Background: The heterogeneity and flexibility of HIV-1 envelope glycoprotein N-glycans interfere with structural and vaccine studies.
Results: HIV-1 envelope trimers can be partially deglycosylated without affecting trimer integrity.
Conclusion: HIV-1 envelope glycoprotein N-glycans do not contribute to trimer integrity once the protein is folded.
Significance: Deglycosylated HIV-1 envelope trimers should be useful for structural and vaccine studies.
Keywords: Glycobiology, Glycoprotein, Glycosylation, Protein Chemistry, Vaccine Development, Viral Immunology, Viral Protein, Virology, Virus Entry, Virus Structure
Abstract
The trimeric envelope glycoprotein complex (Env) is the focus of vaccine development programs aimed at generating protective humoral responses to human immunodeficiency virus type 1 (HIV-1). N-Linked glycans, which constitute almost half of the molecular mass of the external Env domains, produce considerable structural heterogeneity and are a major impediment to crystallization studies. Moreover, by shielding the peptide backbone, glycans can block attempts to generate neutralizing antibodies against a substantial subset of potential epitopes when Env proteins are used as immunogens. Here, we describe the partial deglycosylation of soluble, cleaved recombinant Env trimers by inhibition of the synthesis of complex N-glycans during Env production, followed by treatment with glycosidases under conditions that preserve Env trimer integrity. The partially deglycosylated trimers are stable, and neither abnormally sensitive to proteolytic digestion nor prone to aggregation. Moreover, the deglycosylated trimers retain or increase their ability to bind CD4 and antibodies that are directed to conformational epitopes, including the CD4-binding site and the V3 region. However, as expected, they do not react with glycan-dependent antibodies 2G12 and PGT123, or the C-type lectin receptor DC-SIGN. Electron microscopic analysis shows that partially deglycosylated trimers have a structure similar to fully glycosylated trimers, indicating that removal of glycans does not substantially perturb the structural integrity of the trimer. The glycan-depleted Env trimers should be useful for structural and immunogenicity studies.
Introduction
The Env complex of HIV-1 is a trimer of gp120/gp41 heterodimers. This structure, often referred to as a spike, attaches to receptors on target cells and mediates the fusion of the viral and cell membranes at the start of the infection process. Antibodies that bind to the native spike impede one or more stages in the virus-cell fusion process and thereby neutralize infectivity. The HIV-1 Env complex is, therefore, the focus of vaccine development strategies intended to prevent the establishment of infection via the induction of virus-neutralizing antibodies (NAbs).4
The Env gp120 and gp41 subunits are extensively glycosylated, with N-linked glycans constituting approximately half the gp120 mass. There are between 18 and 33 (median, 25) canonical N-linked carbohydrate attachment sites (NX(S/T)) on gp120 proteins from different isolates (1, 2). Although most of these sites are utilized, the glycosylation can be both isolate-dependent and influenced by the producer cell type (3, 4). The number of glycan sites present on Env, and the extent to which they are glycosylated, tend to increase during the course of HIV-1 infection, reflecting antibody-mediated selection pressures that drive resistance to autologous NAbs (5). Gp41 contains 4 or 5 canonical N-linked glycosylation sites, but there are conflicting reports about the extent to which they are actually occupied by a glycan (4, 6–8). One or a few O-linked carbohydrates might also be present on gp120, but would contribute little to the glycan mass as the individual structures have been reported to be small (9, 10).
Glycosylation is a post-translational modification that contributes to the proper folding of the Env protein in the endoplasmic reticulum. Thus, when N-glycosylation is blocked by enzyme inhibitors or by eliminating the canonical sites, gp120 does not fold properly (8, 11–13). The modification or removal of carbohydrates from virions can affect Env function and virus infectivity (14–17). N-Glycans are also involved in the attachment of virions to certain cell types that express appropriate C-type lectin receptors, such as DC-SIGN or Langerin, events that may be important for the establishment and/or systemic spread of HIV-1 infection in a new host (18, 19). Under some circumstances, the N-glycans can trigger immunosuppressive responses that may interfere with anti-Env antibody production (20–25). The glycan component of Env helps counter host immunity by occluding underlying or topologically adjacent protein epitopes (23, 26, 27), and was long thought to be mostly immunosilent and only rarely recognized as foreign by the host. The glycan shielding can hinder the induction of NAbs against certain epitopes and prevent the binding of NAbs once they have been elicited. Thus, the N-linked glycans of Env constitute an important strategy by which HIV-1 evades the humoral immune response and complicates the design of HIV-1 Env-based vaccines.
Paradoxically, the N-glycans on gp120 are components of the epitopes for a subset of broadly active NAbs (bNAbs); such glycan-dependent bNAbs are much more common than previously appreciated (28–30). The highly unusual domain-exchanged 2G12 antibody recognizes a discontinuous gp120 epitope made up of elements from 3 or 4 individual glycans (31–34), whereas the discontinuous epitopes for bNAbs PG9 and PG16 are dependent on glycosylation at specific N-linked sites in the V1/V2 domain (28, 35, 36). bNAbs in the recently discovered PGT category also have glycan-dependent epitopes (29, 37). Of note is that, whereas 2G12 recognizes only the tips of glycan structures, PG9 and PGT128 bind to composite epitopes containing elements of the glycan stems and the underlying peptide backbone (29, 36, 37).
The heterogeneity and flexibility of carbohydrate chains create substantial impediments to crystallizing gp120 monomers or soluble trimers for atomic structure determinations. The crystal structures of HIV-1 gp120 cores were obtained using enzymatically deglycosylated proteins (38–41). To date, no crystal structure of an Env trimer has been obtained, in part because it has not been possible to produce stable and soluble deglycosylated Env trimers. Deglycosylating trimeric Env on virions has proven problematic, probably because steric factors restrict access of glycosidases to their substrates (17, 42). Partially deglycosylated, uncleaved gp140 trimers have recently been described (43).
To stabilize soluble trimeric complexes (gp140), we previously introduced an intermolecular disulfide bond between gp120 and an I559P point substitution in gp41 (44, 45). The resulting gp140 proteins, termed “SOSIP” gp140, are properly folded, soluble and fully cleaved (46). SOSIP gp140 trimers are better antigenic mimics of the native Env spike, compared with uncleaved trimers (44, 45, 47–49). However, all soluble trimers differ antigenically from native viral Env spikes. For example, bNAbs PG9 and PG16 bind poorly to SOSIP gp140 trimers from the KNH1144 strain yet neutralize the corresponding virus potently (28). In addition, some non-neutralizing Abs directed against the CD4 binding site bind to SOSIP gp140 trimers (47). Trimeric SOSIP gp140s are better than the corresponding monomeric gp120 at inducing NAbs in rabbits, although the magnitude and breadth of the response remains unsatisfactory (49–52).
For use in both immunogenicity and structural studies, we have now identified nondenaturing conditions under which SOSIP gp140 trimers can be partially deglycosylated without compromising their structural integrity. The resulting trimers are stable, highly soluble, resistant to proteolysis, and appear to retain an appropriate conformation as judged by the binding of CD4 and monoclonal antibodies (mAbs) to conformation-sensitive epitopes. Furthermore, electron density maps at 14-Å resolution using negative stain electron microscopy show that, other than the expected reduction in their overall size, deglycosylation causes no detectable structural changes to the trimers. Hence, once the Env trimer has formed, most of its glycans are not involved in maintaining its integrity.
MATERIALS AND METHODS
Constructs
The pPPI4 vector system for expressing Env proteins has been described elsewhere (44). Single amino acid substitutions were made using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). In the current study, we used monomeric gp120s from the subtype B strain JR-FL. The KNH1144 SOSIP.R6 gp140 protein contains the trimer-stabilizing A501C, T605C, and I559P substitutions and a hexa-arginine (R6) motif at the C terminus of gp120 that increases the efficiency of gp120-gp41 proteolytic cleavage (44, 45, 53, 54). The sequence was also modified to contain epitopes for mAbs 2F5, 4E10, and 2G12 (47). The resulting SOSIP variant, SOSIP.681, was used for most of the biochemical studies. A new variant, SOSIP.664, which lacks the C-terminal residues 665–681 of the gp41 ectodomain, was used for electron microscopy (EM) and SEC/MALS analyses.
Reagents
DC-SIGN-Fc was purchased from R&D Systems (Minneapolis, MN). HIVIg was obtained through the AIDS Research and Reference Reagent Program (ARRRP), Division of AIDS, NIAID, National Institutes of Health. mAbs 2G12, 4E10, 2F5, and D-50 were provided by Hermann Katinger through the ARRRP. 447-52D was obtained from Polymun Sciences, Klosterneuburg, Austria. mAb b12 was a gift from Dennis Burton (Scripps Research Institute, La Jolla, CA), mAbs 17b, 5.8c, and 1.4e were from James Robinson (Tulane University, New Orleans, LA). The CD4-IgG2 and sCD4 proteins from Progenics Pharmaceuticals have been described elsewhere (53). PGT123 was obtained through the IAVI NAC repository. Mannosidases, endoglycosidase F (Endo F), and endoglycosidase H (Endo H) were purchased from New England Biolabs (Ipswich, MA). Proteases trypsin, chymotrypsin, and proteinase K were obtained from Sigma.
Cell Culture and Transient Transfection
The human embryonic kidney (HEK) cell line 293T was grown under standard conditions (45). The 293S GnTI−/− line is a HEK 293 variant that does not express N-acetylglucosaminyltransferase I (GlcNAc transferase I; GnTI) (55). Proteins produced in these cells are designated, for example, as SOSIP.681GnTl−/−. Both cell lines were transiently transfected with env genes using linear polyethylenimine (Mr 25,000) as described previously (56). Briefly, DNA encoding an Env protein was diluted in Dulbecco's modified Eagle's medium (DMEM; Invitrogen), to 1/10 of the final culture volume and mixed with polyethylenimine (0.15 mg/ml final concentration). After incubation for 20 min, the DNA/polyethylenimine mixture was added to the cells for 4 h before replacement with culture medium containing 10% fetal bovine serum (FBS; Invitrogen), penicillin, streptomycin, and minimal essential medium nonessential amino acids (0.1 mm, Invitrogen). Supernatants were harvested 48 h after transfection. When appropriate, 100 μm kifunensine, an inhibitor of the glycoprotein processing mannosidase I enzyme (TRC, Toronto, Canada), was added to the cultures 1 h before transfection, and the medium was replaced 4 h after transfection with a fresh batch containing 100 μm kifunensine. All protein samples were kept on ice prior to subsequent analyses.
Affinity Purification of KNH1144 SOSIP gp140 Trimers
Env trimers were purified as described elsewhere with some modifications (47, 48). Transfection supernatants were passed (0.5–1 ml/min flow rate) through CNBr-activated Sepharose 4B beads (GE Healthcare) coupled to mAb 2G12 (Polymun Sciences, Klosterneuburg, Austria). The beads were washed with 15–20 column volumes of buffer (500 mm NaCl, 10 mm Tris, pH 8.0) before eluting the Env trimers with 3–5 column volumes of 3 m MgCl2. The eluted trimers were immediately buffer exchanged into 75 mm NaCl, 10 mm Tris, pH 8.0, using Vivaspin ultrafilters (100 kDa MWCO) (GE Healthcare). Purified proteins were stored at −80 °C and thawed only once before use. Protein concentrations were determined using a bicinchonic acid-based assay (BCA assay; Thermo Scientific, Rockford, IL).
Endo- and Exoglycosidase Digestions
We tested many conditions for endo- and exoglycosidase treatment of gp120 monomers and gp140 trimers (data not shown, and see “Results”). Two final protocols were selected (unless indicated otherwise), depending on the analyses to be performed subsequently. Both methods were equally effective at deglycosylating trimers, while preserving their conformation. The SOSIP.681GnTl−/− deglycosylation reactions were performed using 25 units of Endo H/μg of purified Env trimer for 3 h at room temperature and neutral pH (PBS or culture media, pH 7.2). In control studies, the Env protein was first denatured by incubation for 10 min at 95 °C with 0.02% SDS and 1 mm dithiothreitol (DTT), then treated with Endo H using conditions and a buffer specified by the manufacturer. Purified SOSIP.664GnTl−/− proteins were deglycosylated using 0.3 units of Endo H/μg of purified Env trimer for 3 h at room temperature in 100 mm sodium acetate, pH 6.0.
Limited Proteolysis
Purified trimers (17 μg/ml) produced in 293S GnTI−/− cells and deglycosylated with Endo H (or mock treated) were incubated with serial 10-fold dilutions (1–0.001 μg/ml) of various proteases for 1 h at 37 °C. The proteins were analyzed by SDS-PAGE followed by silver staining or Western blotting.
