this hypothesis, we first cloned mouse Ugt3a1 and Ugt3a2 cDNAs and examined their sugar specificity in the heterologous HEK cell expression system.
As shown in Table 2, mouse Ugt3a1 was active with 4-methylumbelliferone and ursodeoxycholic acid using UDP-Glc as the sugar donor. However, it showed no activity toward either substrate with UDP-GlcNAc. Mouse Ugt3a2 was also able to conjugate 4-methylumbelliferone and ursodeoxycholic acid with UDP-Glc, although to a lesser extent than Ugt3a1, and it was also inactive with UDP-GlcNAc.
TABLE 2.
Activities of wild-type mouse Ugt3a1 and Ugt3a2 with various aglycone substrates and either UDP-Glc or UDP-GlcNAc as sugar donor
| Protein variant | Activity with UDP-Glc |
Activity with UDP-GlcNAc |
||
|---|---|---|---|---|
| 4-MUa | UDCA | 4-MU | UDCA | |
| pmol/min/mg | pmol/min/mg | |||
| Ugt3a1 | 3.08 | 5.56 | ND | ND |
| Ugt3a2 | 4.77 | 2.84 | ND | ND |
a 4-MU, 4-methylumbelliferone; UDCA, ursodeoxycholic acid; ND, not detected.
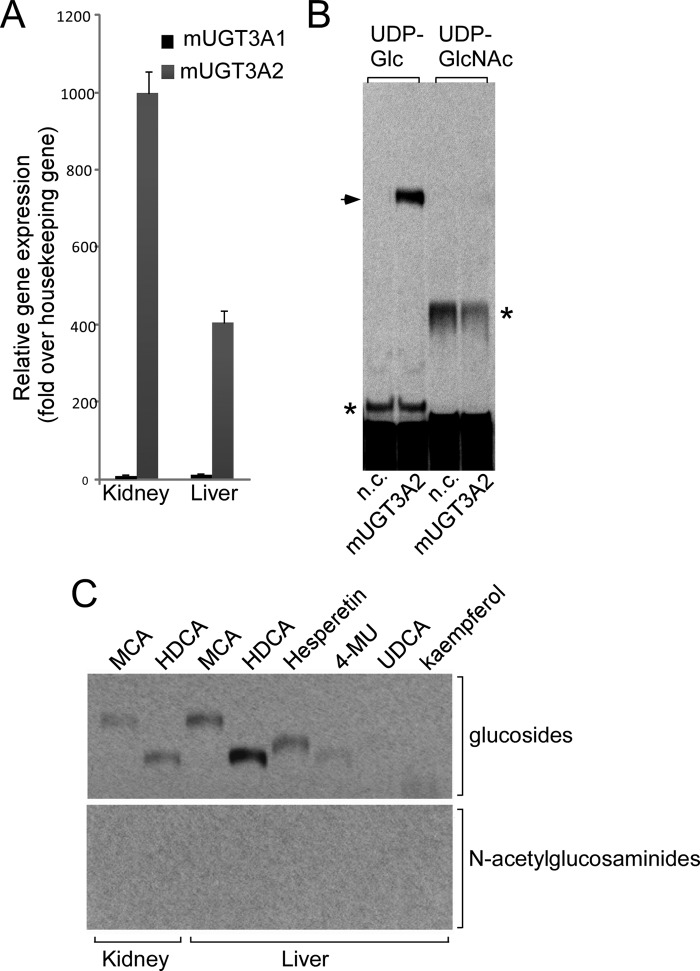
The arrangement of the UGT3A genes and their immediate neighbors in the human genome is LMBRD2-UGT3A2-UGT3A1-CAPSL. However, in the public mouse genome databases, the name Ugt3a1 has been assigned to the gene most proximal to Lmbrd2 (e.g. Lmbrd2-Ugt3a1-Ugt3a2-Capsl) (see Ref. 21). The products of the mouse Ugt3a1 and Ugt3a2 genes show the same degree of sequence similarity to the human UGT3A1 gene product. On the basis of genomic position, we suggest that mouse Ugt3a2 is in fact likely to be the homologue of human UGT3A1. In support of this notion, it was found that mouse Ugt3a1 shows a similar pattern of expression to human UGT3A2 and that mouse Ugt3a2 expression is more similar to human UGT3A1 expression. Specifically, the expression of mouse Ugt3a2 is vastly higher in liver and kidney than that of Ugt3a1 (Fig. 4). We have previously shown that human UGT3A1 mRNA is abundant in human liver and kidney, whereas human UGT3A2 mRNA is barely detectable in liver and is very low in kidney (8, 9).
