Background: Epigenetic mechanisms governing cardiac progenitor cell (CPC) differentiation are not well understood.
Results: Chromatin remodeler BAF250a/SWI/SNF regulates CPC differentiation in vivo and in vitro by modeling the chromatin accessibility at a subset of key genes essential for cardiogenesis.
Conclusion: BAF250a/SWI/SNF plays a key role in CPC differentiation.
Significance: Understanding the SWI/SNF-mediated epigenetic mechanism in CPC differentiation may provide molecular basis for novel heart therapies.
Keywords: Cardiac Development, Chromatin Remodeling, Differentiation, Epigenetics, Stem Cells, BAF250a, SWI/SNF, Cardiac Progenitor Cell Differentiation, Second Heart Field
Abstract
ATP-dependent SWI/SNF chromatin remodeling complexes alter the structure of chromatin at specific loci and facilitate tissue-specific gene regulation during development. Several SWI/SNF subunits are required for cardiogenesis. However, the function and mechanisms of SWI/SNF in mediating cardiac progenitor cell (CPC) differentiation during cardiogenesis are not well understood. Our studies of the SWI/SNF chromatin remodeling complex identified that BAF250a, a regulatory subunit of the SWI/SNF, plays a key role in CPC differentiation. BAF250a ablation in mouse second heart field (SHF) led to trabeculation defects in the right ventricle, ventricular septal defect, persistent truncus arteriosus, reduced myocardial proliferation, and embryonic lethality around E13. Using an embryonic stem cell culture system that models the formation and differentiation of SHF CPCs in vivo, we have shown that BAF250a ablation in CPCs specifically inhibits cardiomyocyte formation. Moreover, BAF250a selectively regulates the expression of key cardiac factors Mef2c, Nkx2.5, and Bmp10 in SHF CPCs. Chromatin immunoprecipitation and DNase I digestion assays indicate that BAF250a regulates gene expression by binding selectively to its target gene promoters and recruiting Brg1, the catalytic subunit of SWI/SNF, to modulate chromatin accessibility. Our results thus identify BAF250a-mediated chromatin remodeling as an essential epigenetic mechanism mediating CPC differentiation.
Introduction
Dissecting the mechanisms of multipotent cardiac progenitor cell (CPC)4 differentiation is essential for understanding the etiology of congenital heart defects and providing the molecular basis for cell-based heart therapies (1–6). Much is known about transcription factors in cardiogenesis and CPC development. However, our basic understanding of epigenetic mechanisms governing the differentiation of CPCs is rather limited. At present, the limited knowledge about the epigenetic regulation of lineage differentiation presents a considerable roadblock to developing cell-based therapies.
The ATP-dependent chromatin remodeling complexes alter chromatin structure that regulates the accessibility of DNA to transcription factors. The SWI/SNF complexes comprise one of the major families of ATP-dependent chromatin remodeling factors. The SWI/SNF complexes consist of 12 different subunits, which form BAF and PBAF complexes in mammals (7) and play a key role in various aspects of development (8). During development the ATP-dependent chromatin remodeling complexes function in a temporal-, spatial- and tissue-specific manner (7–11), and the variations of subunits in the complexes contribute to these specificities.
A series of studies indicate that SWI/SNF subunits are required for cardiogenesis (12–17). RNA interference of BAF60c leads to several heart defects including single ventricle, shorten outflow tract, and poor trabeculation (12). Interestingly, BAF60c can work together with GATA4 and Tbx5 to ectopically activate cardiomyocyte differentiation (13); and in zebrafish, BAF60c can guide CPC migration to the developing heart field (14). Deletion of BAF180, a specific subunit of the PBAF complex, causes cardiac morphogenesis and coronary development defects (15). Conditional KO of Brg1 in early myocardial lineage leads to trabeculation defects, and Brg1 regulates the conversion of α-MHC to β-MHC (16). The function of Brg1 in cardiogenesis has been shown to be dosage-dependent (17). Although several BAF complex subunits have been shown to be required during cardiogenesis, the functions of cardiac specific BAF complex during CPC differentiation are still unclear.
To decipher SWI/SNF-mediated epigenetic mechanisms in CPC differentiation, we have focused on a key regulatory subunit BAF250a (18) that appears essential for lineage specification during embryonic stem cell differentiation (19). We have applied tissue-specific ablation of BAF250a in second heart field (SHF) CPCs to study the roles of BAF250a/SWI/SNF in different stages during cardiac development. Our studies show that BAF250a is required for the proliferation and differentiation of cardiac progenitors both in vivo and in vitro. We identified genes regulated by BAF250a/BAF complex in cardiac cells such as Nkx2.5, Mef2c, and Bmp10. Our biochemical studies indicate that BAF250a binds to the promoter of these targets and regulates the recruitment and activity of the SWI/SNF catalytic subunit Brg1. Our results suggest that the BAF250a-containing SWI/SNF complex plays a key role in regulating the chromatin accessibility of cardiac-specific genes for proper cardiac progenitor differentiation.
EXPERIMENTAL PROCEDURES
Generation of Conditional Cardiac-specific BAF250a-KO Mice and ES Cell Lines
SHF lineage reporter ES cell lines were derived from E3.5 blastocysts. E3.5 blastocysts and cardiac-specific BAF250a-KO embryos were harvested from pregnant BAF250af/+;RosaYFP female mice mated with Mef2c-AHF-Cre (20);BAF250af/+ male mice as described in Ref. 19.
