Background: The function of KLF11/TIEG2 under stressful conditions is undefined.
Results: KLF11 increases brain MAO expression through its promoter and a chromatin partner, which can be enhanced by stress.
Conclusion: This is the first elucidation of mechanisms underlying stress-induced KLF11-MAO up-regulation.
Significance: This novel KLF11-MAO pathway may play an important role in stress-related brain disorders.
Keywords: Animal Models, Chronic Stress, Gene Knockout, Gene Transcription, Glucocorticoids, Kruppel-like Factor (KLF), Neuroblastoma, Serotonin, Monoamine Oxidase A
Abstract
Chronic stress is a risk factor for psychiatric illnesses, including depressive disorders, and is characterized by increased blood glucocorticoids and brain monoamine oxidase A (MAO A, which degrades monoamine neurotransmitters). This study elucidates the relationship between stress-induced MAO A and the transcription factor Kruppel-like factor 11 (KLF11, also called TIEG2, a member of the Sp/KLF- family), which inhibits cell growth. We report that 1) a glucocorticoid (dexamethasone) increases KLF11 mRNA and protein levels in cultured neuronal cells; 2) overexpressing KLF11 increases levels of MAO A mRNA and enzymatic activity, which is further enhanced by glucocorticoids; in contrast, siRNA-mediated KLF11 knockdown reduces glucocorticoid-induced MAO A expression in cultured neurons; 3) induction of KLF11 and translocation of KLF11 from the cytoplasm to the nucleus are key regulatory mechanisms leading to increased MAO A catalytic activity and mRNA levels because of direct activation of the MAO A promoter via Sp/KLF-binding sites; 4) KLF11 knockout mice show reduced MAO A mRNA and catalytic activity in the brain cortex compared with wild-type mice; and 5) exposure to chronic social defeat stress induces blood glucocorticoids and activates the KLF11 pathway in the rat brain, which results in increased MAO A mRNA and enzymatic activity. Thus, this study reveals for the first time that KLF11 is an MAO A regulator and is produced in response to neuronal stress, which transcriptionally activates MAO A. The novel glucocorticoid-KLF11-MAO A pathway may play a crucial role in modulating distinct pathophysiological steps in stress-related disorders.
Introduction
Chronic stress increases the levels of blood glucocorticoids (1–3) and brain monoamine oxidase (MAO)3 A (4, 5). Monoamine oxidases (MAO) are catalytic enzymes prevalent in the brain and peripheral tissue (6). Present as two structurally distinct isoforms, MAO A and MAO B both degrade monoamine neurotransmitters and produce hydrogen peroxide as a toxic byproduct (7). MAO A primarily deaminates serotonin, norepinephrine, and dopamine and, therefore, is implicated in several psychiatric diseases. Elevated brain MAO A levels are present in living patients and post-mortem human subjects with depressive disorders, including major depressive disorder (8–12) and in mothers during postpartum depression (13), supporting the theory that an imbalance in biogenic amines can influence the affective state (14, 15). Such correlations suggest MAO A to be a biochemical link for stress and depression, which often exists comorbidly in clinical studies. Increasing efforts have been made to understand the mechanism of stress-induced MAO A expression (4, 5, 16–18). Dexamethasone, a synthetic glucocorticoid that induces cellular stress, has been shown to increase MAO A mRNA, protein, and enzymatic activity in human skeletal muscle cells (19) and to increase MAO A mRNA levels in the dorsal raphe nucleus in rats (20). In addition, dexamethasone treatment of neuroblastoma cells demonstrated that activation and subsequent binding of glucocorticoid receptors (GR) to the glucocorticoid response DNA element (GRE) sequence on the MAO A promoter-activated MAO A transcription (21). In addition to three consensus GREs in the distal promoter region, the core promoter of MAO A contains four Sp/KLF-binding sites that have been implicated in the transcriptional activation of MAO A (21). Kruppel-like factor 11 (KLF11), a member of the Sp/KLF family of transcription factors, has been shown to up-regulate MAO B transcription via similar Sp/KLF-binding sites (22). However, possible involvement of KLF11 in MAO A transcriptional activation has not been studied. KLF11 is also referred to as transforming growth factor β-inducible early gene 2 (TIEG2). It is currently better known because of its role in metabolism. Indeed, publications from our group demonstrate that KLF11 (TIEG2) regulates genes involved in the metabolism of lipids, glucose, prostaglandins, neurotransmitters, and alcohol/drugs (23–25). We have also published that KLF11 is linked to three types of diabetes (a metabolic disease) (26–28). Thus, a more in-depth investigation of how stress hormones, such as glucocorticoids, affect the function of KLF11, as shown here, is of substantial biomedical relevance. Here we report that KLF11 acts as a direct transcriptional activator in stress-induced MAO A expression. First, cultured neuronal cells treated with dexamethasone show increased KLF11 expression and nuclear translocation and increased MAO A expression and activity. Second, the MAO A expression and activity after dexamethasone administration is influenced by KLF11, as shown by both the overexpression and knockdown of KLF11. Third, dexamethasone-induced KLF11 directly binds and activates transcription of MAO A via the p300 pathway, independently of the GR-induced transcriptional activation. Fourth, KLF11 knockout mice show decreased MAO A expression in brain. Fifth, KLF11 and MAO A levels are increased in the brains of rats exposed to chronic social defeat (CSD) stress, a well established animal model for depression (29–31). Together, these findings suggest that KLF11 is a direct activator for MAO A and that the novel stress-induced KLF11-MAO A pathway may modulate behavioral traits associated with stress or depressive disorders.
EXPERIMENTAL PROCEDURES
Cell Line and Rat Primary Cortical Neurons
The SH-SY5Y human neuroblastoma cell line was purchased from the ATCC. Cells were cultured in DMEM supplemented with 10% FBS. Rat brain cortex (E18, 19) neuronal cells were purchased from Lonza (R-Cx-500) and cultured in poly-D-lysine-coated plates with PNGMTM BulletKitTM medium following the instructions of the manufacturer. After ∼4 days in culture, cells were treated with or without 100 nm dexamethasone for 48–72 h, as described previously (21, 32, 33).
mRNA Extraction and Quantitative Real-time RT-PCR
TRIzol reagent (Invitrogen) was used to extract RNA from SH-SY5Y cells and rat brain tissue. Reverse transcription was performed using the SuperScript III first-strand synthesis system (Invitrogen). Resulting cDNA was quantified using an iCycler MyiQ real-time PCR detection system (Bio-Rad), as described previously (22, 34). Specific primers for human, mouse, and rat were used and designed as follows: human MAO A, 5′-CGTGATCGGAGGTGGCATTTC-3′ (sense) and 5′-AAAGGCGCCCCGAAAGG-3′ (antisense); human KLF11, 5′-CCTGTTGCGGATAAGACCCCTCAC-3′ (sense) and 5′-AAAGCCGGCAATCTGGAGTCTGGA-3′ (antisense); mouse MAO A, 5′-CCGCTCCTTTTCCATGCCTCTCAA-3′ (sense) and 5′-CCCTTGTACCGCCCCTTGACTGAA-3′ (antisense); rat MAO A, 5′-ACGCTCAGGAATGGGACAAGATG-3′ (sense) and 5′-CCCACACTGCCTCACATACCACA-3′ (antisense); rat KLF11, 5′-CTCCTGCAGGGCCGTGATGAC-3′ (sense) and 5′-GGGGAACAGGCCACCAGACTTG-3′ (antisense). The 18 S ribosomal RNA primer was used as an internal control (22, 34).