SDS-PAGE, Blue Native PAGE, and Western Blotting
Env proteins were detected using SDS-PAGE and Western blotting procedures involving the anti-gp120 mAb ARP3119 (0.2 μg/ml; Programme EVA Centre for AIDS Reagents) or the anti-gp41 mAb D50, HRP-labeled goat anti-mouse IgG (1:5,000 dilution) and the Western Lightning ECL system (PerkinElmer Life Sciences). Blue Native (BN)-PAGE was carried out using minor modifications to a published method (57). Purified proteins or cell culture supernatants were diluted with an equal volume of buffer (100 mm MOPS, 100 mm Tris-HCl, pH 7.7, 40% glycerol, and 0.1% Coomassie Blue), immediately prior to loading onto a 4 to 12% BisTris NuPAGE gel (Invitrogen). Usually, the gels were run for 2 h at 150 V (∼0.07 A) using 50 mm MOPS, 50 mm Tris, pH 7.7, as the buffer. Unless indicated otherwise, SDS-PAGE and BN-PAGE analyses were performed immediately after Endo H digestion.
Biophysical Analyses
Purified SOSIP.664 trimers (900 μg) were treated with Endo H (Roche Applied Science) in 100 mm sodium acetate buffer, pH 6.0, at room temperature for 3 h. Endo H-treated or untreated SOSIP.664 trimers were analyzed on a Superose-6 10/30 column (GE Healthcare) using an AKTA Avant FPLC system (GE Healthcare). A size exclusion chromatography (SEC) system was coupled in-line with the following calibrated detection systems: 1) HP1 1050 Hewlett-Packard UV detector (Norwalk, CT); 2) MiniDawn Treos multi-angle light scattering (MALS) detector (Wyatt Corporation, CA); 3) quasi-elastic light scattering detector (Wyatt Corporation, CA); and 4) Optilab T-reX refractive index (RI) detector (Wyatt Corporation). These combined measurements allow both the absolute molar mass of an eluting glycoprotein, and the individual contributions made by protein and carbohydrate components, to be determined (58–60). Briefly, the carbohydrate contribution to the molar mass can be assessed because glycans contribute directly to the RI signal but not the UV signal. Astra V software (Wyatt Corporation) was used to apply the protein conjugate template to the measured data. The extinction coefficient of the SOSIP.664 polypeptide, A2800.1%, was determined to be 1649 ml mg−1 cm−1 from its primary amino acid sequence (EXPASY). The specific refractive index increment values (dn/dc) were estimated to be 0.168 and 0.179 for the mock- and Endo H-treated SOSIP.664 samples, respectively. The hydrodynamic radii, rH, of the mock- and Endo H-treated SOSIP.664 samples were determined from the quasi-elastic light scattering/RI signals of the samples eluting from the SEC column, using Astra V software.
MALDI-TOF MS Analysis
Peptides generated by trypsin digestion of SOSIP.681GnT−/− trimers were detected by MALDI-TOF MS. Briefly, trimers were treated with Endo H under native or denaturing conditions, or with PNGase F under denaturing conditions, and diluted to 0.5 mg/ml before an overnight digestion with trypsin (0.3 μg in 30 μl of 25 mm ammonium bicarbonate) at 37 °C. The reaction products were extracted and analyzed by MALDI-TOF MS in positive reflection mode (Autoflex; Billerica, MA). Glycopeptides and deglycosylated peptides were identified by comparing the MS data with the National Center for Biotechnology Information (NCBI) data base, and with the KNH1144 SOSIP.681 sequence, using Mascot (Matrix Science, London, UK, version 2.2.04). The experimentally identified peptides were matched to ones derived from an in silico (i.e. theoretical) trypsin digest of SOSIP.681, allowing for the following possible peptide modifications: 1) oxidation of methionines; 2) deamidation of asparagines from which an N-glycan was removed by PNGase F; and 3) addition of a HexNac (GlcNAc) residue to asparagines from which an N-glycan was removed by Endo H. No fully glycosylated peptides could be detected in the MALDI-TOF analyses.
Immunoprecipitation Assays
Twenty-fold concentrated supernatants from transiently Env-transfected 293T or 293S GnTI−/− cells were incubated overnight at 4 °C with mAbs or CD4-IgG2 in 500 μl of RIPA buffer (150 mm NaCl, 1 mm EDTA, 50 mm Tris-HCl, pH 7.0). A 50-μl volume of protein G-coated agarose beads (Pierce/Thermo Fisher Scientific) was then added, with rotation mixing, for 2 h at 4 °C. The beads were washed extensively with ice-cold RIPA buffer containing 0.01% Tween 20 before proteins were eluted by heating for 5 min at 100 °C in 50 μl of SDS-PAGE loading buffer supplemented with 100 mm DTT. The immunoprecipitated proteins were analyzed on 8% SDS-PAGE gels (125 V, 2 h).
Electron Microscopy
Purified glycosylated or deglycosylated SOSIP.664 trimers (0.1 mg/ml) were applied to freshly glow-discharged, carbon-coated 400-mesh Cu grids (Ted Pella Inc., Redding, CA) and stained with 2% uranyl formate. The grids were viewed using an FEI Tecnai F20 electron microscope at a magnification of ×100,000 and an accelerating voltage tension of 120 kV. Images were acquired on a Gatan 4k × 4k CCD camera at 0, 15, 30, 45, and 55° tilt angles using a defocus range of 600 to 720 nm. The tilt angles provided additional particle orientations to improve the image reconstructions. The pixel size of the CCD camera was calibrated at this magnification to be 1.09 Å using a two-dimensional catalase crystal with known cell parameters.
Image Processing
Particles were manually selected from micrographs using the DoGPicker package (61, 62). The contrast transfer function estimations for untilted micrographs were completed using the ctffind3 program and applied using the ACE2 program (61, 63). Contrast transfer function estimations for the tilted micrographs and particles were calculated using ctftilt, and phases were flipped using applyctf in EMAN (61, 64, 65). Particles were extracted and binned by four into dimensions of 80 × 80 pixels for calculating reference free class averages with the Sparx package (66). An ab initio three-dimensional image reconstruction was generated from each set of reference free class averages using the EMAN2 package (65, 66). To test for model bias, each ab initio model was refined separately against the glycosylated and deglycosylated reference free class averages. In total, two models were produced for each set of reference free class averages. The two image reconstructions for each set were nearly indistinguishable, suggesting that refinement with the reference free class averages was able to overcome any possible model bias. This procedure was followed through 80 iterations of projection matching using the Sparx package (66). The final three-dimensional image reconstructions were calculated using images of single particles with a spatial resolution of 2.17-Å per pixel and refined using the EMAN package (64). C3 symmetry was imposed throughout the three-dimensional image reconstruction process. According to a Fourier shell correlation of 0.5 the resolution of the three-dimensional reconstruction was 14 Å for the untreated trimer and 20 Å for the Endo H-treated trimer.
RESULTS
Deglycosylation Strategy for Recombinant Env
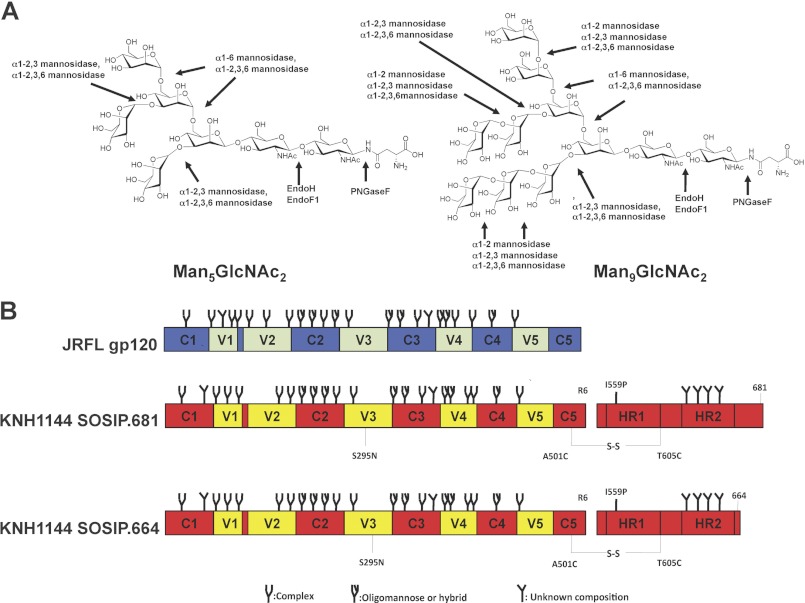
The considerable heterogeneity of the glycan structures present on HIV-1 Env compromises the efficiency of various deglycosylation strategies (17, 42). In previous studies using monomeric gp120 or virions, many different glycosidases were tested under a range of experimental conditions, some of which are harsh enough to probably be incompatible with Env trimer stability (8). To avoid such problems, we adopted a two-stage strategy in which we first made trimers bearing a more homogeneous set of glycans that could then be digested by enzymes under mild conditions of temperature, salt, and pH (67). The specificities of the various enzymes that we evaluated are shown in Fig. 1A.
FIGURE 1.
Schematics of Env constructs and glycosidase specificities. A, two different oligomannose N-glycans (Man5GlcNAc2 and Man9GlcNAc2) that are present on Env proteins produced in GnTl−/− cells are depicted, along with the cleavage sites for the enzymes used in this study. Env produced in the presence of kifunensine would only contain Man9GlcNAc2 glycans. B, schematics of the gp120 and gp140 proteins. JR-FL gp120, conserved regions in blue, variable regions in green; KNH1144 gp140, conserved regions in red, variable regions in yellow. The gp140 was modified to reconstitute the 2G12 epitope (S295N substitution), and the 2F5 and 4E10 epitopes in the MPER (A662E, G664D, S668N, and N671T substitutions) (47). The gp140 also contains the A501C and T605C substitutions to create the SOS disulfide bond between gp120 and gp41 (44), the I559P mutation to promote trimerization (45), and a hexa-arginine cleavage site (R6) to enhance cleavage efficiency (54). The type of N-glycan indicated to be present at each glycosylation site (oligomannose, complex or undefined) is based on glycan characterization of IIIB gp120 (83). However, which type of glycan is actually present at some of these sites may vary between strains (83), and can also depend on the oligomerization status of Env proteins (69, 84). The gp41 glycans have not been characterized.
For optimizing deglycosylation conditions in pilot experiments, we used unpurified monomeric gp120 from the clade B strain JR-FL rather than purified, cleaved, disulfide-stabilized, soluble gp140 trimers. We then used the optimized protocol to deglycosylate the trimers. Because SOSIP gp140 from the clade A strain KNH1144 trimerizes more efficiently than the corresponding JR-FL gp140 (68), we used KNH1144 SOSIP.681 trimers for the deglycosylation studies (see “Materials and Methods” and Fig. 1B for more details). We also used a variant construct from which the MPER was deleted to prevent aggregation in the absence of detergent (SOSIP.664; Fig. 1B).
We previously described two methods for generating recombinant Env monomers and trimers that lack complex carbohydrates and, thus, contain only oligomannose sugars (69). First, we produced SOSIP.681 trimers in 293S GnTI−/− cells in which the attachment of GlcNAc to Man5GlcNAc2 in the cis-Golgi is blocked; Man5GlcNAc2 moieties are the most prevalent carbohydrate structure on the expressed glycoproteins. When we produced gp120 and trimeric SOSIP.681 in 293S GnTI−/− cells, they contained Man5GlcNAc2 glycans, as expected (69). However, a considerable number of Man6–9GlcNAc2 moieties were also present because mannose residues were trimmed inefficiently at some sites (69). In our second approach, we treated 293T cells with the mannosidase inhibitor kifunensine and, thereby, produced Env proteins on which the predominant carbohydrate structure was Man9GlcNAc2 (Fig. 1A) (69).
We investigated whether oligomannose-enriched gp120 and SOSIP.681 gp140 proteins could be efficiently trimmed by a set of mannosidases and endoglycosidases without compromising their structural integrity. Endo H and Endo F1 cleave between the N-acetylglucosamine residues of the diacetylchitobiose core of N-linked glycans, generating a truncated glycan molecule in which one N-acetylglucosamine moiety remains attached to the asparagine moiety and, hence, the protein backbone (70). Oligomannose and most hybrid-type N-glycans, but not complex-type oligosaccharides, are hydrolyzed by Endo H and Endo F1. The α1–2,3,6-mannosidase should remove the outer four α1–3- and α1–6-linked mannose residues from Man5GlcNAc2 structures, and the outer eight α1–2-, α1–3-, and α1–6-linked residues from Man9GlcNAc2 variants (Fig. 1A). We have previously used this enzyme to cleave mannose moieties from gp120 and eliminate the binding sites for mAb 2G12 and DC-SIGN (21, 22, 34). We also tested α1–2-, α1–6-, and α1–2,3-mannosidases that have a more limited specificity (Fig. 1A).