Histological Analysis
Embryos were harvested and fixed in 4% paraformaldehyde and dehydrated, embedded in paraffin, and sectioned. H&E staining was carried out using standard procedures.
Immunostaining
Embryos were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned. Sections were rehydrated. Antigen retrieval was performed in boiled water for 20 min. Sections were then blocked with 10% donkey serum in PBS buffer containing 0.1% Tween 20 for 1 h 37 °C and incubated with mouse anti-BAF250a (Abnova) or mouse anti-Isl1 (DSHB) at 37 °C for 1 h. The sections were then washed in PBS three times before being incubated with donkey anti-mouse Alexa Fluor 488-conjugated secondary antibody at 37 °C for 1 h. After washing, sections were mounted and analyzed by fluorescent microscopy.
Cell Sorting
A BD FACSAria cell sorter was used for quantifying and purifying Mef2c-YFP+ cells. Cells from embryos or day 6 embryoid bodies (EBs) were dissociated with 0.15% trypsin for 10 min at 37 °C. The dissociated cells were filtered in a 40-μm cell strainer before sorting. Lasers that detected PerCP and FITC were used to segregate the YFP+ and YFP− cells. Reanalysis of YFP+ cells was performed to confirm the purity of YFP+ cells.
Quantitative RT-PCR
Total RNA was isolated from FACS-purified ES cell-derived cardiac cells using an RNeasy Mini kit (Qiagen) as described according to the manufacturer's instructions. Quantitative RT-PCR was as described (19).
ES Cell Differentiation Assays
WT and BAF250a-conditional KO ES cells (500 cells) were seeded in one hanging drop in differentiation medium. The procedures for inducing cell differentiation were as described (21). Three independent experiments were performed for each cell type.
Chromatin Immunoprecipitation
Chromatin samples were isolated from E9.5 embryonic hearts and day 6 EB-derived CPCs. ChIP experiments were performed as described (15) using anti-BAF250a (Abnova)/anti-FLAG (mouse IgG; Sigma) or anti-Brg1 (22)/normal rabbit IgG (Santa Cruz Biotechnology). PCR was performed using following primers: Nkx2.5 forward, CCACCCCCAACCCTGCGTTT and reverse, AGGGGCCGCGACACATTTGG; Mef2c forward, GTAGAGGCTTGGGGTGGGGAG and reverse, GGCCAGGGTGCACCTGTTCAT; Gata4 forward, GTAGCGATCGCCTGCGCTGA and reverse, TGGGAAGAGTCCTGCGGGCG; Bmp10 forward, TCCATACTTGTGTCGTGTCCAGTGA and reverse, GCTCAGCTCCCAGGCCAACC.
DNase I Hypersensitivity Assay
Nuclei were purified from day 6 EB-derived CPCs as described (23). DNA samples were treated with different concentrations of DNase I (Worthington) for 3 min at room temperature. DNAs were then recovered by phenol-chloroform extraction and ethanol precipitation. DNase I accessibility was assessed by quantitative RT-PCR using the same primers as for ChIP.
RESULTS
Generation of BAF250a-conditional KO Embryos and ES Cell Lines
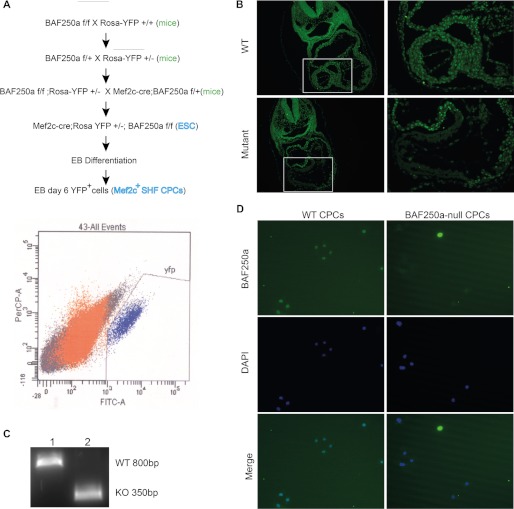
To study the potential functions of BAF250a in CPCs, we generated the BAF250a-conditional KO mice in SHF lineage by crossing BAF250af/f with Mef2c-AHF-Cre mice (19, 20) (Fig. 1A). To confirm the ablation of BAF250a mediated by Cre recombinase, immunofluorescent staining using BAF250a antibody was examined on transverse sections of both wild-type (WT) and BAF250a mutant embryos. In E9.5 WT embryos, BAF250a was highly expressed in heart tissues, whereas in E9.5 mutant hearts, BAF250a was undetectable in the outflow tract, right ventricle, and ventricular septum, which were typically derived from SHF (Fig. 1B), indicating that BAF250a was efficiently deleted by Mef2c-AHF-Cre recombinase.
FIGURE 1.
Establishing in vivo and in vitro systems to examine the function of BAF250a in CPC differentiation. A, diagram shows SHF CPC isolation accompanied by BAF250a deletion in vivo and in vitro. B, BAF250a was efficiently deleted from SHF by Mef2c-Cre in vivo as evidenced with immunostaining in E9.5 WT and mutant hearts. C, genotyping of FACS-purified YFP+ cells from day 6 WT and mutant Ebs is shown. Lane 1, BAF250af/f; lane 2, Mef2c-Cre;RosaYFP;BAF250af/f. D, BAF250a was also efficiently ablated from Mef2c+ SHF CPCs derived in vitro. BAF250a immunostaining was performed with SHF CPCs from day 6 EBs.
