Background: rRNA gene transcription and processing have to be coordinated, as they are intimately linked.
Results: Mybbp1a regulates RNA Pol I transcription and pre-rRNA processing.
Conclusion: Mybbp1a coordinates both processes.
Significance: Mybbp1a was shown to be an important regulator of cellular proliferation, related to RNA Pol II genes. We link its function now as well to regulating RNA Pol I genes and to control ribosome biogenesis.
Keywords: Gene Regulation, Nucleolus, Ribosomal RNA (rRNA), Ribosomal RNA Processing, Transcription Regulation
Abstract
Ribosomal RNA gene transcription, co-transcriptional processing, and ribosome biogenesis are highly coordinated processes that are tightly regulated during cell growth. In this study we discovered that Mybbp1a is associated with both the RNA polymerase I complex and the ribosome biogenesis machinery. Using a reporter assay that uncouples transcription and RNA processing, we show that Mybbp1a represses rRNA gene transcription. In addition, overexpression of the protein reduces RNA polymerase I loading on endogenous rRNA genes as revealed by chromatin immunoprecipitation experiments. Accordingly, depletion of Mybbp1a results in an accumulation of the rRNA precursor in vivo but surprisingly also causes growth arrest of the cells. This effect can be explained by the observation that the modulation of Mybbp1a protein levels results in defects in pre-rRNA processing within the cell. Therefore, the protein may play a dual role in the rRNA metabolism, potentially linking and coordinating ribosomal DNA transcription and pre-rRNA processing to allow for the efficient synthesis of ribosomes.
Introduction
Cellular growth and division is tightly coupled to ribosome synthesis, a major cellular undertaking that is energetically very costly (for review, see Ref. 1). This process requires the matched activity of the three forms of RNA polymerase to produce the building blocks of the ribosomes and the coordinated expression, processing, and assembly of the rRNA precursor (pre-rRNA)4 into ribosomes (2, 3).
Mammalian cells contain 300–400 rRNA gene repeats, most of which are arranged as tandem repeats. The level of rRNA synthesis in the cell is determined by the dynamic regulation of about half of those genes, whereas the remaining genes are constantly repressed (4). Active and inactive gene populations are discriminated by different epigenetic states and different topological conformations (5–7). The expression levels of the active portion of the rRNA genes are tightly regulated in accordance with the physiological condition of the cell. Various signal transduction pathways, including the mTOR pathway and the MAPK cascade, directly target different steps of rRNA synthesis to efficiently regulate the production of ribosomes. Several post-translational modifications of the Pol I-specific transcription initiation factor TIF-IA and the rDNA transcription factor Upstream Binding Factor have been characterized, and their impacts on transcriptional pre-initiation complex (PIC) formation and/or elongation have been shown (8–11).
The localization of ribosome biogenesis to the nucleolus and the close juxtaposition of the factors involved in transcription, processing, and ribosome assembly, as revealed by microscopy techniques, suggest that these processes are directly coupled. Co-transcriptional processing of the pre-rRNA transcript was recently shown by EM visualization of nascent rRNA transcripts. The 5′-terminal knobs containing U3 small nucleolar RNA and associated Utps (U three proteins) increased in size with transcript length, indicating that rRNA processing, RNA folding, RNA modification, and ribosomal protein assembly also occur co-transcriptionally (12–15).
Early rRNA processing is initiated by the assembly of the small ribosome subunit processome complex, which contains U3 small nucleolar RNA (14) that base pairs with sequences near the 5′ end of the pre-rRNA. Small ribosome subunit processome-dependent processing leads to rRNA cleavage and the release of the small subunit rRNA (18 S) from the ribosome (13). A subset of seven small ribosome subunit processome proteins, the t-Utps (transcriptional-Utps), is also required for efficient rDNA transcription in yeast, linking rDNA transcription with rRNA processing and ribosome assembly (2). McStay and Prieto (16) identified the human orthologs of the t-Utps and confirmed their requirement for both efficient transcription and processing of the 47 S pre-rRNA, indicating that this coordinated mechanism is conserved throughout evolution. Their study further revealed that the t-Utps are recruited to rDNA, irrespective of the transcription status of the rDNA gene.