Deglycosylation of Oligomannose-enriched gp120
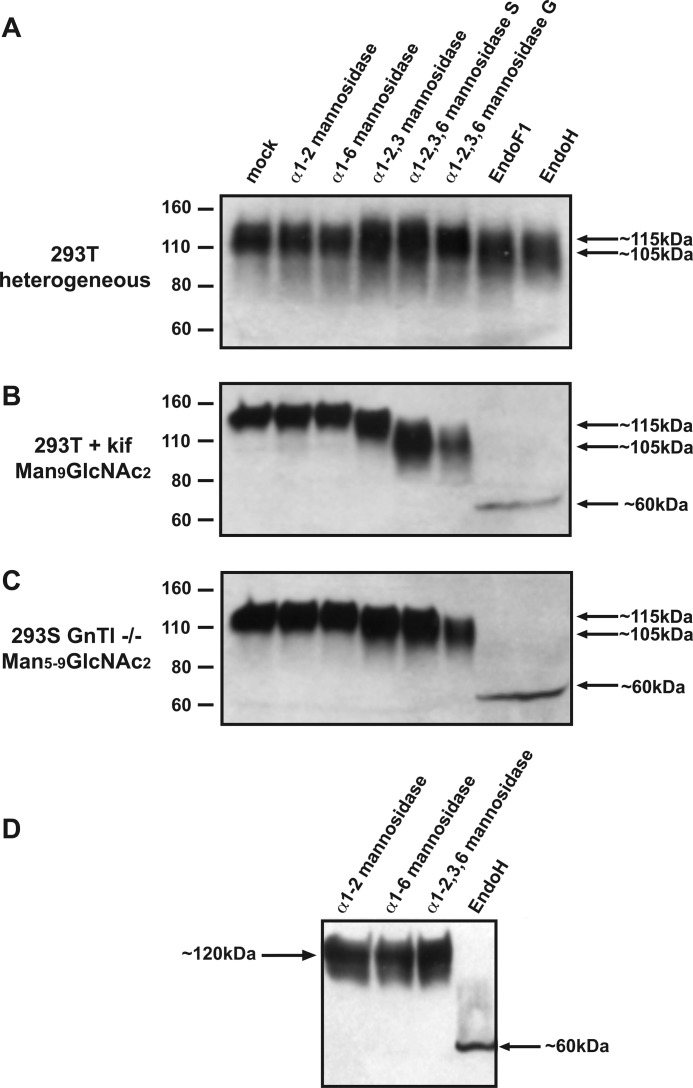
We first studied the effect of various enzymes on 293T cell-expressed JR-FL gp120 that contains a heterogeneous mixture of complex and oligomannose carbohydrates (designated gp120WT). The efficiency of deglycosylation was evaluated by SDS-PAGE and Western blot analysis, using size reduction as an indicator of glycan removal (Fig. 2A). We note that SDS-PAGE analysis does not provide absolute values for glycoprotein sizes, because glycoproteins do not migrate on these gels in a way that is strictly proportional to their molecular weight (Mr) and, hence, to their glycan content. The glycosidase-mediated Mr reductions, as measured on SDS-PAGE, are therefore merely apparent values that generally overestimate the extent of glycan loss.
FIGURE 2.
Deglycosylation of gp120 with endo- and exoglycosidases. The indicated glycosidases were used to treat the following gp120s under mild conditions. A, 293T cell-produced gp120 that contains both complex and oligomannose (i.e. heterogeneous) N-glycans; B, oligomannose-enriched gp120kif from kifunensine-treated 293T cells (i.e. Man9GlcNAc2 N-glycans); C, oligomannose-enriched gp120GnTI−/− from 293S GnTI−/− cells (i.e. Man6Man5–9GlcNAc2 N-glycans). The enzyme-treated gp120 variants were analyzed by reducing SDS-PAGE and Western blot. The positions of the molecular mass marker proteins and the approximate apparent molecular mass of the enzyme-treated gp120s are indicated; D, glycosidase treatment of gp120GnTl−/− under mild conditions. The gp120s were analyzed by SDS-PAGE analysis and Western blotting.
For each enzyme, the buffer conditions recommended by the manufacturers were used, i.e. 50 mm sodium citrate, pH 4.5–5.5 for α1–2,3,6-mannosidase, Endo H and Endo F1. There was no detectable size reduction when gp120WT was treated with α1–2-, α1–6-, or α1–2,3-mannosidase, implying either that the enzymes were inactive or that they removed too little carbohydrate for any size reduction to be visible using SDS-PAGE. In contrast, a small, but clear size reduction (∼5 kDa) was observed when gp120WT was exposed to α1–2,3,6-mannosidase, which is consistent with our earlier findings (21, 22) (Fig. 2A). Endo H or Endo F1 digestion lead to a slightly greater apparent molecular mass decrease of ∼10–15 kDa (Fig. 2A). These enzymes had only limited effects because most sugars on gp120WT are of the complex-type and, hence, not substrates; in addition, steric constraints may impede enzyme access to some N-glycans that would otherwise be targets.
The same set of enzymes were then used to treat JR-FL gp120 proteins that had been expressed in 293T cells in the presence of kifunensine (designated gp120kif); most N-glycans should, therefore, be of the Man9GlcNAc2 form. The gp120kif proteins were of a similar size to gp120WT but migrated as a more compact band, which is consistent with their decreased glycan heterogeneity (compare the first lane of Fig. 2, A and B). There was, again, no detectable size difference when gp120kif was exposed to either α1–2- or α1–6-mannosidase (Fig. 2B). This outcome was expected with α1–6-mannosidase because Man9GlcNAc2 glycans do not contain terminal α1–6-mannose residues. However, we had anticipated that α1–2-mannosidase would be active; each Man9GlcNAc2 glycan theoretically contains four α1–2-linked mannoses, so this enzyme should reduce the size of gp120kif by ∼17 kDa (i.e. four mannoses, each of 0.18 kDa, removed from ∼24 glycans). A reduction of this magnitude would be easily visible on an SDS-PAGE gel, and yet it was not observed. Although α1–2-mannosidase has been effective on other glycoprotein substrates (32), steric hindrances may limit its activity on gp120. We, therefore, abandoned the use of this enzyme. The size of gp120kif was slightly reduced (∼5 kDa) by α1–2,3-mannosidase, and more so (an apparent shift of ∼15 kDa) by α1–2,3,6-mannosidase. Treatment of gp120kif with Endo H or Endo F1 had a much more marked effect; the apparent reductions in the size of gp120kif to ∼60 kDa imply that these enzymes remove a considerable amount of its carbohydrate content.
Broadly comparable results were obtained using gp120GnT−/− proteins produced in 293S GnTI−/− cells (Fig. 2C). α1–2- and α1–6-Mannosidase had no detectable effect on the size of these proteins. This outcome was again unexpected because the gp120GnT−/− glycans should have both α1–6-mannose and α1–2-mannose moieties at their termini (albeit the latter on only a minority of glycans). Again, perhaps steric factors limit access of these enzymes to their substrates on gp120. As seen with gp120kif, there was a small decrease in the size of gp120GnT−/− upon exposure to α1–2,3-mannosidase or α1–2,3,6-mannosidase. The apparent decrease of ∼10 kDa in the size of α1–2,3,6-mannosidase-treated gp120GnT−/− was slightly less than the ∼15 kDa shift seen with gp120kif. The quantitative difference arises because there are fewer susceptible mannose residues on gp120GnT−/− (which contains a mixture of Man5–9GlcNAc2 glycans) than on gp120kif (which contains only Man9GlcNAc2). Finally, Endo H and Endo F1 reduced the size of gp120GnT−/− to ∼60 kDa, an outcome similar to when gp120kif was treated with the same enzymes (Fig. 2, B and C).
Deglycosylation of Oligomannose-enriched gp120 under Mild Conditions
The recommended buffers for optimal function of Endo H, Endo F1, and mannosidases use 50 mm sodium citrate at pH values in the range 4.5–5.5. Although the antigenic structure of gp120 is not compromised under such conditions (34), we considered it possible that gp140 trimers would be more pH-sensitive. We note that KNH1144 SOSIP.664 trimers do indeed dissociate below pH 5.0 (see below). To avoid such problems, we investigated whether endo- and exoglycosidases could still digest carbohydrates at a more physiological pH, by treating gp120GnT−/− proteins with these enzymes for 2 h at room temperature in PBS, pH 7.2 (Fig. 2D).
There was no detectable effect of α1–2,3,6-mannosidase on the size of gp120GnT−/− proteins under these reaction conditions (Fig. 2D), which contrasts with the size decrease seen using the optimal enzyme digestion buffer (Fig. 2, B and C). The differential outcome is consistent with reports that the optimal pH for this enzyme is in the range 3.5–4.5, with no activity at pH 7.2 (71). In contrast, Endo H was still able to reduce the size of gp120GnT−/− from ∼110 to ∼60 kDa when the pH was increased to 7.2 (Fig. 2D). Hence, Endo H can remove a considerable amount of the carbohydrates from the glycan-modified gp120GnT−/− proteins even under apparently suboptimal digestion conditions that better preserve the structure of gp140 trimers. Similar results were obtained when 293T cell culture medium, pH 7.6, was used instead of PBS (data not shown).
Deglycosylation of Oligomannose-enriched gp140 Trimers
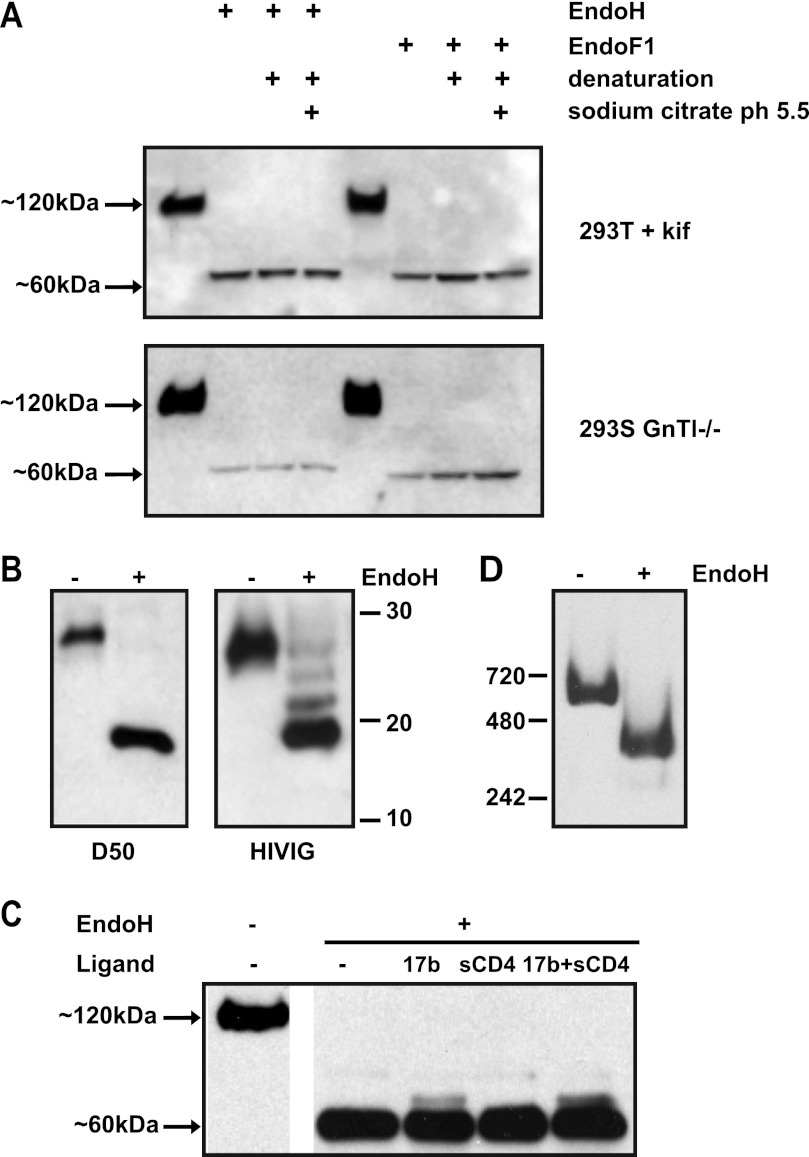
Env trimer glycans on SIV virions are more resistant to glycosidase digestion than those on gp120 monomers, probably because steric factors impede the access of the enzymes to their substrates on the trimers (17). We therefore tested whether the native reaction conditions identified above would also work with soluble gp140 trimers. Cleaved KNH1144 SOSIP.681 gp140 trimers were produced in 293T cells in the absence (SOSIP.681WT) or presence of kifunensine (SOSIP.681kif), or in 293S GnTI−/− cells (SOSIP.681GnT−/−), as previously described (69). The various (unpurified) trimers were then treated with Endo H or Endo F1, fractionated by SDS-PAGE, and probed with the anti-gp120 mAb ARP3119 after Western blotting (Fig. 3A). As observed when monomeric gp120 was similarly treated, the gp120 subunits of both SOSIP.681kif and SOSIP.681GnT−/− (i.e. gp120 in the context of a trimer) were reduced in size to, apparently, ∼60 kDa. For comparison, we denatured the same trimers using SDS plus DTT, and then treated them with Endo H using conditions optimal for enzyme activity (sodium citrate, pH 5.5). The resulting trimers were indistinguishable in size, i.e. ∼60 kDa, from ones that had been exposed to the enzyme under nondenaturing conditions (Fig. 3A). Hence, using nondenaturing conditions at neutral pH appears to be adequate for removing all the glycans from SOSIP.681GnT−/− trimers that are actually Endo H-sensitive under the test conditions. However, we discuss below how biophysical techniques yielded additional information that caused us to refine the above conclusion.