ES cell-based differentiation systems are capable of providing sufficient CPCs for in vitro differentiation models (21). Therefore, we generated ES cell lines that label SHF lineages by Mef2c-AHF-Cre-mediated Rosa YFP expression with both WT and BAF250af/f alleles. Mef2c-AHF-Cre expression should also simultaneously ablate BAF250a expression in BAF250af/f ES cells. Both Mef2c-AHF-Cre;R26-YFP;BAF250 WT and Mef2c-AHF-Cre;R26-YFP; BAF250f/f cell lines were subjected to EB-based in vitro differentiation. YFP+ CPCs were observed at day 6 EB culture, and beating YFP+ cells were observed at day 10. Genotyping and staining of the YFP+ cells derived from Mef2c-AHF-Cre;R26-YFP;BAF250f/f EBs revealed a complete deletion of BAF250a during differentiation (Fig. 1, C and D), indicating that the in vitro EB-based CPC differentiation system could be used to dissect BAF250a function in CPC differentiation.
BAF250a Is Required for SHF Development
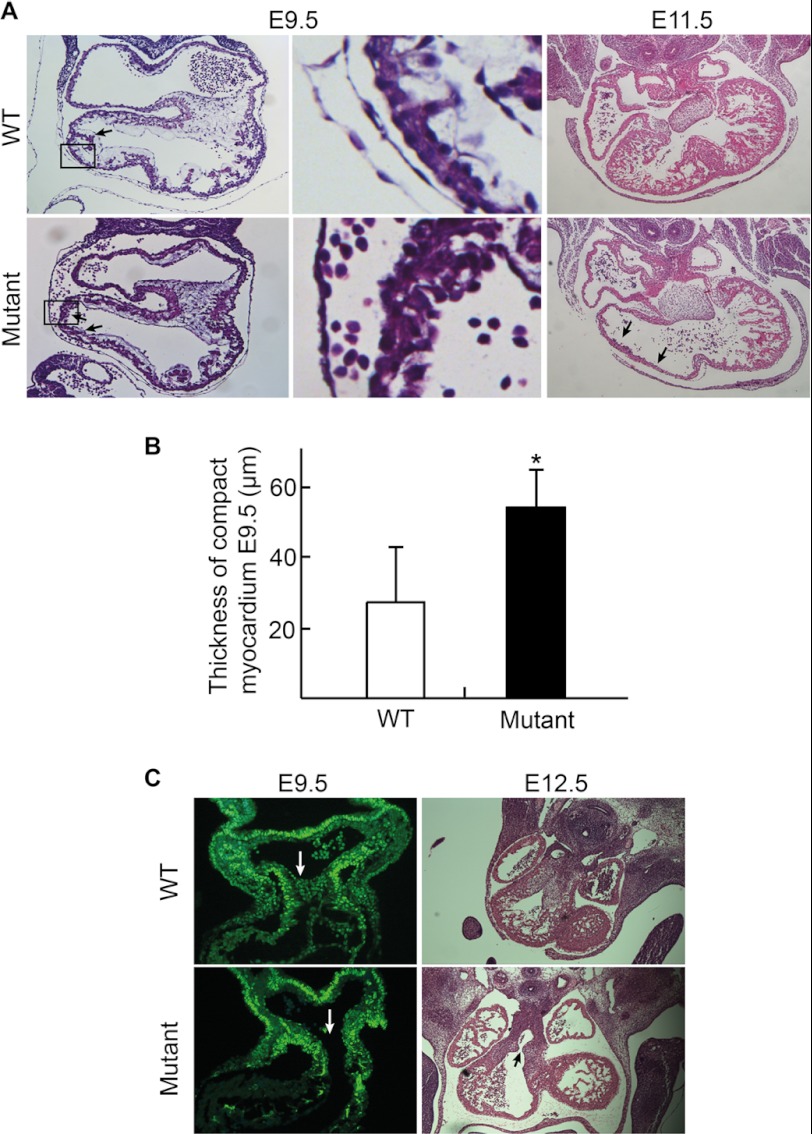
Deletion of BAF250a using Mef2c-AHF-Cre caused embryonic lethality, suggesting that BAF250a has critical functions in cardiac development. To determine the roles of BAF250a in SHF development, we carried out histological analyses on E9.5–E12.5 embryos. At E9.5, both the left and right ventricle developed trabeculae protrusions toward the ventricular lumen in WT embryos. In contrast, the BAF250a mutant hearts only developed few protrusions in the right ventricle whereas the growth of left ventricle was normal (Fig. 2A, left). At E10.5, the Mef2c-Cre;BAF250af/f embryos displayed cardiac hypoplastic wall in the right ventricle. At E11.5 in WT hearts, the trabeculae were extensively developed into the ventricular lumen in both left and right ventricles. However, in the BAF250a mutant embryos, only sparse trabeculae were developed in the right ventricle with normally developed left ventricle (Fig. 2A, right). The BAF250a mutant embryos also failed to form the ventricular septum (Fig. 2A, right). Furthermore, the average thickness of the ventricular compact zone in right ventricle was 26 ± 15 μm in WT, but was 54 ± 10 μm in mutant hearts at E9.5 (Fig. 2, A, middle, and B). Increase of the compact ventricular myocardium in the E9.5 mutants suggests that without BAF250a, cells about to differentiate into trabeculae are kept in the compact myocardium. The sparse trabeculae found in mutant embryos indicate that BAF250a is not required for initiation of cardiac trabeculation, but is crucial for the growth or differentiation of trabeculae myocardium.