Recently, Moss and co-workers (17) suggested a role for transcription elongation in rRNA processing and ribosome assembly. Erk-dependent phosphorylation of UBF directly influences RNA Pol I elongation rates by inducing an UBF-dependent rearrangement of the chromatin environment. This effect could serve as a mechanism to coordinate transcription and the assembly of pre-ribosomal complexes on nascent rRNA by modulating the Pol I elongation rate. Further insights into how these processes might be coordinated came from Nomura and co-workers (18) who demonstrated a physical interaction of Pol I with the elongation factors Spt4 and Spt5 and an rRNA processing defect upon depletion of the latter. A connection between transcription elongation and rRNA processing was further confirmed by using a yeast strain with a mutated RNA Pol I enzyme with reduced elongation rates (3). Reduced elongation rates led to severe defects in rRNA processing and ribosome assembly, suggesting an intimate link between elongation rates and rRNA maturation. Thus, ribosome biogenesis appears to be controlled by feedback mechanisms, which ensure that proper assembly occurs on all synthesis levels, from transcription initiation to ribosome export.
The expression of the rRNA genes is regulated at different levels of the rRNA metabolic pathway at the stage of transcription initiation (for review, see Ref. 19), transcription elongation (17, 18), rRNA processing, and assembly (20–22). To identify novel regulators of rRNA gene synthesis, we screened for factors interacting with RNA Pol I. We identified Myb-binding protein 1a (Mybbp1a), which is ubiquitously expressed and predominantly localizes to the nucleolus (23, 24). The 160-kDa Mybbp1a protein had originally been described as an interacting partner of the proto-oncogene c-Myb and since then it has been shown to interact with and to modulate the activity of several regulators of Pol II-dependent transcription. In certain myeloid cell types, the protein is post-translationally processed to generate an N-terminal fragment of 67 kDa (p67MBP) (23). Mybbp1a, and in some cases also p67MBP, have been described to interact with several other transcriptional regulators, such as peroxisome proliferator-activated receptor-γ coactivator 1α, NF-κB, aromatic hydrocarbon receptor, Prep1, and CRY1 and linked to diverse cellular processes (e.g. mitochondrial biogenesis, cell proliferation, apoptosis etc.) by modulating the activity of these regulators (25–29).
Despite its predominant localization to the nucleolus, the function of Mybbp1a in this cellular compartment remained largely unclear. The data shown here demonstrate two new functions for Mybbp1a in the nucleolus. It associates with the RNA Pol I machinery and is able to repress its transcriptional activity.
EXPERIMENTAL PROCEDURES
DNA Constructs
To generate a FLAG-tagged RPA116 expression construct, the coding sequence of mouse RPA116 was amplified from the cDNA clone pBS-RPA 116 6-2 (kindly provided by I. Grummt (30)) using the following primer pair in a standard PCR with the Expand High Fidelity system (Roche Applied Science). The forward (5′-ATG GAT CCA TGG ACT ACA AGG ACG ACG ATG ACA AGG ATG TCG ACG GCC GG-3′) and reverse primers (5′-GAG GAT CCT CAG ATG ACA TCC AGT TTC ACT-3′) introduced a FLAG coding sequence upstream of the RPA116 ORF. The PCR product was cloned into pEGZ (pEGZ-fRPA116).