Western Blot Analysis
Whole-cell protein extracts were obtained in radioimmune precipitation assay buffer (Sigma), and the lysates were centrifuged at 4 °C (11,500 rpm) for 10 min to pellet and eliminate the cell debris. Brain tissue from each animal was homogenized in a 0.5-ml solution containing 1 mm EDTA, 10 mm Tris-HCl, and fresh protease inhibitor (Sigma) and centrifuged at 4 °C (3,500 rpm) for 10 min. Supernatants were stored at −80 °C. Forty micrograms of total protein were separated in 10.5% SDS-PAGE gel. After transfer, the membranes were probed with primary and secondary antibodies. All band intensities were normalized to those of β-actin using Quantity One analysis software (8, 23).
Immunofluorescence
Cells were plated on a four-well chamber slide (Nalge) with or without 100 nm dexamethasone treatment for 48 h. Cells were then fixed with 4% paraformaldehyde and incubated with mouse anti-KLF11 (1:1000) antibody overnight at 4 °C. After incubation with secondary antibody, stained slides were mounted with Vectashield in the presence of 4,6-diamino-2-phenylindole (DAPI nuclear stain, Vector Lab, Inc.) (17).
Generation of KLF11 and pcDNA Stably Transfected Cell Lines (Overexpressing KLF11)
Cells were plated at a density of 106 cells per 10-cm dish. The next day, the KLF11 expression vector or pcDNA 3.1 control vector was transfected into cells with SuperFect transfection reagent (Qiagen, Inc). After 24 h, cells were treated with the antibiotic Geneticin (G418, 600 μg/ml). Resistant clones were isolated into separate dishes after 6 days and cultured under continuous G418 selection (35).
siRNA-mediated Klf11 Gene Knockdown
Control siRNA or KLF11 siRNA for human KLF11 (Santa Cruz) or for rat KLF11 (Qiagen) was transfected into SH-SY5Y cells or the rat primary cortical neurons with the siPORT amine transfection agent (Ambion) following the protocol of the manufacturer. Briefly, siPORT amine transfection agent and Opti-MEM I medium were mixed with each siRNA for 10 min with a final siRNA concentration of 20 nm per 10-cm dish. The siRNA·siPORT amine transfection agent complex was directly added to the cell culture medium (34).
MAO A Catalytic Activity Assay
SH-SY5Y cells, the rat primary cortical neurons, and animal brain tissue were homogenized in assay buffer (50 mm sodium phosphate buffer). Approximately 100 μg of total protein were incubated with 100 μm [14C]5-hydroxytryptamine in assay buffer at 37 °C for 20 min. The reaction was terminated by addition of 100 μl of 6N HCl. Reaction products were then extracted with benzene/ethyl acetate, and its radioactivity was determined by liquid scintillation spectroscopy (8, 21).
Transient Transfection and Luciferase Activity Assay
KLF11 interaction with the MAO A promoter was determined by transient transfection and luciferase assays using the following luciferase reporter gene constructs: 1) a segment of the MAO A core promoter containing only Sp/KLF-binding sites, 2) the MAO A promoter containing only three GREs (deleted Sp/KLF-binding sites), or 3) the MAO A 2 kb promoter (containing both Sp/KLF-binding sites and GREs) (22). These MAO A promoter-luciferase reporter gene constructs were cotransfected with the KLF11 vector (or the pcDNA3.1 vector) (22) and the p300-expression vector (28) in SH-SY5Y cells using Superfect transfection reagent (Qiagen) following the protocol of the manufacturer (21, 22).
ChIP Assays
SH-SY5Y cells (150-mm dish) were cross-linked by 1% formaldehyde for 10 min, scraped into PBS containing protease inhibitors (Sigma), and centrifuged. Cells were then resuspended in 350 μl of lysis buffer (1% SDS, 10 mm EDTA, and 50 mm Tris-HCl (pH 8.1)). Nuclear protein-DNA complexes were immunoprecipitated by incubation with anti-KLF11 (with BioMag goat anti-mouse) antibody overnight at 4 °C. DNA was recovered from the beads by elution buffer (1% SDS and 0.1 m NaHCO3) and analyzed by real-time PCR as described previously (22).
Klf11−/− Mice
The Klf11 homozygous knockout model was generated at the University of Washington, Seattle following standard homologous recombination techniques to inactivate the endogenous Klf11 gene in embryonic stem cells, generating chimeras, and isolating colony founders carrying the knockout gene (28, 36). This animal was originally generated in a mixed background and subsequently transferred to the Mayo Animal Facilities where it was crossed back into a pure C57BL/6 background for more than 20 generations to produce the inbred strain used in this study. In all of the experiments, male Klf11−/− animals were compared with age-matched male Klf11+/+ littermates.
CSD Experiments
Twenty adult, male Wistar rats (weighing 180–220 g) were provided with free access to Purina rat chow and water. Rats were housed in individual cages in a temperature- and humidity-controlled room with a reversed 12:12-h light/dark cycle (1). The chronic social stress induced in the experimental group was on the basis of the original resident-intruder paradigm (37, 38). Each rat (total = 10) was transferred from its home environment to a cage holding one of ten male, Long Evans “resident” rats (weighing 580–620 g, Harlan). Within 3 min, the intruder was attacked and defeated by the resident, as indicated by freezing behavior and submissive posture. The intruder and resident were then immediately separated, and the intruder was kept in a small plastic wire mesh compartment within the cage of the resident for 1 h. Subsequently, the intruder was released from the small cage back into its home habitat. This procedure was repeated once daily for 4 days during week 1, for 2 days for weeks 2 and 3, and for 4 days during week 4. The rats (control and stressed groups) were sacrificed by decapitation on day 29 (1).
The control group (10 rats) was maintained and handled in the same manner as the social defeated rats except for the stress exposure (CSD). All animal protocols were performed according to the Ethical Guidelines on Animal Experimentation and approved by the Institutional Animal Care and Use Committees.
Radioimmunoassay of Corticosterone Levels
Blood from decapitated rats was collected for determination of individual corticosterone levels by the Radioimmunoassay Laboratory at the University of Mississippi Medical Center using the Coat-A-Count rat corticosterone kit (Diagnostic Products Corp., Los Angeles, CA) (1).
HPLC Measures for Serotonin Levels
Rat brain tissue (100 mg) was homogenized on ice with a 0.5 ml solution containing 0.1 m perchloric acid, 0.3 mm EDTA, and 0.01 mm ascorbic acid. The resulting homogenate was stored on dry ice for ∼10 min, thawed, and centrifuged at 4 °C for 5 min (12,500 rpm). The supernatant was then used to determine serotonin levels.
Serotonin was separated by HPLC analysis using an HPLC system with a Waters 600 pump/controller, 717 autosampler, Waters 2465 electrochemical detector and a PerkinElmer Life Sciences C18 column with a guard column and a combination of isocratic elution. The mobile phase contained 50 mm anhydrous citric acid, 50 mm sodium acetate, 50 mm sodium hydroxide, EDTA, 1 mm sodium octyl sulfate, 7% methanol and 6% acetonitrile (pH 4.5). Ten microliters of each supernatant was injected per sample. The resulting pellets were dissolved in 0.1 m sodium hydroxide, and protein content was determined using the bicinchoninic acid kit (Pierce Biotechnology, Inc). Waters Millennium32 software was used for programming the pump flow rate, controlling the autosampler, and for acquisition of data and analysis.
Statistical Analysis
Statistical significance was evaluated using Student's t test for two group comparison or analysis of variance followed by Bonferroni adjusted tests when comparing more than two groups. A value of p < 0.05 was considered significant.