Given the likelihood that mouse Ugt3a2 is the homologue of human UGT3A1, we sought additional aglycone substrates for this enzyme to confirm its apparent inability to use UDP-GlcNAc. Several bile salts and related chemicals were investigated. Hyodeoxycholic acid was identified as the aglycone with which mouse Ugt3a2 had the highest glucosidation activity. However, even with this substrate, Ugt3a2 showed no activity with UDP-GlcNAc as the sugar donor (Fig. 5B). This result further supports the idea that both mouse Ugt3a enzymes can utilize only UDP-Glc.
FIGURE 5.
A, analysis of Ugt3a1 and Ugt3a2 mRNA levels in neonatal mouse (m) liver and kidney by quantitative RT-PCR. Data were analyzed using a standard curve for each gene and normalized to the housekeeping gene Rps26. B, analysis of the enzymatic activity of mouse Ugt3a2 with its preferred aglycone substrate hyodeoxycholic acid (HDCA) and either UDP-Glc or UDP-GlcNAc as the sugar donor. Assays were chromatographed and visualized by autoradiography. The arrow indicates the hyodeoxycholic acid glucoside, and the asterisks indicate endogenous glycosidation activities in HEK293T cells. n.c., negative control HEK293T lysate. C, analysis of endogenous glucosidation and N-acetylglucosaminidation activities in mouse liver and kidney microsomal preparations. Assays were performed using a range of aglycone substrates and either UDP-Glc or UDP-GlcNAc as the sugar donor and were analyzed as described for B. MCA, mycophenolic acid; 4-MU, 4-methylumbelliferone; UDCA, ursodeoxycholic acid.
To assess whether the mouse has any capacity for N-acetylglucosaminidation of small molecules, we tested microsomal preparations of mouse liver and kidney for endogenous conjugation activity using UDP-Glc or UDP-GlcNAc and a variety of aglycones, including mycophenolic acid, hyodeoxycholic acid, hesperetin, and 4-methylumbelliferone. Only glucose conjugates were detected (Fig. 5C), further supporting the idea that the mouse does not express an UDP-N-acetylglucosaminyltransferase.
Activities of Chimp, Rhesus, Cow, and Rabbit UGT3A Proteins
To further test our hypothesis that only genes that encode Asn-391 may utilize UDP-GlcNAc as a sugar donor in conjugation reactions, we obtained an assortment of clones encoding UGT3A enzymes from various primate and non-primate species and expressed them in HEK293T cell culture. Activity was examined using a variety of aglycone substrates and both UDP-Glc and UDP-GlcNAc as the sugar donor. All recombinant UGT3A enzymes were catalytically active. However, only those UGT3A enzymes that contain Asn-391 were able to use UDP-GlcNAc as a sugar donor (Fig. 6). Those with a phenylalanine in this position were active only with UDP-Glc as the sugar donor.
FIGURE 6.