The human FLAG-Mybbp1a expression construct was generated by PCR amplification of the human Mybbp1a cDNA clone described previously (31) using Pfu DNA Polymerase (Promega). The forward primer (5′-AG TCA AGC TTC ACC ATG GAC TAC AAG GAC GAC GAT GAC AAG GAG AGC CGG GAT CCC GCC-3′) introduced an N-terminal FLAG tag and NcoI restriction site at the initiation codon, and the reverse primer (5′-GCT CTA GAT CAG GGT TTC CCT GCC TTC-3′) introduced an XbaI restriction site at the stop codon to facilitate cloning into the pact expression vector as used previously (23). The mouse FLAG-Mybbp1a (mMybbp1a), mouse FLAG-p67MBP*NLS (FLAG-mp67MBP*), and human FLAG-Tip5 have been previously described (Tavner et al. (23); Strohner et al. (32)). GFP-RPA43 was obtained from Addgene (Addgene plasmid 17659).
Cell Culture and Proliferation Assay
The mouse lymphoblast cell line MB III (ATCC® number CCL-32™) was cultured in minimum Eagle's medium (Earles, with Glutamax) with non-essential amino acids, sodium pyruvate (110 mg/liter), and 10% heat-inactivated newborn calf serum (Invitrogen). To create a MB III cell line stably expressing FLAG-RPA116, cells were transfected with pEGZ-fRPA116, selected with Zeocin (Invitrogen), cloned by serial dilution, and then further maintained with 100 μg/ml Zeocin. HeLa cells were cultured in DMEM containing 10% heat-inactivated fetal calf serum (Invitrogen) and penicillin/streptomycin. Both cell lines were cultured at 37 °C in 5% CO2.
To monitor proliferation rates, HeLa cells were detached with trypsin-EDTA and diluted in PBS. Subsequently cells were stained with trypan blue, and living cells were counted.
Preparation of Cellular Extracts and Immunopurification of Proteins
To prepare nuclear extracts, MB III and FLAG-RPA116-MB III cells were harvested, washed with PBS, and resuspended in three packed cell volumes of buffer A (20 mm HEPES, pH 7.9, 0.2% Nonidet P-40, 10 mm KCl, 1 mm EDTA, 10% glycerol, 1 mm DTT, protease inhibitors) incubated on ice for 10 min. Cell lysis was followed by microscopy. Nuclei were washed in buffer A, resuspended in 3 packed cell volumes of Buffer B (420 mm NaCl, 20 mm HEPES, pH 7.9, 10 mm KCl, 1 mm EDTA, protease inhibitors) containing 2% (v/v) distamycin A hydrochloride (Sigma, D6135), and incubated on a rotating wheel at 4 °C for 40 min. After centrifugation the nuclear fraction was collected and dialyzed against AM100 (100 mm NaCl, 20 mm Tris-HCl, pH 7.9, 5 mm MgCl2, 0.1 mm EDTA, 20% glycerol, 1 mm DTT, protease inhibitors). HEK293T nuclear extracts were prepared 48 h after transfection with FLAG-Mybbp1a or empty vector control as described above.
HeLa nuclear extracts were prepared 48 h after transfection with FLAG-Mybbp1a or vector control. Cells were detached by trypsin-EDTA (Invitrogen), washed with PBS, centrifuged at 2,500 × g for 5 min, and resuspended in 5× packed cell volumes of Buffer C (10 mm KCl, 10 mm HEPES, pH 7.9, 1.5 mm MgCl2, 1 mm DTT, protease inhibitors). After incubation on ice for 15 min, the samples were centrifuged at 2,000 × g for 8 min, and the cell pellet was resuspended in 2× packed cell volumes of Buffer C. The cells were lysed with the help of a Dounce homogenizer (pestle B), and cell lysis was followed by light microscopy. After two centrifugation steps at 1000 × g and 4000 × g each for 10 min, the pellet was resuspended in 3 ml of Buffer D (20 mm HEPES, pH 7.9, 420 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 20% (v/v) glycerol, 1 mm DTT, protease inhibitors) per 109 cells. After incubation at 4 °C for 30 min the extract was sonicated 2 × 20 s in a volume of 200 μl (alternatively 3 × 20 s in a volume of 250 μl) with the Bioruptor (H, Diagenode) and centrifuged at 17,000 × g for 15 min. After centrifugation the nuclear fraction was collected and dialyzed against Buffer D containing 300 mm NaCl. MB III whole cell extracts were prepared according to the Manley protocol (33) and dialyzed against AM100.