RESULTS
Glucocorticoid Exposure Activates the Expression and Nuclear Translocation of KLF11 (TIEG2) in Human Neuronal Cells
Both transcriptional activation and nuclear translocation are hallmarks of the activation of the KLF11 gene. Thus, we initially used the well characterized brain-derived SH-SY5Y cell line as a culture system for performing mechanistic studies to determine whether KLF11 is a glucocorticoid-inducible gene (Fig. 1). SH-SY5Y cells were treated with 100 nm dexamethasone (a synthetic glucocorticoid) for 48 h and KLF11 mRNA levels were determined by quantitative real-time RT-PCR. Approximately a 2.3-fold increase in KLF11 expression (p < 0.01, Fig. 1A) was observed following dexamethasone administration. As detected by Western blot analysis, KLF11 protein levels were increased similarly 1.8-fold in whole cell lysates (p < 0.05, Fig. 1B, lane 2 versus lane 1) upon dexamethasone treatment. Moreover, dexamethasone increased KLF11 protein levels 3.3-fold (p < 0.01, Fig. 1B, lane 4 versus lane 3) in the nuclear fraction, indicating that KLF11 translocated into the nucleus to regulate dexamethasone-induced regulation of its target genes, such as MAO A. We have documented previously that MAO A expression and activity were increased similarly by dexamethasone (21).
FIGURE 1.
Effects of dexamethasone (a synthetic glucocorticoid) on KLF11 (TIEG2) levels and nuclear translocation in a human neuronal cell line (SH-SY5Y). Cells were treated with or without dexamethasone (Dex) for either 48 h or 72 h as indicated. A, KLF11 mRNA levels were determined by quantitative real-time RT-PCR. B, KLF11 protein levels from cell lysates and nuclear extracts were examined and quantified by Western blot analysis. C, nuclear KLF11 protein expression was determined by immunofluorescence microscopy. KLF11 (red) and nuclear (blue, mounted in the presence of DAPI) staining and the superimposition of the two are shown. In addition, the relative distribution of nuclear and cytosolic KLF11 was analyzed by an immunofluorescent microscope, quantified by image analysis software (SlideBook), and expressed as the ratio of nucleus/cytosol as indicated with each group at the right. The cell number (n) is shown for each group.
Next, we visualized the translocation of KLF11 into the nucleus (Fig. 1C) after treatment with dexamethasone using immunofluorescence. The relative distribution of nuclear and cytosolic KLF11 was semiquantified using image analysis software (SlideBook) and expressed as the ratio of nucleus:cytosol. As shown in Fig. 1C, the ratio of nucleus:cytosol for the untreated control group was 1:3.75 and for the dexamethasone-treated group the ratio was 1:1.18 (p < 0.01), indicating that dexamethasone significantly increases KLF11 nuclear translocation at least 3-fold, which is consistent with the Western blot analysis (Fig. 1B). These results are the first to identify inducible cytoplasmic-to-nuclear shuttling of KLF11 in response to corticosteroid treatment, demonstrating the dynamic regulation of KLF11 localization in the nucleus that may underlie the stress-induced regulation of target genes by this important Kruppel-like transcription factor.
KLF11 Mediates Basal and Dexamethasone-induced MAO A mRNA Levels and Enzymatic Activity
Although KLF11 has been shown to up-regulate MAO B transcription (22), its action on MAO A remains unknown. Addressing this question is of significant medical relevance because both MAO isoforms participate in diseases and are targets of psychotropic therapy. Therefore, we quantified the effects of KLF11 on MAO A expression at both the mRNA and protein levels in SH-SY5Y cells stably transfected with KLF11 versus control vector (pcDNA), along with control-siRNA- and KLF11-siRNA-transfected cells (Fig. 2). The overexpressed KLF11 (Fig. 2Aa) (35) and siRNA-mediated KLF11 knockdown (Fig. 2Ba) (34) have been confirmed by Western blot analysis. We find that MAO A mRNA levels in KLF11-transfected samples were increased significantly ∼2-fold compared with pcDNA-transfected cells (p < 0.02, Fig. 2Ab, lane 2 versus lane 1). Moreover, this effect was further increased following 100 nm dexamethasone treatment, upon which KLF11-overexpressing cells showed a 4.2-fold increase in MAO A mRNA levels compared with controls (p < 0.02, Fig. 2Ab, lane 4 versus lane 3). Thus, KLF11 overexpression significantly amplified the dexamethasone-induced increase in MAO A mRNA levels 4-fold compared with untreated KLF11-overexpressing cells (p < 0.05, Fig. 2Ab, lane 4 versus lane 2).
FIGURE 2.
Effects of KLF11 (TIEG2) on MAO A mRNA and catalytic activity in human neuroblastoma cells. A, levels of KLF11 protein (a), MAO A mRNA (b), and catalytic activity levels (c) were determined by Western blot analysis, quantitative real-time RT-PCR, and enzymatic activity assays, respectively, in KLF11-overexpressing SH-SY5Y cells with or without dexamethasone (Dex) treatment. Data are expressed as fold increase relative to the control. *, p < 0.02 or **, p < 0.05 compared with control in each group (for example, Ab, lane 2 versus lane 1 and lane 4 versus lane 3). Ac, p < 0.05 (shown at the top) compares between KLF11-overexpressed groups (lane 4 versus lane 2). B, levels of KLF11 protein (a), MAO A mRNA (b), and catalytic activity (c) levels were determined by Western blot analysis, quantitative real-time RT-PCR, and enzymatic activity assays, respectively, in KLF11-knockdown (siRNA) cells with or without Dex treatment. Data are expressed as fold changes relative to the control group. *, p < 0.02 or **, p < 0.05 compared with the control in each group (Bb, lane 4 versus lane 3).
We complemented this investigation by analyzing the catalytic activity of MAO A in KLF11-overexpressing cells (Fig. 2Ac). Measurement of MAO A activity under steady-state conditions show an increase of ∼40% compared with control cells (p < 0.05, Fig. 2Ac, lane 2 versus lane 1). However, treatment of these cells with 100 nm dexamethasone increased MAO A activity even further, as KLF11-overexpressing cells exhibited twice the level of MAO A catalytic activity compared with control cells (p < 0.02, Fig. 2Ac, lane 4 versus lane 3). The dexamethasone-induced increase in MAO A catalytic activity was amplified significantly 2-fold compared with KLF11-overexpressing cells without dexamethasone treatment (p < 0.05, Fig. 2Ac, lane 4 versus lane 2). To better define the role of KLF11 in basal and dexamethasone-induced MAO A regulation, cells were treated with KLF11-siRNA to deplete endogenous KLF11 and compared with cells treated with control siRNA (Fig. 2B). Without dexamethasone treatment, MAO A mRNA levels were unchanged in KLF11 siRNA-transfected cells compared with control cells (Fig. 2Bb, lane 2 versus lane 1). However, dexamethasone-induced MAO A mRNA levels in KLF11 siRNA-transfected cells were decreased by ∼30% compared with control siRNA-treated cells (p < 0.05 Fig. 2Bb, lane 4 versus lane 3). Furthermore, MAO A enzymatic activity was not altered significantly with or without dexamethasone exposure in KLF11 siRNA-transfected cells compared with control siRNA-transfected cells (Fig. 2Bc, lane 4 versus lane 3 and lane 2 versus lane 1). Thus, these combined experiments, using a well characterized cell model for studying stress responses in SH-SY5Y cells, demonstrate that KLF11 mediates corticoid-induced up-regulation of MAO A at the mRNA, protein, and enzymatic activity levels.