FIGURE 3.
Deglycosylation of oligomannose-enriched SOSIP.681 gp140 trimers. A, SOSIP.681 proteins were produced in either kifunensine-treated 293T cells (upper panel) or 293S GnTI−/− cells (lower panel), treated with endoglycosidases Endo F1 or Endo H under native or denaturing conditions, and analyzed by SDS-PAGE. Deglycosylation reactions were performed in PBS at neutral pH (7.2) or in the buffer recommended by the manufacturer (sodium citrate, pH 5.5), as indicated. B, SDS-PAGE analysis of the gp41 ectodomain of mock- and Endo H-treated SOSIP.681GnTl−/−. The blots were probed with the anti-gp41 mAb D50 (left) or HIVIg (right). The positions of marker proteins are indicated. C, Endo H treatment of SOSIP.681GnTl−/− under mild conditions in the presence and absence of sCD4, 17b, or a combination of both ligands. D, BN-PAGE analysis of SOSIP.681 treated with or without Endo H under native conditions. The molecular masses of reference proteins are indicated.
To monitor the deglycosylation of gp41, which contains potential N-linked glycosylation sites at positions 611, 616, 625, and 637, we used the gp41-specific mAb D50 or HIVIg to probe Western blots after SDS-PAGE analysis. Endo H treatment of SOSIP.681GnT−/− decreased the apparent size of the gp41 component by ∼10 kDa, as detected by Asp-50 (Fig. 3B). However, a ladder of three additional gp41-related bands was detected when HIVIg was used for blotting (Fig. 3B). The different outcome probably reflects the more sensitive detection of trace amounts of partial digestion products when HIVIg is used. Furthermore, the detection of three additional bands implies that only three of the four potential glycosylation sites in gp41 are actually used. This supposition was confirmed by mass spectrometry studies (see below).
To further facilitate crystallization studies, we determined whether Endo H (in PBS at pH 7.2) was active on a complex of purified trimeric KNH1144 SOSIP.681 gp140GnT−/− protein with sCD4 and mAb 17b. The trimers were deglycosylated to similar extents whether sCD4 was present or not (Fig. 3C). A residual, slower migrating band was apparent in the presence of 17b, suggesting that this mAb hinders Endo H access to one or more glycans. Overall, however, the sCD4-gp140–17b ternary complex was efficiently deglycosylated by Endo H.
We used BN-PAGE to study the integrity and stability of mock- or Endo H-treated SOSIP.681GnT−/− trimers, because deglycosylation can increase the tendency of proteins to aggregate, and/or cause oligomeric complexes to dissociate (72). The Endo H-treated, deglycosylated trimers migrated on BN-PAGE gels as a single species, with no indication of aggregate formation or dissociation into monomers (Fig. 3D).
Deglycosylation of Purified Oligomannose-enriched gp140 Trimers
The above studies were carried out using unpurified Env proteins in the supernatants of transiently transfected cells. To further refine the deglycosylation protocol, we next assessed the effectiveness of Endo H treatment of purified trimers. During the course of this study, we found that KNH1144 SOSIP gp140 trimers from which the MPER was deleted (SOSIP.664) adopt a monodisperse distribution in the absence of detergent.5 This property makes them more suitable than SOSIP.681 trimers for BN-PAGE, biophysical analyses, and EM studies. We confirmed that Endo H removed similar amounts of glycans from KNH1144 SOSIP.664 and SOSIP.681 trimers, as judged by SDS-PAGE analysis (not shown). Hence we used SOSIP.664 trimers for most of the following analyses.
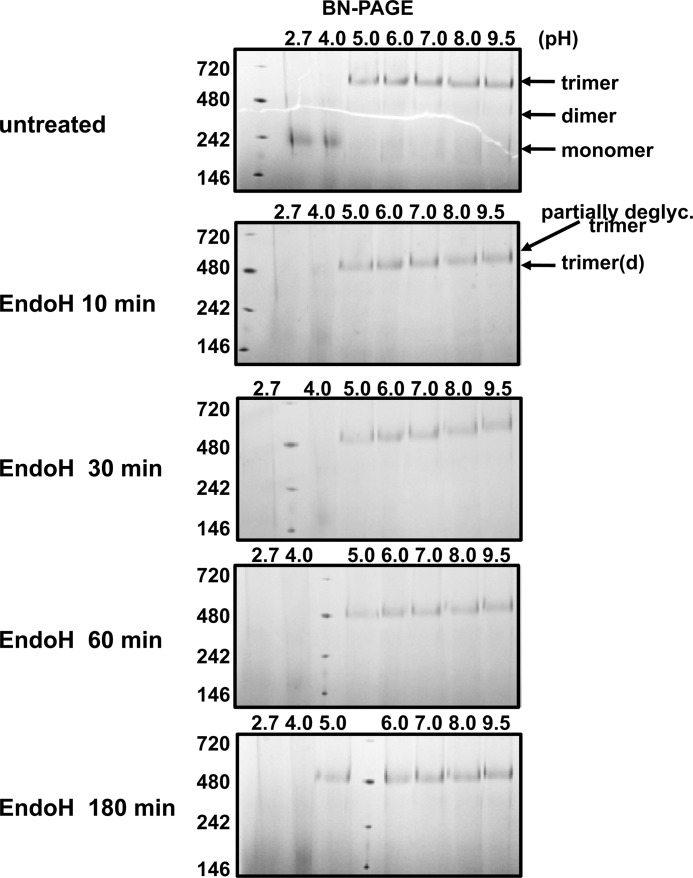
Using purified SOSIP.664GnT−/− trimers as the substrate and Endo H as the enzyme, we evaluated how varying the incubation time, and using buffers of different pH, affected deglycosylation efficiency (Fig. 4). BN-PAGE analysis revealed that purified KNH1144 SOSIP.664GnT−/− trimers were stable in the pH range 5.0–9.5, but dissociated to monomeric gp140 when the pH was below 5.0. The incubation time and the pH both influenced the deglycosylation process, which was faster at pH 5.0 than 7.0. However, deglycosylation proceeded to the same final extent at the higher pH, provided the incubation time was increased (Fig. 4). Accordingly, we chose to perform the subsequent enzyme digestions for 3 h at pH 7.0, unless indicated otherwise.
FIGURE 4.
Time- and pH-dependence of Endo H-mediated deglycosylation of KNH1144 SOSIP.664Gntl−/− trimers. The trimers were treated with Endo H at 37 °C in buffers with varying pH (0.05 m glycine, pH 2.7; 0.075 m sodium citrate, pH 4.0; 0.075 m sodium citrate, pH 5.0; 0.1 m BisTris, pH 6.0; 0.1 m HEPES, pH 7.0; 0.1 m CAPS, pH 9.5) and for varying times, before analysis by BN-PAGE. The species associated with each band are indicated with arrows (d, deglycosylated; partially deglyc., partially deglycosylated).
Stability and Sensitivity of Deglycosylated gp140 Trimers to Proteolysis
Glycans can shield the underlying protein domains from proteases, so a deglycosylated protein might be atypically vulnerable to proteolytic digestion (23, 42, 72). We therefore compared the sensitivities of purified, deglycosylated, and mock-treated SOSIP.681GnT−/− trimers to different concentrations (0.001–1 μg/ml) of three common serine proteases: proteinase K, trypsin, and chymotrypsin. SDS-PAGE analysis followed by silver staining showed that the deglycosylated trimers were more sensitive to proteinase K (Fig. 5A). Thus, whereas the mock-treated trimers were resistant to digestion by 0.1 μg/ml of proteinase K, and were only degraded at 1.0 μg/ml (Fig. 5A, top panel), the deglycosylated trimers were completely degraded at 0.1 μg/ml (Fig. 5A, bottom panel). In contrast, deglycosylation had no effect on trypsin and chymotrypsin sensitivity (Fig. 5, B and C). Overall, whereas glycan removal does increase the vulnerability of SOSIP.681GnT−/− trimers to proteinase K degradation, the effect is not dramatic. Hence, the deglycosylated trimers should be stable enough to be used in immunization and crystallization studies.
FIGURE 5.
Protease sensitivity of deglycosylated gp140 trimers. SOSIP.681GnTl−/− trimers were either Endo H-treated or mock-treated, and then exposed to different serine proteases at a range of concentrations (0.001–1 mg/ml): A, proteinase K; B, trypsin; C, chymotrypsin. The treated proteins were analyzed by SDS-PAGE and silver staining.
Biophysical Characterization of Deglycosylated gp140 Trimers
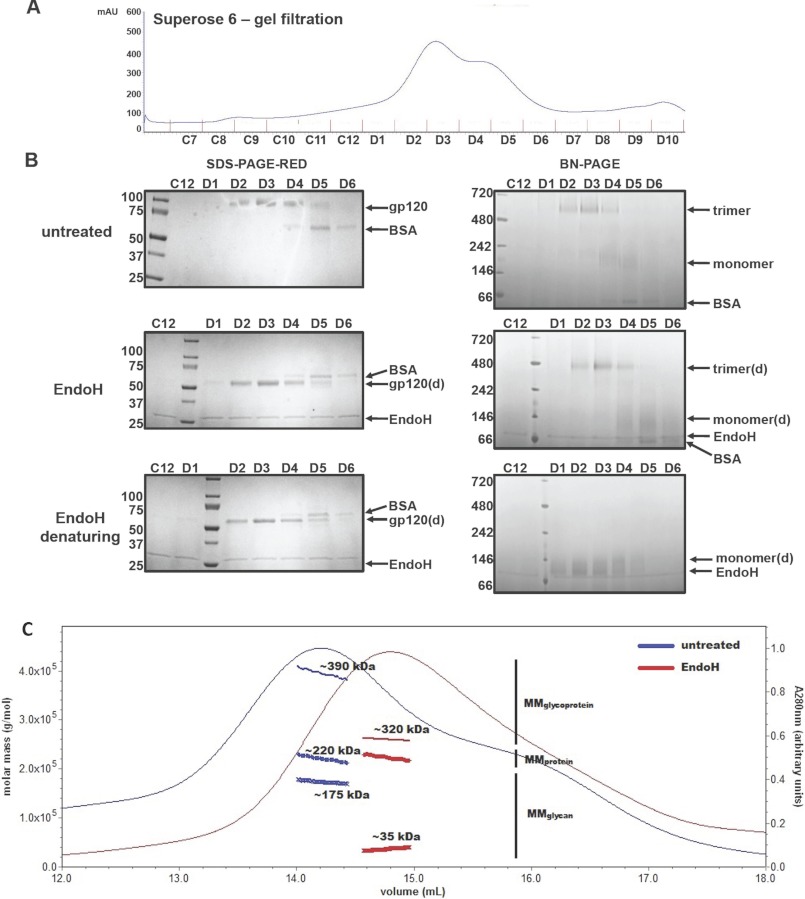
To accurately determine the extent of deglycosylation achieved in our experiments, we separated purified KNH1144 SOSIP.664Gntl−/− gp140 trimers from gp140 monomers by size exclusion chromatography (SEC) on a Superose 6 column (Fig. 6A). A BN-PAGE analysis of the SEC fractions revealed that fractions D2 and D3 contain predominantly trimeric gp140, whereas mostly monomeric gp140 was present in fractions D4 and D5 (Fig. 6B). Fractions D5 and D6 also contained a contaminant, which was identified as BSA by mass spectrometry (not shown).
FIGURE 6.
Biophysical analysis of SOSIP.664GnTl−/− trimers incubated with or without Endo H. A, untreated SOSIP.664GnTl−/− trimers were fractionated using a Superose-6 column. The fractions were treated with or without Endo H and analyzed by reducing SDS-PAGE (B, left panels) or BN-PAGE (B, right panels). B, top panels, untreated trimers; middle panels, Endo H-treated trimers (3 h at 37 °C, pH 5.5); bottom panels, denatured and Endo H-treated trimers. The species associated with each band are indicated. C, overlay of the A280 UV absorbance traces from Superose-6 column fractionations of untreated (blue) and Endo H-treated (3 h at 37 °C, pH 5.5; red) SOSIP.664GnTl−/− trimers. UV/MALS/RI signals were recorded as the samples eluted from the column. Absolute molar masses calculated from these measurements are shown as thick blue and green connected dots under the main peaks for the samples obtained with and without Endo H treatment (see also Table 2). d, deglycosylated.