FIGURE 2.
BAF250a regulates the proper SHF-derived structures in vivo. A, BAF250a ablation severely affects proper cardiac trabeculation. B, BAF250a deletion leads to thickened compact myocardium in E9.5 right ventricle. C, BAF250a ablation also causes defects in Isl1+ CPC migration to outflow tract at E9.5 and outflow tract septation defects at E12.5.
Sections from E9.5–E12.5 embryos also showed that BAF250a was required for outflow tract development. At E9.5, unlike in WT embryos, much fewer Isl1+ cells were present in the outflow tract region in BAF250a mutant heart (Fig. 2C, left). At E12.5 embryos, BAF250a mutant hearts displayed a persistent truncus whereas outflow tract was separated into aorta and pulmonary arties by spiral septum in WT hearts (Fig. 2C, right). These results suggest that expression of BAF250a in SHF CPCs is important for the CPC migration or differentiation into outflow tract myocardium and that BAF250a may regulate the interaction of SHF CPCs and other cell types, such as cardiac neural crest cells (24), for proper outflow tract development.
BAF250a Regulates Proper CPC Differentiation in Vitro
To study the function of BAF250a in CPCs, we examined the differentiation of BAF250a-KO CPCs in vitro. At day 6 of EB differentiation, Mef2c-Cre was expressed in newly formed CPCs which activates YFP expression and also ablates BAF250a from BAF250af/f cells (Fig. 1C). These YFP+ cells were then FACS-purified from day 6 EBs. Similar percentages (1–2%) of YFP+ cells were collected in both WT and BAF250a mutant EBs (supplemental Fig. S1A). Furthermore, cardiac transcripts, such as GATA4 and Isl1, were highly expressed compared with non-YFP cells (supplemental Fig. S1B), indicating that most of these YFP+ cells were SHF CPCs as reported before (21), although a small portion of the cells could represent craniofacial skeletal muscle lineages (20).
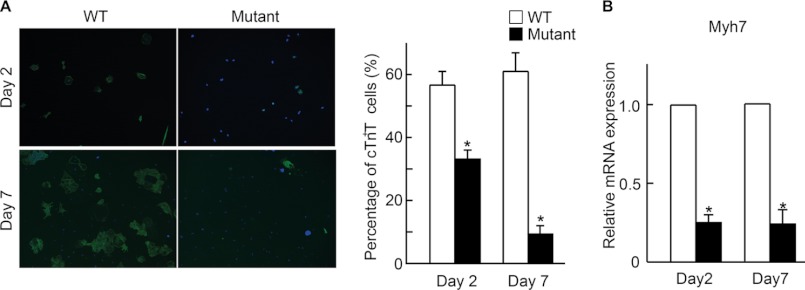
To examine the effect of BAF250a ablation in CPC differentiation potential, FACS-purified CPCs from day 6 EBs were cultured for further differentiation. At differentiation day 2 and day 7, the cardiac structure gene troponin T-positive (cTnT+) cells from BAF250a-KO CPCs formed much fewer cTnT+ cells than WT CPCs (Fig. 3A). The BAF250a-KO CPCs appeared defective in differentiation into functional cardiomyocytes after 7-day culture. Specifically, about one third of the cardiac-like cells from WT progenitors were beating under the culture, whereas very few cells derived from mutant progenitors were beating. Consistent with this, the expression of myocardium marker Myh7 was decreased approximately 4-fold in the BAF250a-KO cardiac progenitors during culture (Fig. 3B). Taken together, it is likely that BAF250a ablation has a specific effect on CPC differentiation into cardiomyocytes.
FIGURE 3.
BAF250a regulates CPC differentiation in an ES cell-based in vitro CPC system. A, lack of BAF250a leading to a significant decrease of cardiomyocyte differentiation from SHF CPCs. Left, cTnT staining of ES cell-derived cardiomyocytes at differentiation day 2 and day 7 after FACS at EB6. Right, comparison of cTnT+ cells derived from WT and mutant SHF CPCs. B, effect of BAF250a ablation on the expression of cardiomyocyte marker Myh7 during SHF CPC differentiation. Error bars, S.D. p value was calculated by two-tailed Student's t test. *, p < 0.05.