Subsequently, we examined whether this pathway is operational in primary neuronal cell culture. We find that the glucocorticoid-KLF11-MAO A pathway is activated in rat primary cortical neurons in a similar fashion to that observed in cell line models (Fig. 3). KLF11 mRNA levels were increased ∼2.6-fold (p < 0.01 Fig. 3Aa) following dexamethasone administration. KLF11 protein levels doubled in the cell lysate from rat primary cortical neurons upon this treatment (p < 0.02, Fig. 3Ab, lane 2 versus lane 1), and the nuclear KLF11 level was increased more than 3-fold (p < 0.01, lane 4 versus lane 3). Also, the catalytic activity of MAO A was significantly increased with the treatment (p < 0.01, Fig. 3Ac, lane 2 versus lane 1). On the other hand, KLF11 siRNA treatment of the rat primary cortical neurons depleted endogenous KLF11 (Fig. 3Ba). Without dexamethasone treatment, MAO A mRNA levels were decreased by 33% in KLF11 siRNA-transfected cells compared with control cells (Fig. 3Bb, lane 2 versus lane 1, p < 0.05). Dexamethasone-induced MAO A mRNA levels in KLF11 siRNA-transfected cells were decreased by ∼47% compared with control siRNA-treated cells (p < 0.05, Fig. 2Bb, lane 4 versus lane 3, p < 0.01). In addition, MAO A enzymatic activity was reduced by ∼27% (3Bc, lane 2 versus lane 1, p < 0.08) without dexamethasone treatment and even more reduced (by ∼36%, lane 4 versus lane 3, p < 0.05) in KLF11 knockdown cells exposed to dexamethasone compared with control siRNA-transfected cells. These results suggest that dexamethasone has more effect on rat primary cortical neurons than on the SH-SY5Y cell line. Our data also demonstrate that KLF11 participates in the dexamethasone-induced transcriptional activation of MAO A.
FIGURE 3.
Effects of KLF11 (TIEG2) on MAO A mRNA and catalytic activity in rat primary cortical neurons. A, levels of KLF11 protein (a), mRNA (b), and MAO A (c) catalytic activity levels were determined by Western blot analysis, quantitative real-time RT-PCR, and enzymatic activity assays, respectively, in rat primary cortical neurons with or without dexamethasone (Dex) treatment. Data are expressed as fold increase relative to the control. B, levels of KLF11 protein (a), MAO A mRNA (b), and MAO A (c) catalytic activity levels were determined by Western blot analysis, quantitative real-time RT-PCR, and enzymatic activity assays, respectively, following siRNA-mediated KLF11 knockdown in rat primary cortical neurons with or without Dex treatment. Data are expressed as fold changes relative to the control group. *, p < 0.02 or **, p < 0.05 compared with the control in each group.
Identification of a p300-KLF11 Pathway That Activates MAO A Transcription at the Promoter Level
To determine whether the KLF11-induced increase in MAO A mRNA is the result of promoter activation, we assessed the activity of luciferase reporter constructs by a MAO A promoter fragment ligated upstream of this luciferase reporter gene vector (Fig. 4). Cells were transiently cotransfected with the MAO A-luciferase reporter construct and KLF11 or control vector (pcDNA3.1). Dexamethasone exposure increased luciferase activity, which is indicative of MAO A promoter activity, 1.8-fold compared with untreated control cells (p < 0.05, Fig. 4A, lane 2 versus lane 1). KLF11 overexpression doubled luciferase activity/MAO A promoter activity (p < 0.05, Fig. 4A, lane 3 versus lane 1). Activation of the MAO A promoter was increased ∼3.4-fold in KLF11 cotransfected cells treated with dexamethasone compared with untreated control cells (p < 0.01, Fig. 4A, lane 4 versus lane 1). MAO A promoter activity was also increased ∼1.8-fold following dexamethasone treatment of KLF11 cotransfected cells compared with transfected cells without dexamethasone treatment (p < 0.02, Fig. 4A, lane 4 versus lane 3). This study was complemented by ChIP assay to define whether the MAO A promoter is a direct target of KLF11 in vivo. Indeed, our results demonstrated that dexamethasone significantly increased the recruitment of KLF11 to the MAO A core promoter, which contains Sp/KLF binding sites (Fig. 4B), suggesting that activation of the MAO A promoter occurs, at least in part, via KLF11 recruitment. However, how KLF11 couples to distinct chromatin remodeling machines to regulate this gene remains unknown. Histone acetyl transferases (HATs), such as p300, have been shown to couple to KLF11 and other KLF transcription factors to regulate transcription in different cell systems (26, 36, 39) However, not much is known about the effect of these HATs in the central nervous system. The potential regulation of MAO A by p300 is of significant clinical relevance because patients who present with mutations in this gene (Rubinstein-Taybi syndrome) are affected by severe mental illnesses (40) and other central nervous system abnormalities (41). Therefore, we investigated the effect of p300 on the KLF11-MAO A pathway using transient cotransfection of the MAO A promoter-luciferase reporter with a p300 and KLF11 expression construct (Fig. 5). Activation of MAO A by KLF11 was greatly increased when cotransfected with p300 (Fig. 5, lane 4 versus lane 2, p < 0.01), revealing that this HAT augments KLF11-mediated transcriptional activation of MAO A. Thus, these results, combined with the rest of our cell biological studies, outline a new pathway that is initiated by glucocorticoids and acts through KLF11 to bind and activate MAO A via a p300-dependent mechanism.
FIGURE 4.
KLF11 (TIEG2) activates MAO A gene expression by interaction with the MAO A core promoter region containing Sp/KLF-binding sites (from −245 bp to −14 bp). A, transient transfection and luciferase assay. Cells were cotransfected with the MAO A 2-kb promoter-luciferase reporter gene and KLF11 expression construct (or empty control vector, pcDNA3.1) with or without dexamethasone (Dex) treatment as indicated. Luciferase activity was then determined. *, p < 0.01 and **, p < 0.05 compared with the control group (lane 1). p < 0.02 (shown at the top) compares lane 4 to lane 3. B, ChIP assays. Binding of KLF11 to the MAO A core promoter or 5′-irrelevant region of MAO A in vivo was determined by ChIP assay followed by quantitative real-time RT-PCR using SH-SY5Y cells. A schematic representation of the MAO A promoter is shown in the top panel. Real-time PCR-targeted regions containing the core promoter (from −245 bp to −14 bp) and 5′-irrelevant locus (from −1666 to −1542 bp) for negative control are indicated. Association of KLF11 with the MAO A core promoter or 5′-irrelevant locus is shown in the bottom panel. Relative differences between the input sample, KLF11 ChIP assay, and negative control were determined. These values are presented as percent input (mean ± S.D.) in which a DNA cross-link input sample was taken as 100% and are from triplicate samples of three independent experiments (22). KLF11 binds to the Sp1-binding sites in the natural promoter of MAO A, and this association is enhanced by dexamethasone (lane 4 versus lane 3; *, p < 0.005).
FIGURE 5.
The effect of a histone acetyltransferase, p300, on KLF11 (TIEG2)-induced MAO A transcriptional activity was determined by transient transfection and luciferase assays. Cells were cotransfected with the MAO A 2-kb promoter-luciferase reporter gene, KLF11 expression construct (or empty control vector, pcDNA3.1), and the wild-type p300-expression construct. Following 48 h incubation, cells were harvested and then assayed for luciferase activity. *, p < 0.02 (lane 2 versus lane 1); p < 0.01 (shown at the top) compares lane 4 to lane 2.