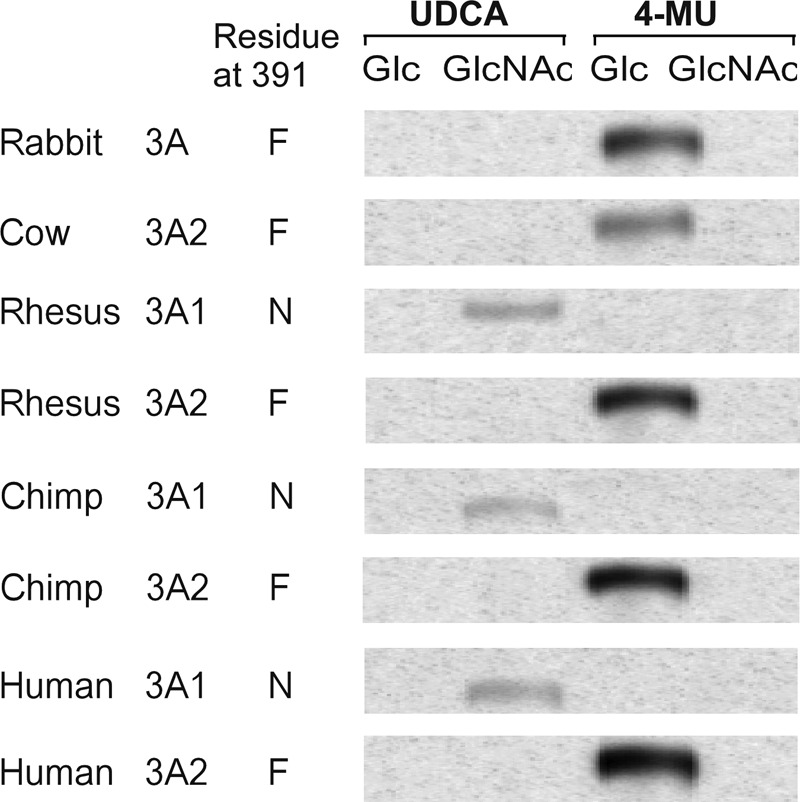
Analysis of enzymatic activities of various primate and non-primate UGT3A isoforms using preferred aglycone substrates and either UDP-Glc or UDP-GlcNAc as sugar donor. Assays were chromatographed, and the glycoside products were visualized by autoradiography. Human UGT3A1 and UGT3A2 served as positive controls. 4-MU, 4-methylumbelliferone; UDCA, ursodeoxycholic acid.
DISCUSSION
UDP-GlcUA recognition by human UGTs has been investigated by several laboratories because early photoaffinity labeling experiments suggested a binding site between amino acids 299 and 466 (23). Several residues within the UGT signature sequence, including His-371 and Glu-379 (numbering from UGT1A6) (24), and the DQ motif that marks the end of the UGT signature sequence (Asp-393–Gln-394 in UGT1A10) (25) have been shown to play important or essential roles in UDP-GlcUA binding (25). In contrast, there has been little progress in understanding the structural basis of differential sugar recognition by UGTs, as most mammalian UGTs show only minor activities with sugars other than UDP-GlcUA. Characterization of the divergent sugar preferences of the UGT3A enzymes provided us with an opportunity to identify residues that may confer selectivity for different UDP-sugars.
By analysis of UGT3A family enzymes, we have shown that Asn-391 within the signature sequence of UGT3A1 is essential for its capacity to utilize UDP-GlcNAc as a sugar donor: substituting phenylalanine at this position blocks its ability to use UDP-GlcNAc and instead promotes utilization of UDP-Glc. Docking of various sugars into a homology model of wild-type UGT3A1 suggested that the catalytic histidine is significantly closer to the anomeric carbon of UDP-GlcNAc than UDP-Glc, which likely explains its greater activity with UDP-GlcNAc than with UDP-Glc.
Although the N391F mutation altered the sugar preference of UGT3A1, the level of activity of mutant N391F with UDP-Glc was lower than that of UGT3A2 with UDP-Glc or of wild-type UGT3A1 with UDP-GlcNAc. This suggests that additional residues determine the efficiency with which the preferred sugar is used. We attempted to identify such residues by swapping signature sequences and adjacent segments between UGT3A1 and UGT3A2 (data not shown). Some of these chimeric enzymes were poorly expressed, suggesting that they may be structurally unstable. However, even among the variant UGT3A1 proteins that were efficiently expressed, we did not find any that had greater activity with UDP-Glc than mutant N391F (data not shown). Thus, the residue at position 391 is the major contributor to sugar specificity within the signature sequence, and other residues that influence enzymatic activity are likely to be located distal to the signature sequence.
Plant UGTs are extremely diverse and have a broader range of UDP-sugar preferences than mammalian UGTs. Several studies involving plant UGTs have identified residues that contribute to sugar specificity, but none of these directly overlap with Phe-391 in mammalian UGTs. Kubo et al. (26) found that the Gln within the DQ motif (Gln-394 in human UGT3A1) is conserved in plant and animal UDP-glucuronosyltransferases and UDP-glucosyltransferases but that this residue is His in plant and animal UDP-galactosyltransferases. Mutation of this residue from His to Gln in a plant UDP-galactosyltransferase lowered the Km of this enzyme for UDP-Glc by ∼40-fold, suggesting that Gln plays a role in glucose recognition. The complementary mutation of Gln to His in a plant UDP-glucosyltransferase impaired its ability to use UDP-Glc but did not confer activity with UDP-Gal (26). Moreover, mutation of Gln to His in another plant glucosyltransferase, UDP-glucose:flavonoid 3-O-glycosyltransferase (VvGT1), abolished activity (27). The Asp and Gln residues were predicted to interact directly with the hydroxyl groups of the sugar moiety of UDP-Glc (27), a conclusion similar to that previously drawn for the DQ motif in binding of UDP-GlcUA (25).