BAF250a Regulates Key Gene Expression during CPC Differentiation and Cardiogenesis
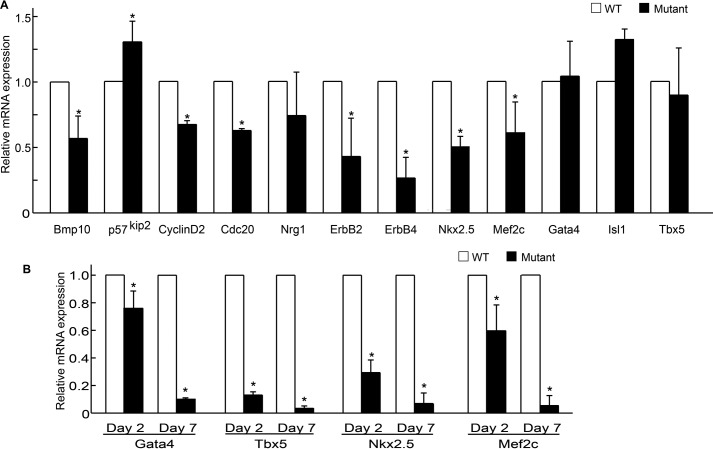
To assess the molecular basis of BAF250a-regulated cardiogenesis in the SHF-derived cell, we applied quantitative RT-PCR analysis to examine the gene expression patterns of E9.5 WT and BAF250a mutant SHF derivatives. It has been reported that Bmp10 regulates cardiac trabeculation via controlling the growth of myocardium (25). In BAF250a mutant we found trabeculation defect and a significant decrease of proliferating cells by ki67 staining (supplemental Fig. S2), which were similar to the Bmp10 mutant. Therefore, we first tested the expression of Bmp10. Quantitative RT-PCR results showed that Bmp10 mRNA was reduced in the BAF250a mutant hearts (Fig. 4A). p57kip2 is a cell cycle inhibitor that has been shown to be regulated by Bmp10. In BAF250a mutant, the p57kip2 mRNA was up-regulated. Furthermore, the expression of cell cycle genes, such as Ccnd2 and cdc20, were down-regulated (Fig. 4A). These data suggest that BAF250a mediates myocardial cell proliferation partially by maintaining the expression of Bmp10 and its target p57kip2 as well as a subset of cell cycle genes.
FIGURE 4.
Effect of BAF250a ablation on gene expression in SHF CPCs and differentiated cells from CPCs. A, BAF250a regulates a subset of genes involved in cardiogenesis. Gene expression changes during cardiac development were analyzed by quantitative RT-PCR. B, BAF250a ablation leads to down-regulation of cardiomyocyte-specific gene expression during SHF CPC differentiation. Error bars, S.D. p value was calculated by two-tailed Student's t test. *, p < 0.05.
One of the most significant defects in BAF250a mutant embryos is the cardiac trabeculation defect. Therefore, we examined the expression of Nrg1 (expressed in endocardium), ErbB2, and ErbB4 (both expressed in myocardium), which are critical for trabeculae development (26–28). The expression of ErbB2 and ErbB4 but not Nrg1 was decreased in the BAF250a mutant embryos (Fig. 4A). ErbB2 in zebrafish has been shown to regulate compact myocardium migrating and differentiating into trabeculae myocardium (29). Taken together with the gene expression analysis and histological observation, it is possible that the down-regulation of ErbB2 and ErbB4 by BAF250a ablation is a major cause of the cardiac trabeculation defect.
Importantly, the expression of Nkx2.5 and Mef2c, which regulates SHF specification and differentiation, was also down-regulated in BAF250 mutant cells (Fig. 4A). The expression of cTnT was decreased as well. However, it appears that the expression of Gata4, Isl1, Tbx5, and Hand2, a number of key transcription factors in cardiogenesis, was not changed in BAF250a mutants (Fig. 4A). Furthermore, we tested the expression of cardiac transcription factors during ES cell-based CPC differentiation. We found that in day 6 ES cell-derived BAF250a mutant CPCs, the expression of Gata4 and Isl1 was not changed (supplemental Fig. S3A). However, during differentiation, the expression of Gata4, Tbx5, Nkx2.5, and Mef2c was down-regulated in BAF250a-mutant cells (Fig. 4B). The down-regulation of these transcription factors may explain the functional differentiation defects found in BAF250a-KO myocardium (Fig. 3A). Altogether, these data indicate that BAF250a regulates specific gene expression during SHF development and suggest that BAF250a is a key epigenetic regulator for proper differentiation of SHF progenitor cells.
BAF250a Is a Key Component of BAF Complexes Regulating the Expression and Chromatin Structure of Their Target Genes
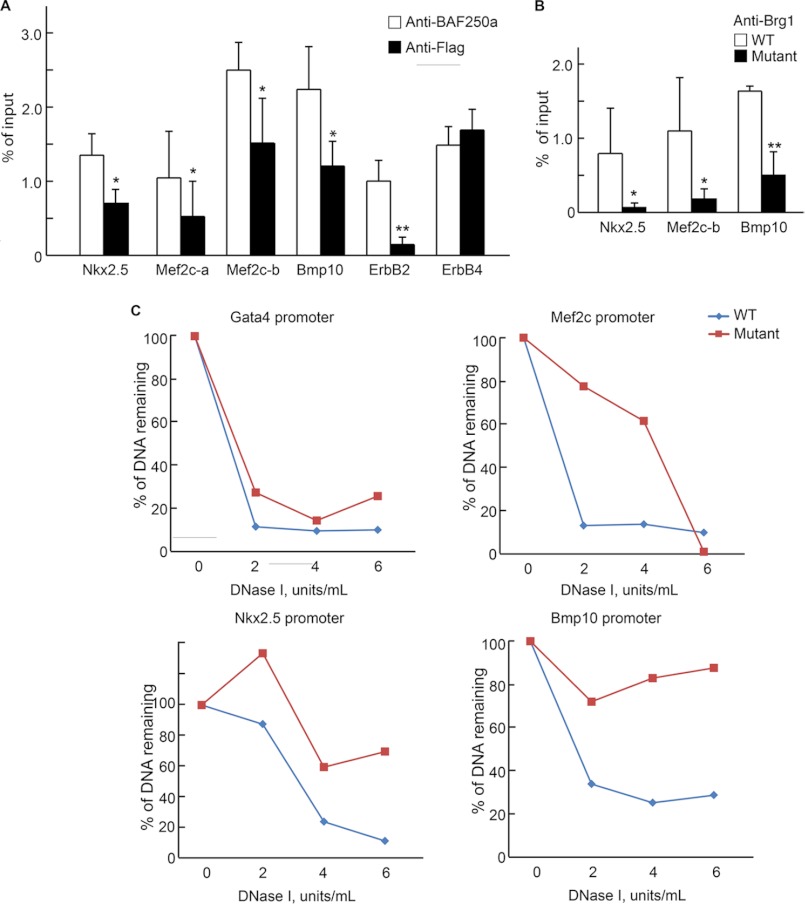
To determine whether BAF250a or BAF complexes directly regulate cardiac specific genes, we examined the binding of BAF250a to the candidate promoters. Brg1 binds to the promoters around the transcription start site region of the target genes in ES cells (30), and it is likely that BAF250a shares a binding pattern similar to Brg1. We identified the conserved promoters of Nkx2.5, Mef2c, Bmp10, ErbB2, and ErbB4 around their transcription start sites. ChIP assays indicated that antibody against BA250a precipitated approximately 2-fold more chromatin fragments at the promoter regions of Nkx2.5, Mef2c, Bmp10, and ErB2 (Fig. 5A). Little enrichment was found in the promoter region of ErbB4, suggesting that BAF250a may regulate cardiac-specific genes directly. To test whether BAF250a was required for proper recruitment of the cardiac-specific BAF complex, we then examined the effect of BAF250a in the binding of Brg1, the major catalytic subunit of SWI/SNF, to the target genes. ChIP assays using in vitro ES cell-derived CPCs with Brg1 antibody showed that the binding of Brg1 to the promoters of Nkx2.5, Mef2c, and Bmp10 dropped dramatically in the absence of BAF250a (Fig. 5B), suggesting that BAF250a is essential for the recruitment of Brg1 to specific targets.