Both Sp/KLF-binding Sites and GREs Independently Contribute to Dexamethasone-induced MAO A Promoter Activity
To elucidate additional mechanisms underlying glucocorticoid-induced MAO A expression, the effects of dexamethasone on the MAO A core promoter (which only contains putative Sp/KLF binding sites), the MAO A promoter (which only contains three GREs) with deleted Sp/KLF-binding sites and the MAO A 2-kb promoter (containing both Sp/KLF-binding sites and GREs) were assessed independently by transient transfection and luciferase assays (Fig. 6, A and B). As shown in Fig. 6B, dexamethasone exposure increases Sp/KLF11-mediated activation of MAO A core promoter activity by 50% (lane 3 versus lane 2, p < 0.05) and MAO A promoter containing the activity of three GREs 2-fold (lanes 7 versus 6; p < 0.05), respectively.
FIGURE 6.
Effects of Sp/KLF-binding sites and GREs on glucocorticoid-induced MAO A activity. A, illustration of the human MAO A 2-kb promoter including both Sp/KLF-binding sites and GREs. B, MAO A promoter activity was determined by transient transfection and luciferase assay in SH-SY5Y cells transfected with the MAO A core promoter (only containing Sp/KLF-binding sites, lanes 1–4), MAO A promoter only containing three GREs (lanes 5–8), or MAO A 2-kb promoter (containing both Sp/KLF-binding sites and GREs, lanes 9–12) and KLF11 expression vector with or without dexamethasone (Dex) and/or GR. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with the basal control in each group (lanes 1, 5, or 9); #, p < 0.02 compared with cells (without GR) in each group (lanes 7 or 11).
However, transfection of KLF11 did not increase the GRE-containing MAO A promoter activity (Fig. 6B, lane 6 versus lane 5), further supporting that KLF11 regulates MAO A gene expression through Sp/KLF-binding sites. Transfection of the GR increased dexamethasone-induced GRE-containing promoter activity 2.0-fold (Fig. 6B, lane 8 versus lane 7, p < 0.02). Conversely, the addition of GR did not significantly increase MAO A core promoter activity (Fig. 6B, lane 4 versus lane 3).
Furthermore, when cells were transfected with the MAO A 2-kb promoter (containing both Sp/KLF-binding sites and three GREs) and treated with dexamethasone, the 2-kb promoter activity was increased by 80% (Fig. 6B, lane 11 versus lane 10. p < 0.05). Following the transfection of GR, the MAO A 2-kb promoter activity was increased further 2.0-fold compared with cells treated with dexamethasone alone (Fig. 6B, lane 12 versus lane 11, p < 0.02), which also showed an additive effect compared with those of MAO A promoters only containing either Sp/KLF-binding sites (lane 12 versus lane 4) or GREs alone (lane 12 versus lane 8). These results suggested that both Sp/KLF sites and GREs exist independently to activate MAO A expression, although quantitatively, GR-induced MAO A expression is regulated via GREs more than through Sp/KLF-binding sites (Fig. 6B, lane 8 versus lane 7, p < 0.02; lane 12 versus lane 11, p < 0.02; and lane 12 versus lane 4, p < 0.02).
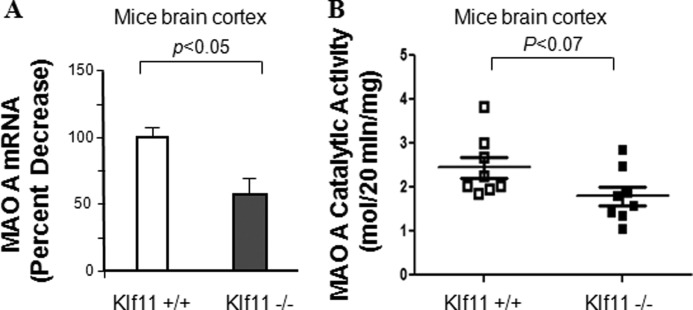
KLF11 Knockout Mice Have Reduced MAO A mRNA Levels and Enzymatic Activity
To verify the importance of KLF11 as a novel transcriptional activator for MAO A gene expression, we investigated whether this pathway is operational in mice carrying a germ line inactivation of KLF11 (Klf11−/− mice). The levels of both MAO A mRNA and catalytic activity were determined in the brain cortex of Klf11 −/− mice and compared with Klf11+/+ littermates (Fig. 7). The mRNA level of MAO A was reduced by 43% in KLF11 knockout mice compared with mice expressing wild-type KLF11 (p < 0.05, Fig. 7A). The enzymatic activity of MAO A was decreased by 26% in the brain cortex of knockout mice compared with the wild type (p < 0.07, Fig. 7B). Together, this data provides in vivo evidence for the role of KLF11 as an upstream transcriptional activator of MAO A gene expression in the central nervous system. These findings are also congruent with the studies in rat primary cortical neurons that were depleted of endogenous KLF11 by siRNA, which resulted in reduced MAO A mRNA levels (Fig. 3Bb, lane 2 versus lane 1, p < 0.05) and MAO A enzymatic activity levels (Bc, lane 2 versus lane 1, p < 0.08) without dexamethasone treatment.
FIGURE 7.
MAO A activity in KLF11 (TIEG2) knockout mouse brains. MAO A mRNA (A) and MAO A catalytic activity levels (B) were evaluated by real-time RT-PCR and enzymatic activity assays, respectively, in brain cortex from knockout (Klf11 −/−) and wild-type (Klf11 +/+) male mice. Data represent the mean ± S.E. of eight male mice (n = 8) in each group.
Activation of the Glucocorticoid-KLF11-MAO A Pathway during CSD Stress
Previous studies have amply demonstrated that many of the effects underlying CSD stress proceeds primarily via corticosteroids (glucocorticoids in rats), making this model an optimal tool to validate whether the new KLF11-MAO A pathway is operational under conditions that simulate the psychosocial stress commonly observed in humans with depression and other stress-related mood disorders. For this purpose, rats were subjected to CSD stress, and MAO A RNA and protein levels were measured. To confirm that the CSD stress was conducted effectively, the levels of blood corticosterone (glucocorticoids) in these rats were determined. As shown in Fig. 8, serum corticosterone levels in rats treated with CSD stress were elevated ∼2.8-fold compared with untreated rats (p < 0.002, Fig. 8A). Because MAO A oxidizes serotonin, brain levels of serotonin were also determined. As expected, serotonin levels were decreased significantly in both the cortex (by 29%, p < 0.001) and thalamus (by 45%, p < 0.03) (Fig. 8B) of CSD-treated rats compared with respective controls. Together, these results demonstrate that the biochemical and physiological parameters of CSD stress are recapitulated in our animal model system. Thus, using this validated model, we measured mRNA and activity levels of MAO A from the brain cortex and thalamus of control and CSD rats (Fig. 9). Our results show that MAO A mRNA expression and catalytic activity were increased significantly in the prefrontal cortex of CSD rats compared with controls, ∼3.2-fold (p < 0.01, Fig. 9Aa) and ∼1.5-fold (p < 0.02, Fig. 9Ab), respectively, in the cortex. Likewise, MAO A mRNA and catalytic activity were increased 3-fold (p < 0.02, Fig. 9Ba) and ∼2-fold (p < 0.05, Fig. 8Bb), respectively, in the rat brain thalamus exposed to CSD stress. Therefore, chronic stress induced by CSD increases MAO A gene expression and enzymatic activity in the rat brain cortex and thalamus.