Human UGT3A1 and UGT3A2 share the DQ motif (Asp-393–Gln-394) with all other mammalian UGTs, suggesting that the motif is not important for sugar specificity. However, the two residues immediately after Gln-394 are divergent between UGT3A1 and other mammalian UGTs (see Fig. 1). We found that mutation of these two residues in UGT3A1 impaired protein stability and activity with both UDP-GlcNAc and UDP-Glc (data not shown). Thus, the C-terminal end of the signature sequence may be involved indirectly in both sugar binding and catalysis by determining correct folding of UGT3A enzymes.
As well as identifying residues in UGT3A enzymes that confer specificity for UDP-GlcNAc or UDP-Glc, it is interesting to consider what sequence or structural features may preclude their use of UDP-GlcUA. Residues that confer UDP-GlcUA specificity have been examined using site-directed mutagenesis and protein modeling of BpUGT94B1, a UGT from red daisy (Bellis perennis) that conjugates glucuronic acid to flavonoids. This analysis identified an arginine outside of the signature sequence (Arg-25) as crucial for activity with UDP-GlcUA, with Arg-25 mutants exhibiting dramatically reduced activity with UDP-GlcUA and 3-fold increased activity with UDP-Glc (28). A conserved arginine is found in the corresponding position relative to the mature N terminus of all human UGTs of the UGT1 and UGT2 families. However, in UGT3A1 and UGT3A2, this residue is histidine (His-49). This may support the notion that arginine at this position is important for utilization of UDP-GlcUA. It should be noted, however, that UGT8, which is known to use only UDP-Gal, also has an arginine at this position; thus, the function of this residue is likely to be context-dependent.
Is UDP-GlcNAc conjugation by UGT3A1 a primate innovation? Our cross-species genomic analysis indicated that Asn-391-containing forms of the UGT3A family occur only in primates. Experimentally, we found that primate UGT3A1 proteins that contain Asn-391 (human, chimp, and rhesus) are all capable of N-acetylglucosaminidation. In contrast, all of the UGT3A proteins from non-primates that we tested (cow, rabbit, and mouse) lack Asn-391 and are incapable of N-acetylglucosaminidation. Although it is not feasible to test all species, this sampling provides strong support for the idea of a primate-specific UGT3A1 enzymatic activity. Additional circumstantial evidence for this notion is provided by the observations that 1) the signature sequences of primate UGT3A gene paralogues generally show lower sequence identity than those of non-primate paralogues, suggesting that the primate UGT3A enzymes are more functionally divergent, and 2) many non-primates have only one UGT3A family gene (e.g. rat, rabbit, and elephant).
It is difficult to determine when the capacity for N-acetylglucosaminidation may have first emerged. Lemurs lack a Asn-391-containing UGT3A isoform, suggesting that this residue may have first appeared around the time that Haplorhini and Strepsirrhini primates diverged 63 million years ago. However, both lemur genomes have lower coverage (<2 times) than the other primate genomes. Thus, future analyses of higher resolution genome assemblies may change this conclusion. There is also evidence for selective gene loss in some primates, e.g. gibbon and orangutan have only one UGT3A gene (both of these genomes have 5–6 times coverage). Similarly, we observed that the entire UGT3A family is absent in sequenced bird genomes, although it is present in reptiles and some species of fish.4 Identification of endogenous substrates for human UGT3A enzymes, as well as their developmental expression patterns, may provide insight into the significance of primate diversification in sugar conjugation capacity.
Supplementary Material
Acknowledgment
We thank Julie-Ann Hulin for technical assistance.
This work was supported by the National Health and Medical Research Council of Australia.

This article contains supplemental Fig. 1.