FIGURE 5.
BAF250a mediates the proper recruitment and activity of SWI/SNF to its target genes to modulate chromatin accessibility. A, ChIP assay of BAF250a binding at candidate loci. B, recruitment of Brg1 to target loci in both presence and absence of BAF250a. C, BAF250a required for the chromatin remodeling at Mef2c, Nkx2.5, and Bmp10 promoter region. Error bars, S.D. p value was calculated by two-tailed Student's t test. *, p < 0.05; **, p < 0.01.
Because the major role of the BAF complex in regulating gene expression is to modulate DNA regulatory elements accessibility, it is very likely that the BAF250a/BAF complex is required to establish the chromatin structure of the target genes. To address this question, we analyzed the chromatin structures at the target promoters in ES cell-derived CPCs. As shown earlier, KO of BAF250a in SHF CPCs did not alter Gata4 mRNA expression but down-regulates Nkx2.5, Mef2c, and Bmp10 mRNA. We examined the DNase I hypersensitivity of these four gene promoters. DNase I-hypersensitive sites represent opened chromatin structure and are often associated with highly expressed genes (31). In the promoters of Nkx2.5, Mef2c, and Bmp10, it was found that much more DNA fragments remained after DNase I digestion in the BAF250a-KO cells (Fig. 5C), indicating a relatively closed chromatin structure in these genes when BAF250a was ablated. In contrast, the Gata4 promoter did not show different resistance to DNase I in WT and BAF250a-KO CPCs (Fig. 5C and supplemental Fig. S4), suggesting that BAF250a ablation did not affect the chromatin structure of the Gata4 promoter. The observed chromatin structure changes are highly correlated with the gene expression patterns (Figs. 4A and 5C). These results suggest that BAF250a is required for the establishment of the open chromatin state at the target promoters during CPC differentiation. Together with the ChIP assay, our studies indicate that BAF250a is a key component for the recruitment of BAF complexes to target genes and is critical for the chromatin remodeling function of BAF complexes during SHF cardiac development.
DISCUSSION
In this study, we demonstrate a key role for BAF250a during mouse cardiogenesis and cardiac progenitor cell differentiation. We have shown that expression of BAF250a in SHF is required for the normal cardiac morphogenesis. Both in vivo and in vitro data showed that BAF250a is required for the proliferation and differentiation of CPCs. Furthermore, we showed that BAF250a regulates the expression of key cardiac factors Nkx2.5, Mef2c, and Bmp10 by modulating the chromatin accessibility around their promoter regions.
Our in vivo and in vitro studies indicate that BAF250a regulates the proper differentiation of SHF CPCs, particularly into beating cardiomyocytes. Using different cardiac lineage-specific Cre recombinase to ablate BAF250a, we demonstrated the role of BAF250a in cardiac development. Deletion of BAF250a in SHF CPCs caused cardiac abnormalities, including trabeculation defects in the right ventricle, ventricular septal defect, persistent truncus arteriosus, and reduced myocardial proliferation. In contrast, ventricular-specific Mlc2v-cre (32)-mediated BAF250a-KO mice did not display cardiac defects (data not shown). These data suggest that BAF250a has a specific function during CPC differentiation but may be dispensable once CPCs differentiate into mature cardiomyocytes.
An interesting question is whether BAF250a specifically regulates the CPC differentiation into mature cardiomyocytes. SHF CPCs have the capacity to differentiate into not only cardiomyocytes, but also smooth muscle and endothelial cells (33). The transcript levels of Flk1 (endothelial marker) and Myh11 (smooth muscle marker) were up-regulated in the BAF250a-KO cardiac progenitors during culture compared with the WT progenitors (data not shown). In contrast to the dramatic decrease of cardiomyocytes, BAF250a-KO did not lead to a decrease in the number of smooth muscle cells (data not shown), suggesting that BAF250a has a rather specific function in the Mef2c+ CPC differentiation into mature cardiomyocytes.