FIGURE 8.
Effects of chronic social stress on rat blood corticosterone (glucocorticoids) and brain serotonin levels after a 28-day exposure to CSD stress (an animal model for depression). A, blood corticosterone levels were determined by radioimmunoassay. B, serotonin levels in the rat cortex or thalamus were measured by HPLC. Data represent the mean ± S.E. of 10 rats (n = 10) in each group.
FIGURE 9.
Effects of chronic social stress on MAO A mRNA and catalytic (enzymatic) activity in the brain tissue of rats exposed to CSD stress (an animal model for depression) compared with unexposed control rats. Stressed rats were exposed to CSD for 28 days. MAO A levels from the brain cortex (A) or brain thalamus (B) were determined by quantitative real-time RT-PCR (for MAO A mRNA levels) (a) and by enzymatic activity assay (for MAO A catalytic activity levels) (b), respectively. Data represent the mean ± S.E. of 10 rats (n = 10) in each group.
Expression of the Transcription Factor, KLF11 (TIEG2), Is Increased Significantly in the Rat Brain Cortex and Thalamus by CSD Stress
Because changes in MAO expression rely on the activity of certain transcription factors, we reasoned that a key regulatory protein may be altered under stressful conditions. To investigate whether the increase of MAO A by CSD was due to changes in KLF11 levels, we determined mRNA and protein levels of KLF11 by quantitative real-time RT-PCR and Western blot analysis, respectively. As shown in Fig. 10, KLF11 mRNA and protein levels were elevated in the cortex of rats exposed to CSD stress compared with controls, ∼3.5-fold (p < 0.005, Fig. 10Aa) and ∼1.7-fold (p < 0.03, Fig. 10Ab), respectively. As expected, KLF11 mRNA and protein levels were also increased in the brain thalamus ∼3-fold (p < 0.01, Fig. 10Ba) and ∼2-fold (p < 0.04, Fig. 10Bb), respectively, compared with unexposed control rats. Thus, these results, which are congruent with our in vitro investigation using isolated neuronal cell lines and primary culture, help to establish that the activation of the glucocorticoid-KLF11-MAO A pathway is operational in the brain cortex and thalamus and is activated under conditions that model the chronic social stress observed in humans.
FIGURE 10.
Effects of chronic social stress on KLF11 (TIEG2) levels in the brain tissue of rats after a 28-day exposure to CSD stress (an animal model for depression). KLF11 levels were determined from the brain cortex (A) or brain thalamus (B). a, KLF11 mRNA levels were quantified by real-time RT-PCR. b, quantitative Western blot analysis of KLF11. Each KLF11 band was evaluated by its relative intensity and normalized to the density of β-actin. Representative Western blot analyses from three untreated controls and three stressed rats are shown in the bottom panels.
DISCUSSION
Abnormalities of MAO A levels and activity have been associated with a number of psychiatric disorders (8–13, 42, 43). It is critical to explore the molecular basis for regulation of MAO A expression and enzymatic function. Through data from relevant cell culture systems under cellular stress, a KLF11-deficient mice model and a rat model for depression (generated by CSD stress), this study reports a novel pathway of stress-induced, KLF11-p300-mediated activation of MAO A expression.
We first found that MAO A and KLF11 expression levels showed a positive correlation. Dexamethasone exposure further augments MAO A activity, which correlates with increased KLF11 expression. ChIP assay results suggest that this up-regulation occurs by the preferential binding of KLF11 to Sp/KLF cis-regulatory sites within the MAO A core promoter, triggering p300-mediated chromatin remodeling and promoter activation.
Our data suggest that both KLF11 and GR contribute to glucocorticoid-induced activation of MAO A through Sp/KLF-binding sites and GREs, respectively. These independent but redundant MAO A transcriptional activation pathways, by KLF11 and GR, may ensure faster and/or greater activation (GR-mediated) in the presence of elevated glucocorticoid levels and may also ensure the long lasting maintenance of MAO A expression (KLF11-mediated) once glucocorticoid levels have decreased during exposure to chronic social stress. Further investigation is needed to fully understand the dynamics of this feed-forward loop.
In rats exposed to CSD stress, this study documents the significant increase in serum corticosterone (glucocorticoids) level along with increased MAO A and KLF11 expression, supporting the results from in vitro experiments. Considering the fact that KLF11 regulates a large number of target genes involved in different processes, such as metabolism, cell cycle, and apoptosis, in addition to MAO A as shown here, this finding implies that stress and stress hormones can alter the expression pattern of a gene network, therefore causing fundamental changes in the cells.
Our model is further reinforced by the observation that dexamethasone-induced nuclear translocation of KLF11 facilitates activation of its target genes. Notably, the translocation of KLF11 into the nucleus parallels the nuclear translocation of the multifunctional protein GAPDH, which has been implicated in cellular apoptosis (44, 45). KLF11 has been shown to associate intranuclearly with GAPDH and to promote neuronal cell degeneration and death via the GAPDH-KLF11-MAO B cascade (23, 34). This interaction may also occur in the cytosol, similar to the complex of the ubiquitin ligase Siah1 and GAPDH (45, 46). Like Siah1, KLF11 contains a putative translocation signal that may facilitate GAPDH-nuclear translocation. KLF11 knockout mice (Klf11−/−) were further used to investigate MAO A mRNA and catalytic activity, which supports the substantial role of KLF11 in the up-regulation of MAO A. These findings are congruent with the decrease in MAO A expression following KLF11 knockdown in primary cortical neurons compared with controls (Fig. 3). Interestingly, the differences in MAO A expression were readily amplified after dexamethasone exposure. Thus, effects of CSD stress in KLF11 knockout mice would need to be investigated in the future. It is expected that KLF11 knockout mice exposed to chronic social stress would exhibit reduced MAO A induction as compared with the wild-type mice.
Additionally, the consistent results of increased KLF11 and elevated MAO A in the CSD chronic stress rat model and in dexamethasone-treated cells (14) validated the importance of stress-mediated KLF11 up-regulation of MAO A in chronic biological stress and depression as it parallels other recent publications using the CSD stress paradigm in rats or mice as a model for depression (1, 29–31).
In summary, our study, using relevant cellular and molecular approaches in models ranging from cells to laboratory rodents, suggests that KLF11 (TIEG2) is a novel transcriptional activator for MAO A gene expression. We have shown that stress increases KLF11 expression and induces its nuclear translocation both in vivo and in vitro and contributes to increase in MAO A expression. Thus, future studies of the interactions of MAO A and its transcription factors could provide insight into novel psychotherapies. Finally, we have demonstrated that KLF11 couples to a HAT, p300, to regulate MAO A. This knowledge, combined with the existence of HAT inhibitors already in advanced phases of clinical trials, suggests that the activity of this transcriptional pathway could be manipulated pharmacologically to combat stress-related disorders such as depression.
This work was supported, in whole or in part, by National Institutes of Health Grant DK 52913 (to R. U.) and by National Institute on Alcohol Abuse and Alcoholism Grant R01AA020103 (to X. O.). This work was also supported by Public Health Service Grant P20 RR 017701 (to C. A. S.), by the National Alliance for Research on Schizophrenia and Depression (to J. H. M. and X. O.), by the Canadian Institutes of Health Research (to P. R. A. and J. H. M.), and by an intramural research support grant from the University of Mississippi Medical Center (to J. W. and X. O.). J. H. M. has applied for a patent to employ MAO measures to diagnose or identify subtypes of major depressive disorder. He has had operating grant support/consultation with GlaxoSmithKline, BristolMyersSquibb, Lundbeck, SK Life Sciences, and Eli-Lilly.