Determining how well gp140 trimers and gp140 monomers are deglycosylated under comparable conditions could reveal whether the trimerization interface influences the accessibility of some glycans to the enzyme. For this purpose, we analyzed fractions D2 and D3 (trimer) and D4 and D5 (monomer). When each fraction was treated with Endo H, BN-PAGE and SDS-PAGE analyses showed that the gp140 trimers and gp140 monomers were both deglycosylated to similar extents (Fig. 6B). The gp140 trimerization interface does not, therefore, shield a substantial subset of glycans from Endo H. A similar conclusion can be drawn from the experiments showing that the efficiency of glycan removal is similar whether the trimers are treated under native conditions or after prior denaturation (Figs. 3A and 6B).
Untreated and deglycosylated trimers had similar SEC elution profiles, with no detectable differences in the distribution of the oligomeric species. However, the deglycosylated trimers eluted slightly later (elution volume 14.8 ml, compared with 14.2 ml), which is consistent with a decrease in their overall mass (Fig. 6C). In a quasi-elastic light scattering analysis, the hydrodynamic radii (rH) of the untreated and Endo H-treated SOSIP.664GnT−/− trimers were determined to be 12.3 and 9.9 nm, respectively, further evidence that an overall decrease in size occurs upon deglycosylation (Table 1).
TABLE 1.
SEC-UV/MALS/RI analysis of Endo H-treated and untreated SOSIP.664GnTl−/−
| UV extinction coefficient A2800.1%a | Estimatedb (dn/dc)glycoprotein d | Expected MMproteinc of monomer | MMproteinfrom SEC-UV/MALS/RIe,f | Calculated stoichiometry | MMglycan from SEC-UV/MALS/RI | MMglycoprotein from SEC-UV/MALS/RI | Hydrodynamic radius, rHf | |
|---|---|---|---|---|---|---|---|---|
| ml mg−1cm−1 | ml/g | g/mol | g/mol | nm | ||||
| Untreated SOSIP.664GnTl−/− | 1,649 | 0.168 | 74,332.6 | 220,600 | 2.97 | 173,500 | 394,200 | 12.3 |
| Endo H-treated SOSIP.664GnTl−/− | 1,649 | 0.179 | 74,332.6 | 223,500 | 3.01 | 36,500 | 260,000 | 9.9 |
a Extinction coefficient at 280 nm measured in water.
b Refractive index increment value.
c Molar mass.
d From the basic principles of molar mass determination by light scattering, it follows that any ± 1% error in the dn/dc value will propagate as a corresponding ± 1% error in the calculated molar mass (59).
e For SEC-UV/MALS/RI analysis of glycoproteins, a typical error is ± 10%. The source of uncertainty predominantly arises from the amount of sample analyzed and the stability of the system (59).
f Errors propagated in the various calculations performed by Astra V range between 0.2 and 50%.
In-line, MALS and RI detectors were used to make additional measurements during the SEC fractionations. Taken together with the A280 nm signal, these biophysical measurements allow the absolute molar mass of the eluting glycoprotein to be determined, as well as the contributions made by the individual protein and carbohydrate components. Together, the SEC-UV/MALS/RI measurements show that the protein components of the untreated and deglycosylated SOSIP.664GnT−/− trimers have the same molar mass of ∼220 kDa (3 × ∼74 kDa for each monomer; Fig. 6C and Table 1). The overall reduction in the molar mass of the trimers (from ∼390 to ∼260 kDa) upon Endo H treatment is, therefore, due to a loss of carbohydrate mass (from ∼175 to ∼35 kDa, which is equivalent to ∼12 kDa remaining carbohydrate per gp140 monomer).
The imprecision of the SEC-UV/MALS/RI technique is such that the ∼35 kDa median estimate of the glycan molar mass remaining on the trimer could actually range from 18 to 52 kDa (corresponding to 0–1 to 6–7 Man9GlcNAc2 glycans per gp140 monomer). This outcome results from intrinsic measurement errors (∼0.2–50%, as detailed in Table 2), and constrains the following discussions about the nature of the ∼35 kDa of glycans that remain on each Endo H-treated gp140 trimer. First, if any O-linked sites are present, they would only contribute perhaps ∼1 kDa of carbohydrate. Second, one GlcNAc moiety (203 Da) remains on every initially glycosylated asparagine residue; assuming every canonical glycosylation site is actually used (see below), the ∼84 GlcNAc moieties would contribute ∼17 kDa to the glycan mass of a SOSIP.664 trimer. Third, some trimer glycans may be inefficiently digested by, and/or inaccessible to, Endo H. Taking into account the ∼17 kDa contributed by the nonremovable GlcNAc moieties, undigested carbohydrates could contribute ∼18 kDa to the Endo H-treated trimer, or ∼6 kDa per monomer. If the remaining glycans are of the Man9GlcNAc2 type, which each have a mass of 1823 Da (excluding the protein-proximal GlcNAc already accounted for above), then Endo H fails to remove ∼0–7 of the 28 glycans on each monomer. If some of the glycans contain fewer mannose moieties, that estimate would increase slightly. Again, we emphasize that this discussion is constrained by the considerable experimental error inherent in the measurement of what glycans remain undigested.
TABLE 2.
MALDI-TOF analysis of Endo H-treated and untreated SOSIP.681GnTl−/−
| N-Glycan postitiona | Env region | Peptide sequenceb | Mockc native conditions | PNGaseFd denaturing conditions | EndoHe denaturing conditions | EndoHe native conditions |
|---|---|---|---|---|---|---|
| 88 | C1 | HNVWATHACVPTDPNPQEIPLENVTEEFNMWK | − | − | HexNAc | HexNAc |
| 130 and 136 | V1 | LAPLCVTLNCTDVTNVTDVSGTR | − | 2N→D | 2HexNAc | 2HexNAc |
| 146 and 156 | V2 | GNITIMKEMEGEIKNCSFNMATEIR | − | 2N→D | 2HexNAc | 2HexNAc |
| 188 and 193 | V2 | LDVVPINQGNSSSKNSSEYR | − | 2N→D | 2HexNAc | 2HexNAc |
| 197 | C2 | LISCNTSAITQACPK | + | N→D | HexNAc | HexNAc |
| 234 | C2 | DKEFNGTGECK | − | N→D | HexNAc | HexNAc |
| 241 | C2 | NVSTVQCTHGIKPVVSTQLLLNGSLAEEK | − | 2N→D | 2HexNAc | 2HexNAc |
| 276 | C2 | IRTENITNNAK | − | N→D | HexNAc | HexNAc |
| 301 | V3 | ISCTRPNNNTR | − | N→D | HexNAc | HexNAc |
| 448 | C4 | CESNITGLLLTR | − | N→D | HexNAc | HexNAc |
| 463 | V5 | DGGEEKNSTNEIFRPGGGDMR | − | N→D | HexNAc | HexNAc |
| 611 and 616 | gp41 loop | LICTTNVPWNSSWSNK | − | N→D | HexNAc | HexNAc |
| 625 | HR2 | SQDEIWNNMTWLQWDK | − | N→D | HexNAc | HexNAc |
| 637 | HR2 | EISNYTNLIYSLIEESQNQQEK | − | N→D | HexNAc | HexNAc |
a Numbering according to the HxB2 sequence.
b Potential N-linked glycosylation sites are indicated in bold and underlined.
c Mock-treated SOSIP.681GnTl−/−; “−” indicates that the peptide was not found, which suggests the peptide is glycosylated and hence not detectable in the MALDI-TOF analyses; “+” indicates that an (unglycosylated) peptide variant was detected.
d PNGaseF-treated SOSIP.681GnTl−/−; “N→D” indicates that one asparagine in the peptide was deamidated, indicative of prior glycosylation and subsequent removal by PNGaseF; “2N→D” indicates that two asparagines in the peptide were deamidated.
e EndoH-treated SOSIP.681GnTl−/−; “HexNAc” indicates that the peptide was found with an additional HexNAc (i.e. GlcNAc) attached, indicative of glycosylation and subsequent glycan removal by EndoH; “2HexNAc” indicates that two HexNAc moieties were present on the peptide.
Mass Spectrometric Analysis of Deglycosylated Gp140 Trimers
Mass spectrometry analysis of tryptic peptides is a useful tool for assessing the utilization of glycosylation sites and the efficiency of deglycosylation protocols (73). We used MALDI-TOF to identify what N-glycans were removed by Endo H treatment. Because N-glycans are degraded during the course of MALDI-TOF spectrometry, glycan-containing peptides cannot be identified directly. However, the efficiency of deglycosylation can still be investigated by studying recovered peptides that contain modifications consistent with deglycosylation-related events. Thus, peptides from which PNGase F has removed an N-glycan can be distinguished from nonglycosylated peptides because they are deamidated and so have an aspartic acid in place of the original asparagine. Similarly, peptides from which Endo H has removed an N-glycan can be identified because one GlcNAc moiety remains attached to the asparagine residue.
We studied tryptic peptides derived from purified SOSIP.681GnT−/− trimers that had been Endo H-treated either under mild conditions (see above) or after denaturation by heating for 10 min at 95 °C in SDS plus DTT. As a further comparison, we also generated a third set of tryptic peptides from denatured SOSIP.681GnT−/− trimers that had been treated with PNGase F. In each case, approximately half of the SOSIP.681GnT−/− tryptic peptides that contain a glycosylation site could be identified (Table 2).
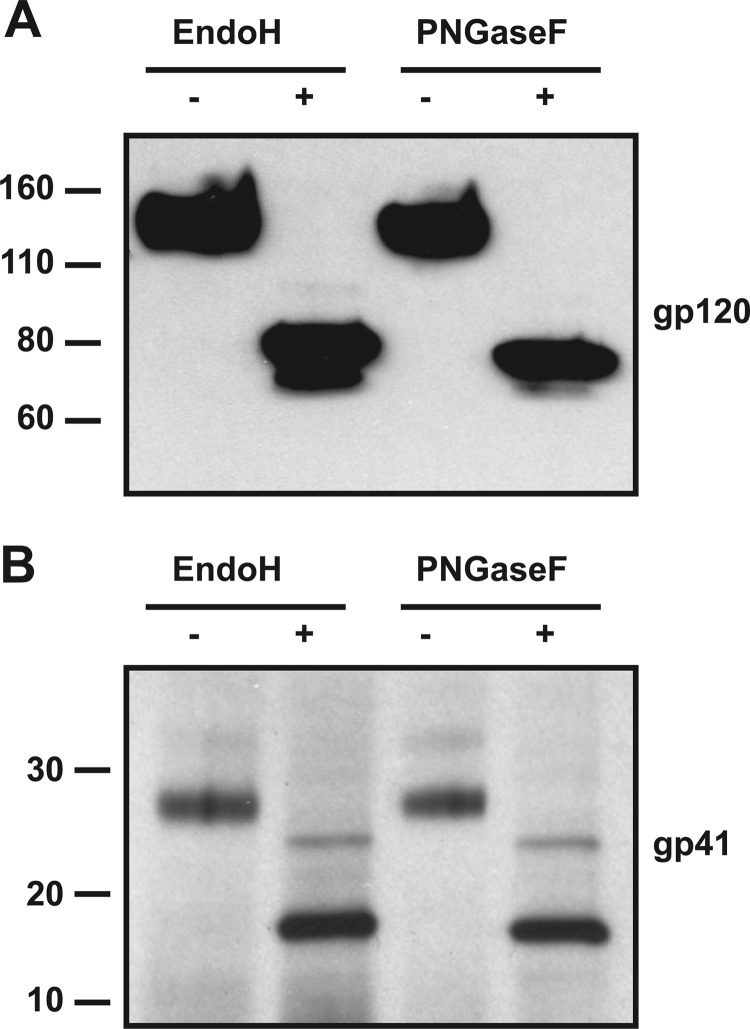
A notable finding from the tryptic peptide comparison is that all the glycans removed from denatured trimers by PNGase F or Endo H were also lost when Endo H was used under native conditions (Table 2). Thus, Endo H was able to deglycosylate trimers with similar efficiencies under either nondenaturing or denaturing conditions, and to an extent comparable with what was achieved using PNGase F under denaturing conditions (Fig. 7). In other words, every glycan moiety on SOSIP.681GnT−/− trimers that is identifiable in the MALDI-TOF analyses and that is vulnerable to Endo H, is removed under mild conditions compatible with maintaining trimer integrity; prior denaturation of the trimers makes no discernible difference.
FIGURE 7.
Comparative analysis of the extent of deglycosylation of SOSIP.681GnTl−/− trimers treated either with Endo H under mild conditions or with PNGase F after prior denaturation. The SDS-PAGE gels were blotted with either (A) the anti-gp120 mAb ARP3119 or (B) the anti-gp41 mAb D50.
The tryptic peptides from the untreated SOSIP.681GnT−/− sample suggested that the Asn-197 canonical site was not glycosylated. However, the presence of modified variants of the corresponding peptide in the PNGase F- and Endo H-treated samples implies that this site is, in fact, sometimes glycosylated. Moreover, when a glycan is present at the Asn-197 site, it is sensitive to these enzymes. The peptide containing the two potential gp41 glycosylation sites at positions Asn-611 and Asn-616 was found to contain only one new aspartic acid in the PNGase F-treated sample, and only a single added GlcNAc moiety in the Endo H-treated samples. Hence only one of the two sites at Asn-611 and Asn-616 is actually glycan-occupied, but we cannot identify which one. The glycan-free status of one of the four canonical sites on gp41 is consistent with the outcome of the SDS-PAGE analyses (Fig. 3B).