R. Meech and P. I. Mackenzie, unpublished data.
- UGT
- UDP-glycosyltransferase
- UDP-GlcUA
- UDP-glucuronic acid.
REFERENCES
- 1. Mackenzie P. I., Owens I. S., Burchell B., Bock K. W., Bairoch A., Bélanger A., Fournel-Gigleux S., Green M., Hum D. W., Iyanagi T., Lancet D., Louisot P., Magdalou J., Chowdhury J. R., Ritter J. K., Schachter H., Tephly T. R., Tipton K. F., Nebert D. W. (1997) The UDP-glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7, 255–269 [DOI] [PubMed] [Google Scholar]
- 2. Meech R., Mackenzie P. I. (1997) Structure and function of uridine diphosphate glucuronosyltransferases. Clin. Exp. Pharmacol. Physiol. 24, 907–915 [DOI] [PubMed] [Google Scholar]
- 3. Tukey R. H., Strassburg C. P. (2000) Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu. Rev. Pharmacol. Toxicol. 40, 581–616 [DOI] [PubMed] [Google Scholar]
- 4. Radominska-Pandya A., Czernik P. J., Little J. M., Battaglia E., Mackenzie P. I. (1999) Structural and functional studies of UDP-glucuronosyltransferases. Drug Metab. Rev. 31, 817–899 [DOI] [PubMed] [Google Scholar]
- 5. Miners J. O., Mackenzie P. I. (1991) Drug glucuronidation in humans. Pharmacol. Ther. 51, 347–369 [DOI] [PubMed] [Google Scholar]
- 6. Mackenzie P. I., Bock K. W., Burchell B., Guillemette C., Ikushiro S., Iyanagi T., Miners J. O., Owens I. S., Nebert D. W. (2005) Nomenclature update for the mammalian UDP-glycosyltransferase (UGT) gene superfamily. Pharmacogenet. Genomics 15, 677–685 [DOI] [PubMed] [Google Scholar]
- 7. Ritter J. K., Chen F., Sheen Y. Y., Tran H. M., Kimura S., Yeatman M. T., Owens I. S. (1992) A novel complex locus UGT1 encodes human bilirubin, phenol, and other UDP-glucuronosyltransferase isozymes with identical carboxyl termini. J. Biol. Chem. 267, 3257–3261 [PubMed] [Google Scholar]
- 8. Mackenzie P. I., Rogers A., Treloar J., Jorgensen B. R., Miners J. O., Meech R. (2008) Identification of UDP-glycosyltransferase 3A1 as a UDP-N-acetylglucosaminyltransferase. J. Biol. Chem. 283, 36205–36210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mackenzie P. I., Rogers A., Elliot D. J., Chau N., Hulin J. A., Miners J. O., Meech R. (2011) The novel UDP-glycosyltransferase 3A2: cloning, catalytic properties, and tissue distribution. Mol. Pharmacol. 79, 472–478 [DOI] [PubMed] [Google Scholar]
- 10. Mackenzie P., Little J. M., Radominska-Pandya A. (2003) Glucosidation of hyodeoxycholic acid by UDP-glucuronosyltransferase 2B7. Biochem. Pharmacol. 65, 417–421 [DOI] [PubMed] [Google Scholar]
- 11. Tang C., Hochman J. H., Ma B., Subramanian R., Vyas K. P. (2003) Acyl glucuronidation and glucosidation of a new and selective endothelin ETA receptor antagonist in human liver microsomes. Drug Metab. Dispos. 31, 37–45 [DOI] [PubMed] [Google Scholar]
- 12. Senafi S. B., Clarke D. J., Burchell B. (1994) Investigation of the substrate specificity of a cloned expressed human bilirubin UDP-glucuronosyltransferase: UDP-sugar specificity and involvement in steroid and xenobiotic glucuronidation. Biochem. J. 303, 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bosio A., Binczek E., Le Beau M. M., Fernald A. A., Stoffel W. (1996) The human gene CGT encoding the UDP-galactose:ceramide galactosyltransferase (cerebroside synthase): cloning, characterization, and assignment to human chromosome 4, band q26. Genomics 34, 69–75 [DOI] [PubMed] [Google Scholar]
- 14. Hobbs S., Jitrapakdee S., Wallace J. C. (1998) Development of a bicistronic vector driven by the human polypeptide chain elongation factor 1α promoter for creation of stable mammalian cell lines that express very high levels of recombinant proteins. Biochem. Biophys. Res. Commun. 252, 368–372 [DOI] [PubMed] [Google Scholar]