For immunoprecipitation experiments, extracts were incubated with anti-FLAG M2-agarose in the presence of 0.02% Nonidet P-40 or 0.25% Triton X-100, protease inhibitors, and DTT (1 mm). FLAG-RPA116-containing complexes were additionally washed with AM300 buffer (300 mm NaCl). Proteins were eluted with 0.25 mg/ml FLAG peptide. If needed proteins were concentrated with Strata Clean resin (Stratagene) or by TCA precipitation. Proteins were separated by SDS-PAGE and either analyzed by Western blotting or silver or Coomassie staining, cut out of the gel, and subjected to MALDI-TOF mass spectrometry.
Fractionation by Glycerol Gradient and Superose 6 Gel Filtration
500 μl of MB III whole cell extract was applied on a 12-ml 10–45% glycerol gradient (AM100) and centrifuged at 35,000 × g (Sw 41 Ti rotor) in a Optima-LE 80K ultracentrifuge (Beckman Coulter) at 4 °C for 12 h (no break). Subsequently 500-μl fractions were collected, and proteins were separated by SDS-PAGE and subjected to Western blotting.
HeLa nuclear extract was incubated with or without 10 μg/ml RNase A (Sigma) at 4 °C for 2 h before Superose 6 HR 10/30 (GE Healthcare) fractionation. 500-μl fractions were collected, and proteins were concentrated with the Strata Clean resin (Stratagene) and analyzed by Western blotting.
Antibodies
The following antibodies were used for Western blotting, ChIP, and immunostaining: rabbit polyclonal α-RPA116 (30), rabbit polyclonal α-PAF53 (34), and α-TIF-IA (Ref. 35; kindly provided by I. Grummt), rabbit polyclonal α-Mybbp1a (generated to a GST fusion protein containing amino acid residues 976–1263 of human Mybbp1a) and α-mouse Mybbp1a (23), mouse monoclonal α-FLAG (M2, Sigma), rabbit polyclonal α-FLAG (Sigma), mouse monoclonal α-tubulin (Dm1a, Sigma), rabbit polyclonal α-RPA194 (sc-28174, Santa Cruz Biotechnology), α-BrdU (Roche Applied Science), α-Pes1 (36), α-TTF-I (37), α-fibrillarin (P2G3 (38)), α-B23 (sc-56622, Santa Cruz Biotechnology), α-EBP2 (kindly provided by L. Frappier (39)), α-rpS2 (kindly provided by M. Bedford (40)), α-DDX21 (Proteintech Group), α-Nol1, α-GFP-Alexa488 (GBA-488, ChromoTek), goat-α-mouse-Alexa488 F(ab′)2 (Molecular Probes), and goat-α-rabbit-Alexa594 (Molecular Probes).
RNA Pol I Reporter Assay
HeLa cells were transfected with Polyfect reagent (Qiagen) according to the manufacturer's instructions with the indicated amounts of expression construct together with empty vector DNA to adjust DNA concentration and the pHrD-IRES firefly reporter construct (a kind gift of S. T. Jacob (41)) as a reporter for RNA Pol I transcription activity. A plasmid carrying the renilla luciferase gene under the control of a thymidine kinase promoter (transcribed by RNA polymerase II) was co-transfected in each experiment to normalize for differences in transfection efficiency. 48 h after transfection, luciferase activity was measured using the Dual Luciferase Assay kit (Promega). The firefly luciferase counts of the RNA polymerase I reporter constructs were divided by the Renilla luciferase counts and compared with control transfections with empty vector. Average and S.D. values of three biological replicates (each in technical duplicates) are shown. Protein levels were monitored by Western blotting.