The MALDI-TOF analyses show that, of the 28 canonical glycosylation sites on each protomer of a SOSIP.681GnT−/− trimer, one is not used at all (either Asn-611 or Asn-616 in gp41), and one only sometimes (Asn-197 in gp120). Endo H can remove glycans from 17 sites, those at gp120 positions 88, 130, 136, 146, 156, 188, 193, 197, 234, 241, 276, 301, 448, 463, and gp41 positions 611 or 616, 625 and 637 (Table 2). We can draw no definite conclusions about the remaining 10 sites, i.e. those at positions 262, 295, 332, 339, 355, 362, 386, 392, 396, and 411, which form a glycan cluster on the outer domain of gp120. The SEC-UV/MALS/RI analyses implied that between 0 and 7 sites are insensitive to Endo H. These two independent estimates are, therefore, in reasonable agreement.
Antigenic Structure of Deglycosylated gp140 Trimers
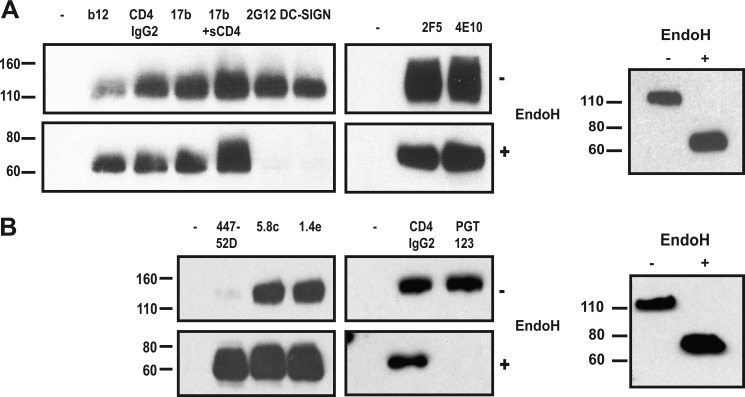
To study whether deglycosylation affects the antigenic structure of trimers, we used a panel of mAbs against gp120 and gp41 neutralization epitopes, and also reagents that bind glycans. Purified deglycosylated and mock-treated SOSIP.664GnT−/− trimers were immunoprecipitated with the various reagents, and then analyzed by SDS-PAGE and Western blot. Most mAbs and the CD4 receptor analog CD4-IgG2 reacted equally efficiently with the untreated and deglycosylated trimers (Fig. 8A). There was an ∼50% increase in b12 binding, in agreement with previous studies showing that glycans, in particular the one at position 386, can restrict access to the b12 epitope (74, 75). Deglycosylation had no effect on basal or sCD4-induced interactions between the trimers and the coreceptor binding site-directed antibody 17b. There were also no apparent differences in the binding of the gp41-MPER directed mAbs 2F5 and 4E10 (Fig. 8A, lower panel). However, as expected, the glycan-dependent ligands 2G12 and DC-SIGN did not bind to the deglycosylated trimers (Fig. 8A, upper panel).
FIGURE 8.
mAb reactivity of deglycosylated SOSIP.664GnTl−/− trimers. The trimers were either Endo H-treated or mock-treated, and then immunoprecipitated with various mAbs (left lanes, no mAb controls). The precipitated trimers were analyzed by SDS-PAGE and Western blotted using the anti-gp120 antibody ARP3119. A, gp120-binding agents b12, CD4-IgG2, 17b ± sCD4, 2G12, and DC-SIGN; and anti-gp41 MAbs 2F5 and 4E10; B, V3 mAbs 447–52D, 5.8c and 1.4e; and CD4-IgG2 and PGT123. The right panels represent loading controls for the immunoprecipitation reactions in A and B.
The V3 region of gp120 and its base are important determinants of coreceptor usage that also contain neutralization epitopes (76). Three glycosylation sites are located in or close to V3 (Fig. 1A). Eliminating the glycan from residue 301 at the base of V3 via mutagenesis (77–79), or generating smaller glycans by producing gp120 in GnT−/− cells (14), increases the accessibility of V3 neutralization epitopes to antibodies. Consistent with these observations, mAb 447–52D and 19b both bound more strongly to deglycosylated trimers than untreated ones (Fig. 8B and data not shown). The epitope for 19b includes residues in both flanks of V3, whereas 447–52D recognizes the relatively conserved tip (80, 81). There was also a modest increase in the binding of two other V3-directed mAbs, 5.8c and 1.4e (Fig. 8B). Finally, we tested the recently described glycan-dependent mAb PGT123 (29). This mAb interacted efficiently with untreated SOSIP.664GnT−/− trimers, but not Endo H-treated ones, consistent with the loss of the glycan from site 332 that is a component of the PGT123 epitope (Fig. 8C).
Negative Stain Electron Microscopy Analysis of SOSIP.664GnT−/− Trimer Morphology
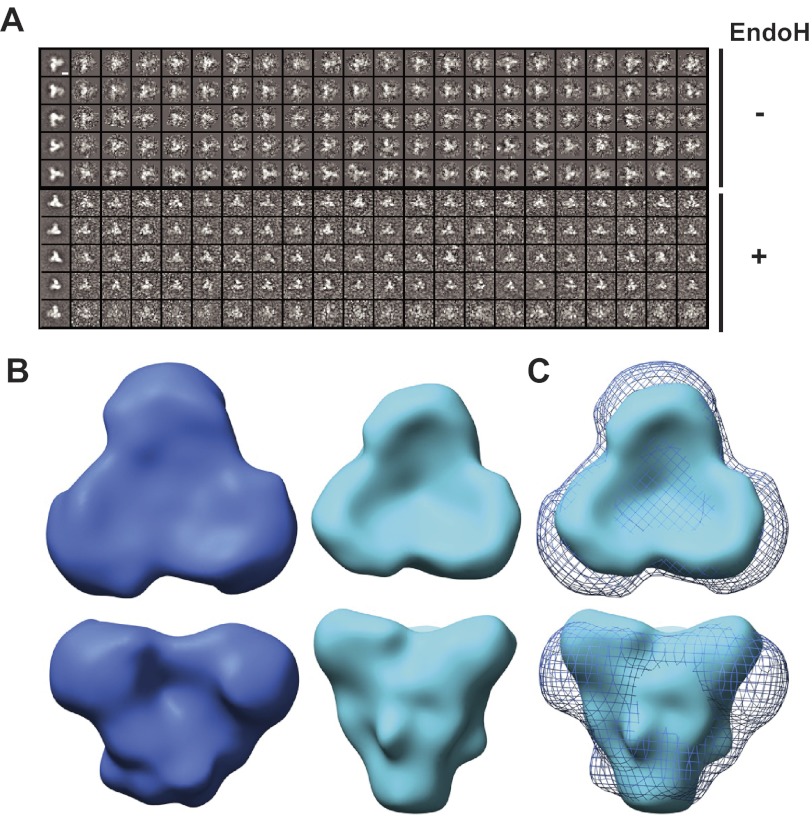
We used negative stain electron microscopy (Fig. 9A) and image reconstruction to visualize untreated and Endo H-treated SOSIP.664GnT−/− trimers (Fig. 9B). Both trimers exhibit a similar morphology, with the principal differences arising at the periphery of the head domains. Thus, additional density, likely corresponding to glycans, protrudes from the head of the glycosylated untreated SOSIP.664GnT−/− trimer, where the glycan-rich gp120 outer domains are expected to be located. Overall, the electron microscopy images demonstrate that the trimers are morphologically similar to one another, but with a clear size reduction between the fully glycosylated trimer and its deglycosylated counterpart (Fig. 9C).
FIGURE 9.
Single particle image reconstructions of negatively stained SOSIP.664GnTl−/− trimers. A, five representative, reference-free two-dimensional class averages (left images) and the corresponding images of oriented raw particles are shown. The top five class averages belong to the fully glycosylated trimers, the lower five to the Endo H-treated trimers. The white scale bar, located in the top left image, corresponds to 50 Å. B, top and side views of trimer reconstructions of untreated, fully glycosylated (blue) and Endo H-treated trimers (cyan) that have been low-pass filtered to 20-Å resolution. Contour levels corresponding to experimentally determined masses by SEC-UV/MALS/RI (see “Materials and Methods”) were calculated using the “volume” algorithm in EMAN. Untreated trimers were contoured using a volume of ∼390 kDa, whereas the Endo H-treated trimers were contoured with a volume of ∼260 kDa. The overall shapes of the trimers are conserved. C, overlay of untreated (blue mesh) and Endo H-treated (cyan surface) trimers, highlighting the conserved morphology and the size difference between the trimers.
DISCUSSION
Our goal was to devise a way to produce conformationally intact, deglycosylated soluble HIV-1 Env trimers to facilitate crystallography and immunogenicity studies. By using a combination of a cell line (293S GnTI−/−) in which the addition of complex sugars does not occur, followed by enzymatic deglycosylation with Endo H under mild conditions, we could make gp140 trimers that lack most of their original N-linked glycan content. The remaining glycan structures are positioned on the trimer in orientations that appear to make them inaccessible to Endo H and other glycosidases.
The intrinsic flexibility and heterogeneity associated with carbohydrate moieties on glycoproteins make it challenging to crystallize such material, but we believe not impossible. In general, any reduction in the glycan content should, to some extent, increase the likelihood of a glycoprotein crystallizing, provided its structural integrity and stability are still maintained. The partially deglycosylated Env trimers are, therefore, more suitable than their fully glycosylated counterparts for crystallization studies. It is relevant that the Endo H-resistant glycans remaining on the partially deglycosylated trimers are relatively homogeneous, as they are all inaccessible to various carbohydrate-processing enzymes of the glycosylation pathway (and also produced in a cell line with a block in the glycosylation pathway). Overall, we believe that the partial deglycosylation of the trimers represents progress toward a crystallization substrate. Whether the procedures we have described here are sufficient remains to be determined, but we think we have characterized a necessary part of the process. For successful crystallization of the partially deglycosylated trimer, we are now testing whether it is necessary to also use ligands, such as antibodies, to provide additional crystal packing contacts in regions where glycans are still present.
Glycoprotein deglycosylation can cause several adverse effects, including protein aggregation, the dissociation of oligomeric complexes, an increased susceptibility to proteolytic degradation, and a loss of native conformation (72). However, we found that the partially deglycosylated SOSIP.664 trimers neither dissociated nor aggregated. Hence the Endo H-accessible glycans are not major contributors to intersubunit interactions within the trimer. Moreover, their loss does not expose hydrophobic surfaces that can cause proteins to aggregate, something that has been suggested to occur with Env trimers (32, 82). Glycan removal did increase protease sensitivity to a modest extent, implying that some scissile sites in the peptide backbone become more accessible. However, the partially deglycosylated trimers are robust enough to be used for various purposes without taking extraordinary precautions to prevent degradation.
The electron microscopy studies gave no indication that glycan removal adversely affected the overall conformation of the trimers. This is further evidence that the Endo H-accessible N-glycans do not contribute to the structural integrity of trimeric Env once the structure is assembled into its native conformation. The efficient binding of mAbs to conformation-sensitive, protein-only epitopes also suggests that the structural integrity of the partially deglycosylated trimers is preserved. Indeed, epitopes associated with the CD4 binding site (e.g. bNAb b12) and the V3 region (e.g. MAbs 19b and 447–52D) were more accessible on the deglycosylated trimers. The elimination of topologically proximal glycans by mutagenesis is known to increase the accessibility of the b12 and V3 epitopes on monomeric gp120. More distant glycans may also be involved in shielding these epitopes on the trimer, however. As expected, the binding sites for the glycan-dependent bNAbs 2G12 and PGT123 (as well as the C-type lectin DC-SIGN) were lost upon glycan removal. Clearly, NAbs of this particular specificity would not be induced by immunization with glycan-depleted trimers. Whether the favorable effects on the exposure of protein-only bNAb epitopes outweigh the loss of glycan-dependent bNAb epitopes remains to be determined.
Although there is likely to be some heterogeneity between individual trimers, the mass spectrometry data show that glycans could be removed from positions 88, 130, 136, 146, 156, 188, 193, 197, 234, 241, 276, 301, 448, and 463 on gp120, and from positions 611 or 616, 625, and 637 on gp41. The loss of glycan-301 would account for the increase in V3 mAb binding (14), whereas the loss of glycan-448 may explain why 2G12 no longer binds (34).