- 15. Udomuksorn W., Elliot D. J., Lewis B. C., Mackenzie P. I., Yoovathaworn K., Miners J. O. (2007) Influence of mutations associated with Gilbert and Crigler-Najjar type II syndromes on the glucuronidation kinetics of bilirubin and other UDP-glucuronosyltransferase 1A substrates. Pharmacogenet. Genomics 17, 1017–1029 [DOI] [PubMed] [Google Scholar]
- 16. Eddy S. R. (1998) Profile hidden Markov models. Bioinformatics 14, 755–763 [DOI] [PubMed] [Google Scholar]
- 17. Radominska-Pandya A., Bratton S. M., Redinbo M. R., Miley M. J. (2010) The crystal structure of human UDP-glucuronosyltransferase 2B7 C-terminal end is the first mammalian UGT target to be revealed: the significance for human UGTs from both the 1A and 2B families. Drug Metab. Rev. 42, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Powell M. J. D. (1977) Beyond symmetric Broyden for updating quadratic models in minimization without derivatives. Mathematical Programming 12, 241–254 [Google Scholar]
- 19. Mayo S. L., Olafson B. D., Goddard W. A. (1990) DREIDING: a generic force field for molecular simulations. J. Phys. Chem. 94, 8897–8909 [Google Scholar]
- 20. Bowalgaha K., Elliot D. J., Mackenzie P. I., Knights K. M., Swedmark S., Miners J. O. (2005) S-Naproxen and desmethylnaproxen glucuronidation by human liver microsomes and recombinant human UDP-glucuronosyltransferases (UGT): role of UGT2B7 in the elimination of naproxen. Br. J. Clin. Pharmacol. 60, 423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meech R., Mackenzie P. I. (2010) UGT3A: novel UDP-glycosyltransferases of the UGT superfamily. Drug Metab. Rev. 42, 45–54 [DOI] [PubMed] [Google Scholar]
- 22. Miley M. J., Zielinska A. K., Keenan J. E., Bratton S. M., Radominska-Pandya A., Redinbo M. R. (2007) Crystal structure of the cofactor-binding domain of the human phase II drug-metabolism enzyme UDP-glucuronosyltransferase 2B7. J. Mol. Biol. 369, 498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Battaglia E., Terrier N., Mizeracka M., Senay C., Magdalou J., Fournel-Gigleux S., Radominska-Pandya A. (1998) Interaction of periodate-oxidized UDP-glucuronic acid with recombinant human liver UDP-glucuronosyltransferase 1A6. Drug Metab. Dispos. 26, 812–817 [PubMed] [Google Scholar]
- 24. Patana A. S., Kurkela M., Goldman A., Finel M. (2007) The human UDP-glucuronosyltransferase: identification of key residues within the nucleotide-sugar binding site. Mol. Pharmacol. 72, 604–611 [DOI] [PubMed] [Google Scholar]
- 25. Xiong Y., Patana A. S., Miley M. J., Zielinska A. K., Bratton S. M., Miller G. P., Goldman A., Finel M., Redinbo M. R., Radominska-Pandya A. (2008) The first aspartic acid of the DQxD motif for human UDP-glucuronosyltransferase 1A10 interacts with UDP-glucuronic acid during catalysis. Drug Metab. Dispos. 36, 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kubo A., Arai Y., Nagashima S., Yoshikawa T. (2004) Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch. Biochem. Biophys. 429, 198–203 [DOI] [PubMed] [Google Scholar]
- 27. Offen W., Martinez-Fleites C., Yang M., Kiat-Lim E., Davis B. G., Tarling C. A., Ford C. M., Bowles D. J., Davies G. J. (2006) Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25, 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Osmani S. A., Bak S., Imberty A., Olsen C. E., Møller B. L. (2008) Catalytic key amino acids and UDP-sugar donor specificity of a plant glucuronosyltransferase, UGT94B1: molecular modeling substantiated by site-specific mutagenesis and biochemical analyses. Plant Physiol. 148, 1295–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.