Short interfering RNAs (siRNA)-mediated Protein Depletion
The day before transfection 2 × 105 HeLa cells were seeded per well (6 well format). 20 μm siRNA were transfected using Oligofectamine (Invitrogen) according to the manufacturer's instruction. The following siRNAs (Thermo or MWG) were used: Control (ctrl; Luciferase), 5′-CUU ACG CUG AGU ACU UCG AdTdT; Mybbp1a.1, 5′-GCC GAC UUG AAU AUA AUA CdTdT; Mybbp1a.2, 5′-UGG AUC AUC UUU CGA UUG GdTdT; Mybbp1a.3, 5′-AUA CGC AAG CUG UUU CUA AdTdT; PES1 (ORF), 5′-AGG UCU UCC UGU CCA UCA AdTdT; TIF-IA, 5′- CAA AGG ATC TAT ATC GCG AdTdT. The three different siRNAs directed against Mybbp1a had similar effects on rRNA gene transcription and pre-rRNA processing (supplemental Fig. S3), ruling out off-target effects.
RNA Extraction and Quantitative Real-time PCR
RNA was extracted from HeLa cells with the RNeasy kit (Qiagen) according to the manufacturer's instructions. 1 μg of total RNA was used for reverse transcription. A 20-μl reaction contained 2 μl of random primer (500 μg/ml, Promega), 2 μl of dNTPs (2.5 mm each, Bioline), and sterile water as added to a total volume of 12 μl. RNA was added and incubated for 5 min at 65 °C. After cooling down, the reaction was supplemented with 5× First Strand Buffer (Invitrogen), 2 μl of 0.1 m RNase-free DTT (Invitrogen), and 1 μl of RNasin (Promega) and incubated for 2 min at 37 °C. 200 units of Moloney murine leukemia virus reverse transcriptase (Invitrogen) were added, and the reactions were incubated for 1 h at 37 °C and subsequently heat-inactivated.
Quantitative PCR was carried out using the ABI PRISM 7000 Sequence detection system (Applied Biosystems). Taqman 2× PCR Master Mix (Applied Biosystems) was used according to the manufacturer's instructions. To detect 47 S rRNA precursor, the following oligos were used: forward, +132/+149 CCT GCT GTT CTC TCG CGC; reverse +198/+181, GGT CAG AGA CCC GGA CCC; Taqman, +155/+174 AGC GTC CCG ACT CCC GGT GC. A β-actin-mRNA amplicon was used for normalization (forward, HBacF +927/+945, TGC CGA CAG GAT GCA GAA G; reverse, +1026/+1006, GCC GAT CCA CAC GGA GTA CTT; Taqman, +980/+1003 TCA AGA TCA TTG CTC CTC CTG AGC). For both amplicons an annealing temperature of 60 °C was applied.
Northern Blot
Total RNA of transfected HeLa cells was isolated using RNeasy (Qiagen). 5 μg of RNA was separated by electrophoresis on 1% agarose-formaldehyde gel and blotted onto a Hybond N+ membrane (Amersham Biosciences). The filter was washed with 2× SSC and dried, and RNA was UV-cross-linked. The filter was prehybridized for 1 h at 65 °C with Church buffer (0.5 m Na-phosphate, pH 7.2; 7% SDS; 1 mm EDTA) and hybridized for 4 h with a specific 32P-labeled probe: 5′ external transcribed spacer (5′ETS) probe, CGG AGG CCC AAC CTC TCC GAC GAC AGG TCG CCA GAG GAC AGC GTG TCA GC; ITS1 prob, CCT CCG CGC CGG AAC GCG CTA GGT ACC TGG ACG GCG GGG GGG CGG ACG. The DNA oligonucleotides were 5′ end-labeled using T4 polynucleotide kinase in the presence of [γ-32P]ATP. After hybridization, the filters were washed with 3 changes (30 min each) of 0.2× SSC, 0.1% SDS at 65 °C. Radioactive signals were detected using phosphoimaging and quantified using the MultiGauge software.