By implication, the remaining 10 sites on each protomer of the trimer may still contain glycans after Endo H treatment, provided of course that they were actually glycosylated in the first place. These sites are 262, 295, 332, 339, 355, 362, 386, 392, 396, and 411, most of which are part of an oligomannose patch on the outer domain of gp120. It is possible that some of the corresponding peptides went undetected in the MALDI-TOF analyses for technical reasons, and not because they were glycosylated. SEC-UV/MALS/RI assays independently imply that ∼0–7 (or slightly more if the glycans in question are smaller than Man9GlcNAc2) sites remain glycosylated after Endo H digestion. The estimates from these two techniques seem reasonably concordant, with any discrepancies being attributable to the lack of detection by MALDI-TOF and/or measurement imprecisions in the SEC-UV/MALS/RI analyses.
One specific discrepancy relates to the glycan at site Asn-332, which is critical for binding of the PGT123 mAb. Indeed, alanine scanning mutagenesis suggests that no other glycan contributes to the PGT123 epitope (29). No peptide corresponding to the Asn-332 site was detected in the MALDI-TOF analyses, so this technique did not provide confirmation that Endo H removes the glycan from residue 332. Nonetheless, the lack of binding of PGT123 to the deglycosylated trimers implies that the Asn-332 glycan is, in fact, Endo H-sensitive.
We found that only three of the four canonical glycosylation sites in gp41 are utilized. In the MALDI-TOF analyses, the peptide containing the two canonical sites at residues 611 and 616 was occupied by only one glycan (which was removed by Endo H). Thus, the gp41 ectodomain on SOSIP.681GnT−/− trimers contains glycans at positions 625 and 637 and a third that is located at either position 611 or 616. All three are removed by Endo H treatment under mild conditions, although some minor, partially deglycosylated forms of gp41 were detected (Fig. 4B).
In general, our findings are consistent with a recent report on PNGase F-mediated deglycosylation of uncleaved JR-FL and CON-S gp140 trimers (43). Thus, in both studies, a subset of the total glycans could not be removed. However, there are also some differences. For example, the asparagine at gp41 position 637 was not occupied on the uncleaved JR-FL and CON-S gp140 trimers, but is used on the cleaved KNH1144 trimers. In addition, we found that the glycans at positions 301 and 448 were removed, whereas they were not efficiently digested away from the uncleaved trimers (43). Whether differences in the design of the trimers (cleaved versus uncleaved, with or without the gp41 fusion peptide), the HIV-1 sequences on which they are based (KNH1144 versus JR-FL/CON-S), the deglycosylation protocol (Endo H versus PNGase F), or the analytical method used to assess deglycosylation, underlie these differences remains unknown.
In summary, we report partial deglycosylation of cleaved HIV-1 Env gp140 trimers under conditions that conserve their most fundamental features. The glycan-depleted trimers may be useful for both structural and immunogenicity studies.
Acknowledgments
We thank James Robinson and Dennis Burton for donating reagents and Kenneth Kang for technical assistance. The electron microscopy data were collected by the authors at the National Resource for Automated Molecular Microscopy (NRAMM), which is supported by the National Institutes of Health through the National Center for Research Resources' P41 program Grant RR017573.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 AI82362 and R01 AI36082 and the International AIDS Vaccine Initiative (IAVI).
R. S. Depetris, J.-P. Julien, R. Khayat, J. H. Lee, R. Pejchal, U. Katpally, N. Cocco, M. Kachare, E. Massi, K. B. David, A. Cupo, A. J. Maroszan, W. C. Olson, A. B. Ward, I. A. Wilson, R. W. Sanders, and J. P. Moore, unpublished results.
- NAb
- neutralizing antibodies
- bNAb
- broadly active NAb
- Endo F
- endoglycosidase F
- Endo H
- endoglycosidase H
- GnTI
- N-acetylglucosaminyltransferase I
- MALS
- multi-angle light scattering
- SEC
- size exclusion chromatography
- BN
- Blue Native
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid
- RI
- refractive index.
REFERENCES
- 1. Gojobori T., Moriyama E. N., Ina Y., Ikeo K., Miura T., Tsujimoto H., Hayami M., Yokoyama S. (1990) Evolutionary origin of human and simian immunodeficiency viruses. Proc. Natl. Acad. Sci. U.S.A. 87, 4108–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kornfeld S., Li E., Tabas I. (1978) The synthesis of complex-type oligosaccharides. II. Characterization of the processing intermediates in the synthesis of the complex oligosaccharide units of the vesicular stomatitis virus G protein. J. Biol. Chem. 253, 7771–7778 [PubMed] [Google Scholar]
- 3. Go E. P., Irungu J., Zhang Y., Dalpathado D. S., Liao H. X., Sutherland L. L., Alam S. M., Haynes B. F., Desaire H. (2008) Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes' accessibility. J. Proteome Res. 7, 1660–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Go E. P., Chang Q., Liao H. X., Sutherland L. L., Alam S. M., Haynes B. F., Desaire H. (2009) Glycosylation site-specific analysis of clade C HIV-1 envelope proteins. J. Proteome Res. 8, 4231–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sagar M., Wu X., Lee S., Overbaugh J. (2006) Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J. Virol. 80, 9586–9598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Go E. P., Rebecchi K. R., Dalpathado D. S., Bandu M. L., Zhang Y., Desaire H. (2007) GlycoPep DB. A tool for glycopeptide analysis using a “Smart Search.” Anal. Chem, 79, 1708–1713 [DOI] [PubMed] [Google Scholar]
- 7. Perrin C., Fenouillet E., Jones I. M. (1998) Role of gp41 glycosylation sites in the biological activity of human immunodeficiency virus type 1 envelope glycoprotein. Virology 242, 338–345 [DOI] [PubMed] [Google Scholar]
- 8. Fenouillet E., Clerget-Raslain B., Gluckman J. C., Guétard D., Montagnier L., Bahraoui E. (1989) Role of N-linked glycans in the interaction between the envelope glycoprotein of human immunodeficiency virus and its CD4 cellular receptor. Structural enzymatic analysis. J. Exp. Med. 169, 807–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stansell E., Canis K., Haslam S. M., Dell A., Desrosiers R. C. (2011) Simian immunodeficiency virus from the sooty mangabey and rhesus macaque is modified with O-linked carbohydrate. J. Virol. 85, 582–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stansell E., Desrosiers R. C. (2010) Functional contributions of carbohydrate on AIDS virus glycoprotein. Yale J. Biol. Med. 83, 201–208 [PMC free article] [PubMed] [Google Scholar]
- 11. Fischer E., Brossmer R. (1995) Sialic acid-binding lectins. Submolecular specificity and interaction with sialoglycoproteins and tumor cells. Glycoconj. J. 12, 707–713 [DOI] [PubMed] [Google Scholar]
- 12. Fenouillet E., Gluckman J. C., Bahraoui E. (1990) Role of N-linked glycans of envelope glycoproteins in infectivity of human immunodeficiency virus type 1. J. Virol. 64, 2841–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fenouillet E., Gluckman J. C., Jones I. M. (1994) Functions of HIV envelope glycans. Trends Biochem. Sci. 19, 65–70 [DOI] [PubMed] [Google Scholar]
- 14. Binley J. M., Ban Y. E., Crooks E. T., Eggink D., Osawa K., Schief W. R., Sanders R. W. (2010) Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. J. Virol. 84, 5637–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Montefiori D. C., Robinson W. E., Jr., Mitchell W. M. (1988) Role of protein N-glycosylation in pathogenesis of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U.S.A. 85, 9248–9252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Montefiori D. C., Stewart K., Ahearn J. M., Zhou J., Zhou J. (1993) Complement-mediated binding of naturally glycosylated and glycosylation-modified human immunodeficiency virus type 1 to human CR2 (CD21). J. Virol. 67, 2699–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Means R. E., Desrosiers R. C. (2000) Resistance of native, oligomeric envelope on simian immunodeficiency virus to digestion by glycosidases. J. Virol. 74, 11181–11190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geijtenbeek T. B., Torensma R., van Vliet S. J., van Duijnhoven G. C., Adema G. J., van Kooyk Y., Figdor C. G. (2000) Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 100, 575–585 [DOI] [PubMed] [Google Scholar]
- 19. Marzi A., Mitchell D. A., Chaipan C., Fisch T., Doms R. W., Carrington M., Desrosiers R. C., Pöhlmann S. (2007) Modulation of HIV and SIV neutralization sensitivity by DC-SIGN and mannose-binding lectin. Virology 368, 322–330 [DOI] [PubMed] [Google Scholar]
- 20. Banerjee K., Michael E., Eggink D., van Montfort T., Lasnik A. B., Palmer K. E., Sanders R. W., Moore J. P., Klasse P. J. (2012) Occluding the mannose moieties on human immunodeficiency virus type 1 gp120 with griffithsin improves the antibody responses to both proteins in mice. AIDS Res. Hum. Retroviruses 28, 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Banerjee K., Andjelic S., Klasse P. J., Kang Y., Sanders R. W., Michael E., Durso R. J., Ketas T. J., Olson W. C., Moore J. P. (2009) Enzymatic removal of mannose moieties can increase the immune response to HIV-1 gp120 in vivo. Virology 389, 108–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shan M., Klasse P. J., Banerjee K., Dey A. K., Iyer S. P., Dionisio R., Charles D., Campbell-Gardener L., Olson W. C., Sanders R. W., Moore J. P.(2007) HIV-1 gp120 mannoses induce immunosuppressive responses from dendritic cells. PLoS Pathog. 3, e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reitter J. N., Means R. E., Desrosiers R. C. (1998) A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4, 679–684 [DOI] [PubMed] [Google Scholar]
- 24. Klasse P. J., Sanders R. W., Cerutti A., Moore J. P. (2012) How can HIV type 1-Env immunogenicity be improved to facilitate antibody-based vaccine development? AIDS Res. Hum. Retroviruses 28, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martinelli E., Cicala C., Van Ryk D., Goode D. J., Macleod K., Arthos J., Fauci A. S. (2007) HIV-1 gp120 inhibits TLR9-mediated activation and IFN-α secretion in plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 104, 3396–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wei X., Decker J. M., Wang S., Hui H., Kappes J. C., Wu X., Salazar-Gonzalez J. F., Salazar M. G., Kilby J. M., Saag M. S., Komarova N. L., Nowak M. A., Hahn B. H., Kwong P. D., Shaw G. M. (2003) Antibody neutralization and escape by HIV-1. Nature 422, 307–312 [DOI] [PubMed] [Google Scholar]
- 27. Pantophlet R., Burton D. R. (2006) GP120. Target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24, 739–769 [DOI] [PubMed] [Google Scholar]
- 28. Walker L. M., Phogat S. K., Chan-Hui P. Y., Wagner D., Phung P., Goss J. L., Wrin T., Simek M. D., Fling S., Mitcham J. L., Lehrman J. K., Priddy F. H., Olsen O. A., Frey S. M., Hammond P. W., Protocol G Principal Investigators, Kaminsky S., Zamb T., Moyle M., Koff W. C., Poignard P., Burton D. R. (2009) Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Walker L. M., Huber M., Doores K. J., Falkowska E., Pejchal R., Julien J. P., Wang S. K., Ramos A., Chan-Hui P. Y., Moyle M., Mitcham J. L., Hammond P. W., Olsen O. A., Phung P., Fling S., Wong C. H., Phogat S., Wrin T., Simek M. D., Protocol G Principal Investigators, Koff W. C., Wilson I. A., Burton D. R., Poignard P. (2011) Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walker L. M., Simek M. D., Priddy F., Gach J. S., Wagner D., Zwick M. B., Phogat S. K., Poignard P., Burton D. R. (2010) A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6, e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Calarese D. A., Lee H. K., Huang C. Y., Best M. D., Astronomo R. D., Stanfield R. L., Katinger H., Burton D. R., Wong C. H., Wilson I. A. (2005) Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc. Natl. Acad. Sci. U.S.A. 102, 13372–13377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scanlan C. N., Pantophlet R., Wormald M. R., Saphire E. O., Calarese D., Stanfield R., Wilson I. A., Katinger H., Dwek R. A., Burton D. R., Rudd P. M. (2003) The carbohydrate epitope of the neutralizing anti-HIV-1 antibody 2G12. Adv. Exp. Med. Biol. 535, 205–218 [DOI] [PubMed] [Google Scholar]