Our data strongly suggest that BAF250a mediates CPC differentiation into cardiomyocytes by selectively regulating the expression of a subset of genes. In this study, we showed that the expression of Gata4, Isl1, and Tbx5 was not changed at the SHF CPCs, whereas the expression of Nkx2.5, Mef2c, Bmp10, ErbB2, and ErbB4 as well as several cell cycle genes was down-regulated. The function of Gata4 and Tbx5 is dependent on BAF complex (12); it is likely that BAF250a cooperates with these cardiac transcription factors to regulate specific target genes, such as Nkx2.5 and Mef2c. Moreover, the specific chromatin structure changes found in the promoter of Nkx2.5/Mef2c/Bmp10 but not in that of Gata4 further reinforce the notion that BAF250a regulates only a subset of genes in CPCs. Interestingly, following the course of CPC differentiation in vitro (Fig. 4B), even the expression of Gata4 and Tbx5 decreases due to BAF250a-KO. This is presumably due to the self-reinforcing nature of the cardiac transcription network (1). Down-regulation of a few key transcription factors in the network eventually promotes the collapse of the whole network.
It is shown that BAF complexes achieve their specificities by different composition assemblies (7). The requirement of various BAF complex subunits during cardiac development suggests that there is a cardiac-specific BAF complex (cBAF) (10, 12, 16, 34). Our results show that BAF250a, BAF60c, and Brg1 are highly expressed in the early developing heart and down-regulated during development (supplemental Fig. S3B). Together with the function of BAF250a we found, it is likely that BAF250a is an essential component of cBAF complex and functions in early cardiac development. One key question is how the cBAF complex is recruited to its target genes to regulate CPC differentiation. One possibility is that BAF250a regulates the proper incorporation of the SWI/SNF subunits. If that were the case, subunits not assembled into the complex would be expected to be degraded quickly (35). However, our quantitative RT-PCR assay showed that the expression level of Brg1 did not change in the absence of BAF250a in E9.5 embryonic heart (supplemental Fig. S3), suggesting that BAF250a may not regulate the core subunit assembly. On the other hand, ChIP analysis shows that the recruitment of Brg1 to the target genes is significantly reduced in the absence of BAF250a. As a consequence, the chromatin remodeling at target promoters is also affected. These results indicate that BAF250a plays a key role in the proper recruitment of cBAF to its target genes for chromatin remodeling and gene activation during CPC differentiation. Further studies with affinity purification, genome-wide nucleosome density/position mapping, and ChIP-seq assays will reveal systematically the regulatory role of BAF250a in the assembly and function of cBAF in CPCs.
Supplementary Material
Acknowledgments
We thank Dr. Brian L. Black for the Mef2c-AHF-Cre mice and Dr. Weidong Wang for the antibody against Brg1.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL109054-01 (to Z. W.). This work was also supported by a seed grant from the Harvard Stem Cell Institute (to Z. W.), an internal grant from Massachusetts General Hospital (to Z. W.), and the Stem Cell and Regenerative Medicine Consortium at Hong Kong University (to M. H. S.).

This article contains supplemental Figs. S1–S4.
- CPC
- cardiac progenitor cell
- cTnT
- cardiac structure gene troponin T
- E
- embryonic day
- EB
- embryoid body
- SHF
- second heart field.
REFERENCES
- 1. Black B. L. (2007) Transcriptional pathways in second heart field development. Semin. Cell Dev. Biol. 18, 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murry C. E., Keller G. (2008) Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132, 661–680 [DOI] [PubMed] [Google Scholar]
- 3. Wu S. M., Chien K. R., Mummery C. (2008) Origins and fates of cardiovascular progenitor cells. Cell 132, 537–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martin-Puig S., Wang Z., Chien K. R. (2008) Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell 2, 320–331 [DOI] [PubMed] [Google Scholar]
- 5. Bruneau B. G. (2008) The developmental genetics of congenital heart disease. Nature 451, 943–948 [DOI] [PubMed] [Google Scholar]
- 6. Hansson E. M., Lindsay M. E., Chien K. R. (2009) Regeneration next: toward heart stem cell therapeutics. Cell Stem Cell 5, 364–377 [DOI] [PubMed] [Google Scholar]
- 7. Wu J. I., Lessard J., Crabtree G. R. (2009) Understanding the words of chromatin regulation. Cell 136, 200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ho L., Crabtree G. R. (2010) Chromatin remodelling during development. Nature 463, 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lessard J., Wu J. I., Ranish J. A., Wan M., Winslow M. M., Staahl B. T., Wu H., Aebersold R., Graef I. A., Crabtree G. R. (2007) An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55, 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lange M., Kaynak B., Forster U. B., Tönjes M., Fischer J. J., Grimm C., Schlesinger J., Just S., Dunkel I., Krueger T., Mebus S., Lehrach H., Lurz R., Gobom J., Rottbauer W., Abdelilah-Seyfried S., Sperling S. (2008) Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 22, 2370–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bajpai R., Chen D. A., Rada-Iglesias A., Zhang J., Xiong Y., Helms J., Chang C. P., Zhao Y., Swigut T., Wysocka J. (2010) CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463, 958–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lickert H., Takeuchi J. K., Von Both I., Walls J. R., McAuliffe F., Adamson S. L., Henkelman R. M., Wrana J. L., Rossant J., Bruneau B. G. (2004) Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432, 107–112 [DOI] [PubMed] [Google Scholar]