Polysome Gradient
A total of 2 × 107 293 cells grown in rich medium and pretreated with 50 μg/ml cycloheximide for 5 min at 37 °C were harvested and washed 2 times with cold phosphate-buffered saline containing 50 μg/ml cycloheximide. Cells were resuspended in 600 μl of polysome buffer (20 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 50 mm KCl, 1 mm DTT). Nonidet P-40 was added to a final concentration of 0.3%, and total cell lysates were sonicated 3 times for 30 s. A total of 250 μg RNA was loaded onto a 10-ml 10–50% linear sucrose gradient in polysome buffer. Subsequently, sucrose gradients were centrifuged at 38 krpm for 2 h in a SW40Ti rotor. Fractions of 500 μl each were collected, and the A254 was measured continuously. Proteins were concentrated by TCA precipitation and analyzed by Western blot.
Chromatin Immunoprecipitations
ChIP experiments were performed using rabbit polyclonal α-RPA116, rabbit polyclonal α-Mybbp1a, and rabbit polyclonal α-FLAG and α-TTF-I antibodies as described (5, 42). Purified DNA of the immunoprecipitates and of input DNA were analyzed by real-time PCR using the SYBR Green (Applied Biosystems) quantitation method according to the manufacturer's instruction on an ABI PRISM 7000 Sequence detection system (Applied Biosystems). Results were calculated relative to standard curve values, corrected for nonspecific binding (normal rabbit IgG ChIP), and presented as the percentage of input DNA. The following primer sequences for the rDNA loci were used; for the promoter, forward (5′-ATG GTG GCG TTT TTG GGG) and reverse (5′-AGG CGG CTC AAG GCA GGA G; product −133 to +116 relative to the transcription start site) and for the intergenic spacer, forward (5′-CGC TGT CCA TCT CTG TCT TTC TAT G) and reverse (5′-ATA CAC CGA GTG GGG AAG CC; product +22730 to +22906 relative to the transcription start site). In ChIP experiments shown in supplemental Fig. S3 a rabbit polyclonal α-RPA194 antibody was used, and real-time quantitative PCR experiments were performed in a Rotor-Gene Q (Qiagen) instrument with the following primers and SYBR Green detection: Hr42857F (promoter forward) 5′ ATG GTG GCG TTT TTG GGG AC; Hr42964R (promoter reverse) 5′ CGA AAG ATA TAC CTC CCC CG; Hr5232F (18 S forward) GGT AAC CCG TTG AAC CCC ATT C; Hr5375R (18 S reverse) GCC TCA CTA AAC CAT CCA ATC G; Hr9661F (28 S forward) 5′ CGA ATG ATT AGA GGT CTT GGG GC; Hr9860R (28 S reverse) 5′ TGG GGT CTG ATG AGC GTC GG; Hr36189F (intergenic spacer forward) 5′ TCG CCG ACT CTC TCT TGA CTT G; Hr36399R (intergenic spacer reverse) 5′ TGG AGC ACA GTG ACA CAA CTA TGG. The numbers of human rDNA (Hr) oligos indicate the position of the 5′ end relative to the transcription start site (+1) in the repeat unit (GenBankTM accession number U13369).
Immunofluorescent Staining and Image Capture
Cells were grown on coverslips or in Labtech chambers overnight and fixed in ice-cold methanol/acetone (1:1) for 1 min. Alternatively, cells were fixed in 2% paraformaldehyde, PBS on ice for 15 min and permeabilized with 0.25% Triton X-100 in the presence of 1% paraformaldehyde, PBS on ice for 10 min. Unspecific binding was blocked with PBS-Tween 0.01% (PBS-T) containing 2% BSA and 5% goat serum for 1 h. Primary antibodies were incubated in blocking solution at room temperature for 1 h or at 4 °C in a humidified chamber overnight. Cells were washed with PBS-T and incubated with Cy2- or Cy3-labeled secondary antibodies (Dianova) in blocking solution at room temperature for 1 h. DNA was counterstained with Hoechst 33342 stain (Sigma) and mounted in Vectashield (Axxora). Fluorescence images were acquired with a Zeiss Axiovert 200 inverse microscope.