- 33. Scanlan C. N., Pantophlet R., Wormald M. R., Ollmann Saphire E., Stanfield R., Wilson I. A., Katinger H., Dwek R. A., Rudd P. M., Burton D. R. (2002) The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1 → 2 mannose residues on the outer face of gp120. J. Virol. 76, 7306–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanders R. W., Venturi M., Schiffner L., Kalyanaraman R., Katinger H., Lloyd K. O., Kwong P. D., Moore J. P. (2002) The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76, 7293–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Doores K. J., Burton D. R. (2010) Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J. Virol. 84, 10510–10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McLellan J. S., Pancera M., Carrico C., Gorman J., Julien J. P., Khayat R., Louder R., Pejchal R., Sastry M., Dai K., O'Dell S., Patel N., Shahzad-ul-Hussan S., Yang Y., Zhang B., Zhou T., Zhu J., Boyington J. C., Chuang G. Y., Diwanji D., Georgiev I., Kwon Y. D., Lee D., Louder M. K., Moquin S., Schmidt S. D., Yang Z. Y., Bonsignori M., Crump J. A., Kapiga S. H., Sam N. E., Haynes B. F., Burton D. R., Koff W. C., Walker L. M., Phogat S., Wyatt R., Orwenyo J., Wang L. X., Arthos J., Bewley C. A., Mascola J. R., Nabel G. J., Schief W. R., Ward A. B., Wilson I. A., Kwong P. D. (2011) Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pejchal R., Doores K. J., Walker L. M., Khayat R., Huang P. S., Wang S. K., Stanfield R. L., Julien J. P., Ramos A., Crispin M., Depetris R., Katpally U., Marozsan A., Cupo A., Maloveste S., Liu Y., McBride R., Ito Y., Sanders R. W., Ogohara C., Paulson J. C., Feizi T., Scanlan C. N., Wong C. H., Moore J. P., Olson W. C., Ward A. B., Poignard P., Schief W. R., Burton D. R., Wilson I. A. (2011) A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334, 1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kwong P. D., Wyatt R., Desjardins E., Robinson J., Culp J. S., Hellmig B. D., Sweet R. W., Sodroski J., Hendrickson W. A. (1999) Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1). J. Biol. Chem. 274, 4115–4123 [DOI] [PubMed] [Google Scholar]
- 39. Huang C. C., Lam S. N., Acharya P., Tang M., Xiang S. H., Hussan S. S., Stanfield R. L., Robinson J., Sodroski J., Wilson I. A., Wyatt R., Bewley C. A., Kwong P. D. (2007) Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317, 1930–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang C. C., Tang M., Zhang M. Y., Majeed S., Montabana E., Stanfield R. L., Dimitrov D. S., Korber B., Sodroski J., Wilson I. A., Wyatt R., Kwong P. D.(2005) Structure of a V3-containing HIV-1 gp120 core. Science 310, 1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou T., Xu L., Dey B., Hessell A. J., Van Ryk D., Xiang S. H., Yang X., Zhang M. Y., Zwick M. B., Arthos J., Burton D. R., Dimitrov D. S., Sodroski J., Wyatt R., Nabel G. J., Kwong P. D. (2007) Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445, 732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Crooks E. T., Tong T., Osawa K., Binley J. M. (2011) Enzyme digests eliminate nonfunctional Env from HIV-1 particle surfaces, leaving native Env trimers intact and viral infectivity unaffected. J. Virol. 85, 5825–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ma B. J., Alam S. M., Go E. P., Lu X., Desaire H., Tomaras G. D., Bowman C., Sutherland L. L., Scearce R. M., Santra S., Letvin N. L., Kepler T. B., Liao H. X., Haynes B. F. (2011) Envelope deglycosylation enhances antigenicity of HIV-1 gp41 epitopes for both broad neutralizing antibodies and their unmutated ancestor antibodies. PLoS Pathog. 7, e1002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Binley J. M., Sanders R. W., Clas B., Schuelke N., Master A., Guo Y., Kajumo F., Anselma D. J., Maddon P. J., Olson W. C., Moore J. P. (2000) A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74, 627–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sanders R. W., Vesanen M., Schuelke N., Master A., Schiffner L., Kalyanaraman R., Paluch M., Berkhout B., Maddon P. J., Olson W. C., Lu M., Moore J. P. (2002) Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76, 8875–8889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harris A., Borgnia M. J., Shi D., Bartesaghi A., He H., Pejchal R., Kang Y. K., Depetris R., Marozsan A. J., Sanders R. W., Klasse P. J., Milne J. L., Wilson I. A., Olson W. C., Moore J. P., Subramaniam S. (2011) Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc. Natl. Acad. Sci. U.S.A. 108, 11440–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dey A. K., David K. B., Lu M., Moore J. P. (2009) Biochemical and biophysical comparison of cleaved and uncleaved soluble, trimeric HIV-1 envelope glycoproteins. Virology 385, 275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Iyer S. P., Franti M., Krauchuk A. A., Fisch D. N., Ouattara A. A., Roux K. H., Krawiec L., Dey A. K., Beddows S., Maddon P. J., Moore J. P., Olson W. C. (2007) Purified, proteolytically mature HIV type 1 SOSIP gp140 envelope trimers. AIDS Res. Hum. Retroviruses 23, 817–828 [DOI] [PubMed] [Google Scholar]
- 49. Beddows S., Franti M., Dey A. K., Kirschner M., Iyer S. P., Fisch D. C., Ketas T., Yuste E., Desrosiers R. C., Klasse P. J., Maddon P. J., Olson W. C., Moore J. P. (2007) A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology 360, 329–340 [DOI] [PubMed] [Google Scholar]
- 50. Kang Y. K., Andjelic S., Binley J. M., Crooks E. T., Franti M., Iyer S. P., Donovan G. P., Dey A. K., Zhu P., Roux K. H., Durso R. J., Parsons T. F., Maddon P. J., Moore J. P., Olson W. C.(2009) Structural and immunogenicity studies of a cleaved, stabilized envelope trimer derived from subtype A HIV-1. Vaccine 27, 5120–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Beddows S., Schülke N., Kirschner M., Barnes K., Franti M., Michael E., Ketas T., Sanders R. W., Maddon P. J., Olson W. C., Moore J. P. (2005) Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 79, 8812–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Melchers M., Bontjer I., Tong T., Chung N. P., Klasse P. J., Eggink D., Montefiori D. C., Gentile M., Cerutti A., Olson W. C., Berkhout B., Binley J. M., Moore J. P., Sanders R. W. (2012) Targeting HIV-1 envelope glycoprotein trimers to B cells by using APRIL improves antibody responses. J. Virol. 86, 2488–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Allaway G. P., Davis-Bruno K. L., Beaudry G. A., Garcia E. B., Wong E. L., Ryder A. M., Hasel K. W., Gauduin M. C., Koup R. A., McDougal J. S. (1995) Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res. Hum. Retroviruses 11, 533–539 [DOI] [PubMed] [Google Scholar]
- 54. Binley J. M., Sanders R. W, Master A., Cayanan C. S., Wiley C. L., Schiffner L., Travis B., Kuhmann S., Burton D. R., Hu S. L., Olson W. C., Moore J. P. (2002) Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 76, 2606–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reeves P. J., Kim J. M., Khorana H. G. (2002) Structure and function in rhodopsin. A tetracycline-inducible system in stable mammalian cell lines for high-level expression of opsin mutants. Proc. Natl. Acad. Sci. U.S.A. 99, 13413–13418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kirschner M., Monrose V., Paluch M., Techodamrongsin N., Rethwilm A., Moore J. P. (2006) The production of cleaved, trimeric human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein vaccine antigens and infectious pseudoviruses using linear polyethylenimine as a transfection reagent. Protein Expr. Purif. 48, 61–68 [DOI] [PubMed] [Google Scholar]
- 57. Schägger H., von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 58. Wen J., Arakawa T., Philo J. S. (1996) Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal. Biochem. 240, 155–166 [DOI] [PubMed] [Google Scholar]
- 59. Arakawa T., Wen J. (2001) Determination of carbohydrate contents from excess light scattering. Anal. Biochem. 299, 158–161 [DOI] [PubMed] [Google Scholar]
- 60. Kendrick B. S., Kerwin B. A., Chang B. S., Philo J. S. (2001) Online size-exclusion high-performance liquid chromatography light scattering and differential refractometry methods to determine degree of polymer conjugation to proteins and protein-protein or protein-ligand association states. Anal. Biochem. 299, 136–146 [DOI] [PubMed] [Google Scholar]
- 61. Lander G. C., Stagg S. M., Voss N. R., Cheng A., Fellmann D., Pulokas J., Yoshioka C., Irving C., Mulder A., Lau P. W., Lyumkis D., Potter C. S., Carragher B. (2009) Appion. An integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Voss N. R., Yoshioka C. K., Radermacher M., Potter C. S, Carragher B. (2009) DoG Picker and TiltPicker. Software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mindell J. A., Grigorieff N. (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 [DOI] [PubMed] [Google Scholar]
- 64. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 65. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J. (2007) EMAN2. An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 [DOI] [PubMed] [Google Scholar]
- 66. Hohn M., Tang G., Goodyear G., Baldwin P. R., Huang Z., Penczek P. A., Yang C., Glaeser R. M., Adams P. D., Ludtke S. J. (2007) SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 157, 47–55 [DOI] [PubMed] [Google Scholar]
- 67. Chang V. T., Crispin M., Aricescu A. R., Harvey D. J., Nettleship J. E., Fennelly J. A., Yu C., Boles K. S., Evans E. J., Stuart D. I., Dwek R. A., Jones E. Y., Owens R. J., Davis S. J. (2007) Glycoprotein structural genomics. Solving the glycosylation problem. Structure 15, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Beddows S., Kirschner M., Campbell-Gardener L., Franti M., Dey A. K., Iyer S. P., Maddon P. J., Paluch M., Master A., Overbaugh J., VanCott T., Olson W. C., Moore J. P. (2006) Construction and characterization of soluble, cleaved, and stabilized trimeric Env proteins based on HIV type 1 Env subtype A. AIDS Res. Hum. Retroviruses 22, 569–579 [DOI] [PubMed] [Google Scholar]
- 69. Eggink D., Melchers M., Wuhrer M., van Montfort T., Dey A. K., Naaijkens B. A., David K. B., Le Douce V., Deelder A. M., Kang K., Olson W. C., Berkhout B., Hokke C. H., Moore J. P., Sanders R. W. (2010) Lack of complex N-glycans on HIV-1 envelope glycoproteins preserves protein conformation and entry function. Virology 401, 236–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tarentino A. L., Plummer T. H., Jr., (1994) Enzymatic deglycosylation of asparagine-linked glycans. Purification, properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Methods Enzymol. 230, 44–57 [DOI] [PubMed] [Google Scholar]
- 71. Snaith S. M., Levvy G. A. (1968) Purification and properties of α-d-mannosidase from Jack bean meal. Biochem. J. 110, 663–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Varki A. (1993) Biological roles of oligosaccharides. All of the theories are correct. Glycobiology 3, 97–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Go E. P., Hewawasam G. S., Ma B. J., Liao H. X., Haynes B. F., Desaire H. (2011) Methods development for analysis of partially deglycosylated proteins and application to an HIV envelope protein vaccine candidate. Int. J. Mass Spectrom. 305, 209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sanders R. W., van Anken E., Nabatov A. A., Liscaljet I. M., Bontjer I., Eggink D., Melchers M., Busser E., Dankers M. M., Groot F., Braakman I., Berkhout B., Paxton W. A. (2008) The carbohydrate at asparagine 386 on HIV-1 gp120 is not essential for protein folding and function but is involved in immune evasion. Retrovirology 5, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pantophlet R., Ollmann Saphire E., Poignard P., Parren P. W., Wilson I. A., Burton D. R. (2003) Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77, 642–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rucker J., Edinger A. L., Sharron M., Samson M., Lee B., Berson J. F., Yi Y., Margulies B., Collman R. G., Doranz B. J., Parmentier M., Doms R. W. (1997) Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J. Virol. 71, 8999–9007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Back N. K., Smit L., De Jong J. J., Keulen W., Schutten M., Goudsmit J., Tersmette M. (1994) An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology 199, 431–438 [DOI] [PubMed] [Google Scholar]
- 78. Losman B., Biller M., Olofsson S., Schønning K., Lund O. S., Svennerholm B., Hansen J. E., Bolmstedt A. (1999) The N-linked glycan of the V3 region of HIV-1 gp120 and CXCR4-dependent multiplication of a human immunodeficiency virus type 1 lymphocyte-tropic variant. FEBS Lett. 454, 47–52 [DOI] [PubMed] [Google Scholar]
- 79. Malenbaum S. E., Yang D., Cavacini L., Posner M., Robinson J., Cheng-Mayer C. (2000) The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J. Virol. 74, 11008–11016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Stanfield R. L., Gorny M. K., Williams C., Zolla-Pazner S., Wilson I. A. (2004) Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447–52D. Structure 12, 193–204 [DOI] [PubMed] [Google Scholar]
- 81. Moore J. P., Trkola A., Korber B., Boots L. J., Kessler J. A., 2nd, McCutchan F. E., Mascola J., Ho D. D., Robinson J., Conley A. J. (1995) A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J. Virol. 69, 122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Scanlan C. N., Offer J., Zitzmann N., Dwek R. A.(2007) Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature 446, 1038–1045 [DOI] [PubMed] [Google Scholar]
- 83. Leonard C. K., Spellman M. W., Riddle L., Harris R. J., Thomas J. N., Gregory T. J. (1990) Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265, 10373–10382 [PubMed] [Google Scholar]
- 84. Bonomelli C., Doores K. J., Dunlop D. C., Thaney V., Dwek R. A., Burton D. R., Crispin M., Scanlan C. N. (2011) The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS One 6, e23521. [DOI] [PMC free article] [PubMed] [Google Scholar]