- MAO
- monoamine oxidase
- GR
- glucocorticoid receptor
- GRE
- glucocorticoid response DNA element
- KLF
- Kruppel-like factor
- CSD
- chronic social defeat
- HAT
- histone acetyl transferase.
REFERENCES
- 1. Kieran N., Ou X. M., Iyo A. H. (2010) Chronic social defeat down-regulates the 5-HT1A receptor but not Freud-1 or NUDR in the rat prefrontal cortex. Neurosci. Lett. 469, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen Y., Wang Q., Wang F. F., Gao H. B., Zhang P. (2012) Stress induces glucocorticoid-mediated apoptosis of rat Leydig cells in vivo. Stress 15, 74–84 [DOI] [PubMed] [Google Scholar]
- 3. Dietz D. M., Laplant Q., Watts E. L., Hodes G. E., Russo S. J., Feng J., Oosting R. S., Vialou V., Nestler E. J. (2011) Paternal transmission of stress-induced pathologies. Biological Psychiatry 70, 408–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Y., Wang H. D., Xia X., Kung H. F., Pan Y., Kong L. D. (2007) Behavioral and biochemical studies of total furocoumarins from seeds of Psoralea corylifolia in the chronic mild stress model of depression in mice. Phytomedicine 14, 523–529 [DOI] [PubMed] [Google Scholar]
- 5. Mao Q. Q., Ip S. P., Ko K. M., Tsai S. H., Xian Y. F., Che C. T. (2009) Effects of peony glycosides on mice exposed to chronic unpredictable stress. Further evidence for antidepressant-like activity. J. Ethnopharmacol. 124, 316–320 [DOI] [PubMed] [Google Scholar]
- 6. Thorpe L. W., Westlund K. N., Kochersperger L. M., Abell C. W., Denney R. M. (1987) Immunocytochemical localization of monoamine oxidases A and B in human peripheral tissues and brain. J. Histochem. Cytochem. 35, 23–32 [DOI] [PubMed] [Google Scholar]
- 7. Youdim M. B., Edmondson D., Tipton K. F. (2006) The therapeutic potential of monoamine oxidase inhibitors. Nat. Rev. Neurosci 7, 295–309 [DOI] [PubMed] [Google Scholar]
- 8. Johnson S., Stockmeier C. A., Meyer J. H., Austin M. C., Albert P. R., Wang J., May W. L., Rajkowska G., Overholser J. C., Jurjus G., Dieter L., Johnson C., Sittman D. B., Ou X. M. (2011) The reduction of R1, a novel repressor protein for monoamine oxidase A, in major depressive disorder. Neuropsychopharmacology 36, 2139–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Du L., Faludi G., Palkovits M., Sotonyi P., Bakish D., Hrdina P. D. (2002) High activity-related allele of MAO-A gene associated with depressed suicide in males. Neuroreport 13, 1195–1198 [DOI] [PubMed] [Google Scholar]
- 10. Du L., Bakish D., Ravindran A., Hrdina P. D. (2004) MAO-A gene polymorphisms are associated with major depression and sleep disturbance in males. Neuroreport 15, 2097–2101 [DOI] [PubMed] [Google Scholar]
- 11. Meyer J. H., Wilson A. A., Sagrati S., Miler L., Rusjan P., Bloomfield P. M., Clark M., Sacher J., Voineskos A. N., Houle S. (2009) Brain monoamine oxidase A binding in major depressive disorder. Relationship to selective serotonin reuptake inhibitor treatment, recovery, and recurrence. Arch. Gen. Psychiatry 66, 1304–1312 [DOI] [PubMed] [Google Scholar]
- 12. Meyer J. H., Ginovart N., Boovariwala A., Sagrati S., Hussey D., Garcia A., Young T., Praschak-Rieder N., Wilson A. A., Houle S. (2006) Elevated monoamine oxidase A levels in the brain. An explanation for the monoamine imbalance of major depression. Arch. Gen. Psychiatry 63, 1209–1216 [DOI] [PubMed] [Google Scholar]
- 13. Sacher J., Wilson A. A., Houle S., Rusjan P., Hassan S., Bloomfield P. M., Stewart D. E., Meyer J. H. (2010) Elevated brain monoamine oxidase A binding in the early postpartum period. Arch. Gen. Psychiatry 67, 468–474 [DOI] [PubMed] [Google Scholar]
- 14. Morilak D. A., Frazer A. (2004) Antidepressants and brain monoaminergic systems. A dimensional approach to understanding their behavioural effects in depression and anxiety disorders. Int. J. Neuropsychopharmacol. 7, 193–218 [DOI] [PubMed] [Google Scholar]
- 15. Schildkraut J. J., Kety S. S. (1967) Biogenic amines and emotion. Science 156, 21–37 [DOI] [PubMed] [Google Scholar]
- 16. Filipenko M. L., Beilina A. G., Alekseyenko O. V., Dolgov V. V., Kudryavtseva N. N. (2002) Repeated experience of social defeats increases serotonin transporter and monoamine oxidase A mRNA levels in raphe nuclei of male mice. Neurosci. Lett. 321, 25–28 [DOI] [PubMed] [Google Scholar]
- 17. Ou X. M., Chen K., Shih J. C. (2006) Monoamine oxidase A and repressor R1 are involved in apoptotic signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 103, 10923–10928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doyle A., Hucklebridge F., Evans P., Clow A. (1996) Salivary monoamine oxidase A and B inhibitory activities correlate with stress. Life Sci. 59, 1357–1362 [DOI] [PubMed] [Google Scholar]
- 19. Manoli I., Le H., Alesci S., McFann K. K., Su Y. A., Kino T., Chrousos G. P., Blackman M. R. (2005) Monoamine oxidase-A is a major target gene for glucocorticoids in human skeletal muscle cells. FASEB J. 19, 1359–1361 [DOI] [PubMed] [Google Scholar]
- 20. Jahng J. W., Kim N. Y., Ryu V., Yoo S. B., Kim B. T., Kang D. W., Lee J. H. (2008) Dexamethasone reduces food intake, weight gain and the hypothalamic 5-HT concentration and increases plasma leptin in rats. Eur. J. Pharmacol. 581, 64–70 [DOI] [PubMed] [Google Scholar]
- 21. Ou X. M., Chen K., Shih J. C. (2006) Glucocorticoid and androgen activation of monoamine oxidase A is regulated differently by R1 and Sp1. J. Biol. Chem. 281, 21512–21525 [DOI] [PubMed] [Google Scholar]
- 22. Ou X. M., Chen K., Shih J. C. (2004) Dual functions of transcription factors, transforming growth factor-β-inducible early gene (TIEG)2 and Sp3, are mediated by CACCC element and Sp1 sites of human monoamine oxidase (MAO) B gene. J. Biol. Chem. 279, 21021–21028 [DOI] [PubMed] [Google Scholar]
- 23. Ou X. M., Stockmeier C. A., Meltzer H. Y., Overholser J. C., Jurjus G. J., Dieter L., Chen K., Lu D., Johnson C., Youdim M. B., Austin M. C., Luo J., Sawa A., May W., Shih J. C. (2010) A novel role for glyceraldehyde-3-phosphate dehydrogenase and monoamine oxidase B cascade in ethanol-induced cellular damage. Biol. Psychiatry 67, 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cao S., Fernandez-Zapico M. E., Jin D., Puri V., Cook T. A., Lerman L. O., Zhu X. Y., Urrutia R., Shah V. (2005) KLF11-mediated repression antagonizes Sp1/sterol-responsive element-binding protein-induced transcriptional activation of caveolin-1 in response to cholesterol signaling. J. Biol. Chem. 280, 1901–1910 [DOI] [PubMed] [Google Scholar]