To assess RNA-dependent localization of proteins, HeLa cells were washed with PBS and incubated with PBS containing 0.1% (v/v) Triton X-100 for 10 min and subsequently incubated with 1 mg/ml RNase A (Sigma) in PBS at room temperature for 20 min. Control cells were incubated with either 3 μl/ml RNase-free DNase I (Roche Applied Science) and 3 mm MgCl2 or both RNase A and DNase I. Cells were washed with PBS, fixed, and prepared for immunofluorescence microscopy as described above.
HeLa cells were incubated in DMEM containing 10 μm BrdU at 37 °C for 30 min, washed with PBS-T, and fixed with 4% formaldehyde, PBS at room temperature for 10 min. Cells were washed, permeabilized with 0.2% Triton X-100/PBS, and incubated with 2 n HCl, PBS for 10 min. After two washing steps, cells were analyzed by immunostaining for the presence of BrdU incorporation.
Super-resolution Fluorescence Microscopy
HeLa cells were grown on coverslips and transfected with EGFP-RPA43 (43). Non-transfected and transfected cells were fixed with 4% paraformaldehyde in 1× PBS for 10 min permeabilized with 1% Triton X-100 in 1× PBS for 10 min, and immunofluorescence detections of Mybbp1a, fibrillarin, B23/nucleophosmin, and GFP-RPA43 were performed. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Super-resolution fluorescence microscopy was performed with a DeltaVision OMX V3 3D-SIM system (Applied Precision) equipped with a 100×/1.40 NA PlanApo oil immersion objective (Olympus), Cascade II:512 EMCCD cameras (Photometrics), and 405-, 488-, and 593-nm diode lasers. Image stacks of 8–10 μm were recorded with z-distances of 125 nm. Iterative three-dimensional deconvolution, structured illumination reconstruction, and image processing was performed with the SoftWoRx 3.7 imaging software package (Applied Precision).