- 13. Takeuchi J. K., Bruneau B. G. (2009) Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459, 708–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lou X., Deshwar A. R., Crump J. G., Scott I. C. (2011) Smarcd3b and Gata5 promote a cardiac progenitor fate in the zebrafish embryo. Development 138, 3113–3123 [DOI] [PubMed] [Google Scholar]
- 15. Wang Z., Zhai W., Richardson J. A., Olson E. N., Meneses J. J., Firpo M. T., Kang C., Skarnes W. C., Tjian R. (2004) Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 18, 3106–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hang C. T., Yang J., Han P., Cheng H. L., Shang C., Ashley E., Zhou B., Chang C. P. (2010) Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 466, 62–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takeuchi J. K., Lou X., Alexander J. M., Sugizaki H., Delgado-Olguín P., Holloway A. K., Mori A. D., Wylie J. N., Munson C., Zhu Y., Zhou Y. Q., Yeh R. F., Henkelman R. M., Harvey R. P., Metzger D., Chambon P., Stainier D. Y., Pollard K. S., Scott I. C., Bruneau B. G. (2011) Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat. Commun. 2, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nie Z., Xue Y., Yang D., Zhou S., Deroo B. J., Archer T. K., Wang W. (2000) A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20, 8879–8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao X., Tate P., Hu P., Tjian R., Skarnes W. C., Wang Z. (2008) ES cell pluripotency and germ layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc. Natl. Acad. Sci. U.S.A. 105, 6656–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Verzi M. P., McCulley D. J., De Val S., Dodou E., Black B. L. (2005) The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev. Biol. 287, 134–145 [DOI] [PubMed] [Google Scholar]
- 21. Domian I. J., Chiravuri M., van der Meer P., Feinberg A. W., Shi X., Shao Y., Wu S. M., Parker K. K., Chien K. R. (2009) Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science 326, 426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang W., Côté J., Xue Y., Zhou S., Khavari P. A., Biggar S. R., Muchardt C., Kalpana G. V., Goff S. P., Yaniv M., Workman J. L., Crabtree G. R. (1996) Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15, 5370–5382 [PMC free article] [PubMed] [Google Scholar]
- 23. Wurster A. L., Pazin M. J. (2008) BRG1-mediated chromatin remodeling regulates differentiation and gene expression of T helper cells. Mol. Cell. Biol. 28, 7274–7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu W., Selever J., Wang D., Lu M. F., Moses K. A., Schwartz R. J., Martin J. F. (2004) Bmp4 signaling is required for outflow-tract septation and branchial-arch artery remodeling. Proc. Natl. Acad. Sci. U.S.A. 101, 4489–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen H., Shi S., Acosta L., Li W., Lu J., Bao S., Chen Z., Yang Z., Schneider M. D., Chien K. R., Conway S. J., Yoder M. C., Haneline L. S., Franco D., Shou W. (2004) BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 131, 2219–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gassmann M., Casagranda F., Orioli D., Simon H., Lai C., Klein R., Lemke G. (1995) Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 378, 390–394 [DOI] [PubMed] [Google Scholar]
- 27. Lee K. F., Simon H., Chen H., Bates B., Hung M. C., Hauser C. (1995) Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature 378, 394–398 [DOI] [PubMed] [Google Scholar]
- 28. Meyer D., Birchmeier C. (1995) Multiple essential functions of neuregulin in development. Nature 378, 386–390 [DOI] [PubMed] [Google Scholar]
- 29. Liu J., Bressan M., Hassel D., Huisken J., Staudt D., Kikuchi K., Poss K. D., Mikawa T., Stainier D. Y. (2010) A dual role for ErbB2 signaling in cardiac trabeculation. Development 137, 3867–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ho L., Jothi R., Ronan J. L., Cui K., Zhao K., Crabtree G. R. (2009) An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. U.S.A. 106, 5187–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gross D. S., Garrard W. T. (1988) Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57, 159–197 [DOI] [PubMed] [Google Scholar]
- 32. Chen J., Kubalak S. W., Chien K. R. (1998) Ventricular muscle-restricted targeting of the RXRα gene reveals a non-cell-autonomous requirement in cardiac chamber morphogenesis. Development 125, 1943–1949 [DOI] [PubMed] [Google Scholar]
- 33. Moretti A., Caron L., Nakano A., Lam J. T., Bernshausen A., Chen Y., Qyang Y., Bu L., Sasaki M., Martin-Puig S., Sun Y., Evans S. M., Laugwitz K. L., Chien K. R. (2006) Multipotent embryonic Isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127, 1151–1165 [DOI] [PubMed] [Google Scholar]
- 34. Huang X., Gao X., Diaz-Trelles R., Ruiz-Lozano P., Wang Z. (2008) Coronary development is regulated by BAF180, a unique subunit in ATP-dependent chromatin remodeling complex PBAF. Dev. Biol. 319, 258–266 [DOI] [PubMed] [Google Scholar]
- 35. Keppler B. R., Archer T. K. (2010) Ubiquitin-dependent and ubiquitin-independent control of subunit stoichiometry in the SWI/SNF complex. J. Biol. Chem. 285, 35665–35674 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.