- 25. Buttar N. S., DeMars C. J., Lomberk G., Rizvi S., Bonilla-Velez J., Achra S., Rashtak S., Wang K. K., Fernandez-Zapico M. E., Urrutia R. (2010) Distinct role of Kruppel-like factor 11 in the regulation of prostaglandin E2 biosynthesis. J. Biol. Chem. 285, 11433–11444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fernandez-Zapico M. E., van Velkinburgh J. C., Gutiérrez-Aguilar R., Neve B., Froguel P., Urrutia R., Stein R. (2009) MODY7 gene, KLF11, is a novel p300-dependent regulator of Pdx-1 (MODY4) transcription in pancreatic islet β cells. J. Biol. Chem. 284, 36482–36490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garin I., Edghill E. L., Akerman I., Rubio-Cabezas O., Rica I., Locke J. M., Maestro M. A., Alshaikh A., Bundak R., del Castillo G., Deeb A., Deiss D., Fernandez J. M., Godbole K., Hussain K., O'Connell M., Klupa T., Kolouskova S., Mohsin F., Perlman K., Sumnik Z., Rial J. M., Ugarte E., Vasanthi T., Neonatal Diabetes International Group, Johnstone K., Flanagan S. E., Martínez R., Castaño C., Patch A. M., Fernández-Rebollo E., Raile K., Morgan N., Harries L. W., Castaño L., Ellard S., Ferrer J., Perez de Nanclares G., Hattersley A. T. (2010) Recessive mutations in the INS gene result in neonatal diabetes through reduced insulin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 3105–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bonnefond A., Lomberk G., Buttar N., Busiah K., Vaillant E., Lobbens S., Yengo L., Dechaume A., Mignot B., Simon A., Scharfmann R., Neve B., Tanyolaç S., Hodoglugil U., Pattou F., Cavé H., Iovanna J., Stein R., Polak M., Vaxillaire M., Froguel P., Urrutia R. (2011) Disruption of a novel Kruppel-like transcription factor p300-regulated pathway for insulin biosynthesis revealed by studies of the c.-331 INS mutation found in neonatal diabetes mellitus. J. Biol. Chem. 286, 28414–28424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vialou V., Maze I., Renthal W., LaPlant Q. C., Watts E. L., Mouzon E., Ghose S., Tamminga C. A., Nestler E. J. (2010) Serum response factor promotes resilience to chronic social stress through the induction of ΔFosB. J. Neurosci. 30, 14585–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Covington H. E., 3rd, Lobo M. K., Maze I., Vialou V., Hyman J. M., Zaman S., LaPlant Q., Mouzon E., Ghose S., Tamminga C. A., Neve R. L., Deisseroth K., Nestler E. J. (2010) Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. J. Neurosci. 30, 16082–16090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. LaPlant Q., Vialou V., Covington H. E., 3rd, Dumitriu D., Feng J., Warren B. L., Maze I., Dietz D. M., Watts E. L., Iñiguez S. D., Koo J. W., Mouzon E., Renthal W., Hollis F., Wang H., Noonan M. A., Ren Y., Eisch A. J., Bolaños C. A., Kabbaj M., Xiao G., Neve R. L., Hurd Y. L., Oosting R. S., Fan G., Morrison J. H., Nestler E. J. (2010) Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat. Neurosci. 13, 1137–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson S., Tazik S., Lu D., Johnson C., Youdim M. B., Wang J., Rajkowska G., Ou X. M. (2010) The new inhibitor of monoamine oxidase, M30, has a neuroprotective effect against dexamethasone-induced brain cell apoptosis. Front. Neurosci. 4, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ou X. M., Storring J. M., Kushwaha N., Albert P. R. (2001) Heterodimerization of mineralocorticoid and glucocorticoid receptors at a novel negative response element of the 5-HT1A receptor gene. J. Biol. Chem. 276, 14299–14307 [DOI] [PubMed] [Google Scholar]
- 34. Ou X. M., Lu D., Johnson C., Chen K., Youdim M. B., Rajkowska G., Shih J. C. (2009) Glyceraldehyde-3-phosphate dehydrogenase-monoamine oxidase B-mediated cell death-induced by ethanol is prevented by rasagiline and 1-R-aminoindan. Neurotox. Res. 16, 148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu D., Johnson C., Johnson S., Tazik S., Ou X. M. (2008) The neuroprotective effect of antidepressant drug via inhibition of TIEG2-MAO B-mediated cell death. Drug Discov. Ther. 2, 289–295 [PMC free article] [PubMed] [Google Scholar]
- 36. Song C. Z., Keller K., Murata K., Asano H., Stamatoyannopoulos G. (2002) Functional interaction between coactivators CBP/p300, PCAF, and transcription factor FKLF2. J. Biol. Chem. 277, 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miczek K. A., Thompson M. L., Shuster L. (1982) Opioid-like analgesia in defeated mice. Science 215, 1520–1522 [DOI] [PubMed] [Google Scholar]
- 38. Miczek K. A., Nikulina E. M., Shimamoto A., Covington H. E., 3rd. (2011) Escalated or suppressed cocaine reward, tegmental BDNF, and accumbal dopamine caused by episodic versus continuous social stress in rats. J. Neurosci. 31, 9848–9857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Urvalek A. M., Wang X., Lu H., Zhao J. (2010) KLF8 recruits the p300 and PCAF co-activators to its amino terminal activation domain to activate transcription. Cell Cycle 9, 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Verhoeven W. M., Tuinier S., Kuijpers H. J., Egger J. I., Brunner H. G. (2010) Psychiatric profile in Rubinstein-Taybi syndrome. A review and case report. Psychopathology 43, 63–68 [DOI] [PubMed] [Google Scholar]
- 41. Janknecht R. (2002) The versatile functions of the transcriptional coactivators p300 and CBP and their roles in disease. Histol. Histopathol. 17, 657–668 [DOI] [PubMed] [Google Scholar]
- 42. Shih J. C., Chen K., Ridd M. J. (1999) Monoamine oxidase. From genes to behavior. Annu. Rev. Neurosci. 22, 197–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cases O., Seif I., Grimsby J., Gaspar P., Chen K., Pournin S., Müller U., Aguet M., Babinet C., Shih J. C. (1995) Aggressive behavior and altered amounts of brain serotonin and norepinephrine in mice lacking MAOA. Science 268, 1763–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sawa A., Khan A. A., Hester L. D., Snyder S. H. (1997) Glyceraldehyde-3-phosphate dehydrogenase. Nuclear translocation participates in neuronal and nonneuronal cell death. Proc. Natl. Acad. Sci. U.S.A. 94, 11669–11674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hara M. R., Agrawal N., Kim S. F., Cascio M. B., Fujimuro M., Ozeki Y., Takahashi M., Cheah J. H., Tankou S. K., Hester L. D., Ferris C. D., Hayward S. D., Snyder S. H., Sawa A. (2005) S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7, 665–674 [DOI] [PubMed] [Google Scholar]
- 46. Sen N., Hara M. R., Kornberg M. D., Cascio M. B., Bae B. I., Shahani N., Thomas B., Dawson T. M., Dawson V. L., Snyder S. H., Sawa A. (2008) Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat. Cell Biol. 10, 866–873 [DOI] [PMC free article] [PubMed] [Google Scholar]