RESULTS
RNA Polymerase I Is Associated with Mybbp1a
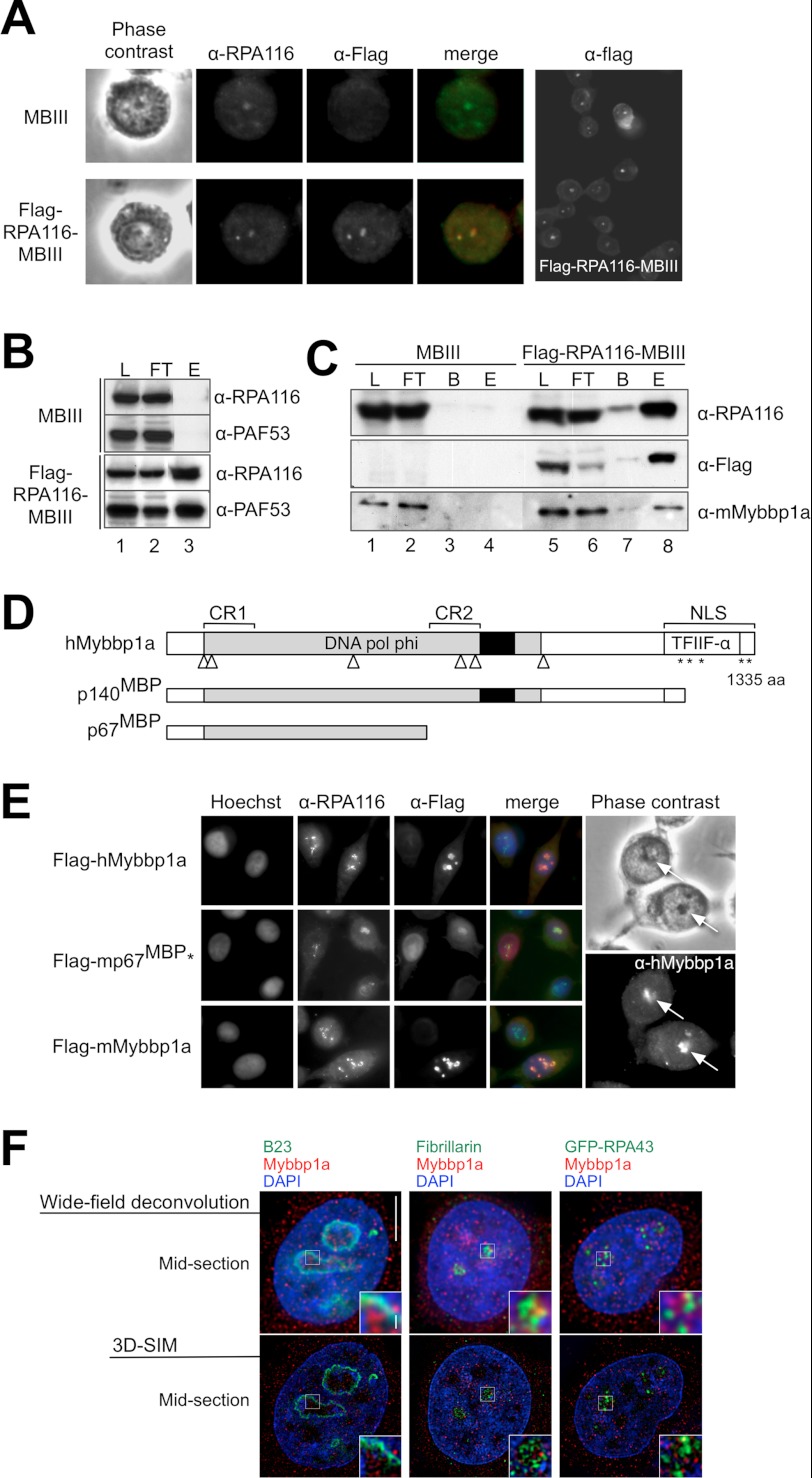
To reveal potential regulators that act at the level of transcription initiation and elongation, we generated a murine MB III cell line stably expressing FLAG-tagged RPA116 (FLAG-RPA116-MBIII), the second-largest subunit of RNA Pol I (30). The N-terminal-tagged protein was constitutively expressed at low levels in this cell line and localized similarly to the endogenous protein (Fig. 1A). FLAG-RPA116 was fully incorporated into the RNA Pol I complex, as revealed by co-immunoprecipitation analysis. FLAG-RPA116 co-purified with PAF53, a known subunit of the RNA Pol I complex (Fig. 1B).
FIGURE 1.
Characterization of the subcellular localization of Mybbp1a and its interaction with RNA Pol I. A, control MB III cells and MB III cells stably expressing FLAG-RPA116 (FLAG-RPA116-MB III) were incubated with α-RPA116 (green) and α-FLAG (red) antibodies to detect the endogenous and tagged RPA116 proteins and analyzed by phase contrast and immunofluorescence microscopy. A larger field is shown (right). B, FLAG-RPA116-containing complexes were immunoprecipitated with α-FLAG-agarose from nuclear extracts of control MB III and FLAG-RPA116-MB III cells. Load (L; 6%), flow-through (FT; 6%), and eluted proteins (E) were analyzed by Western blotting with the indicated antibodies. C, FLAG-RPA116-containing complexes were immunoprecipitated with α-FLAG-agarose from whole cell extracts of MB III and FLAG-RPA116-MB III cells. For the detection with α-RPA116 and α-FLAG load (L, 40%), flow-through (FT, 40%), bead-bound (B), and eluted (E) proteins were analyzed by Western blotting. For the detection with α-Mybbp1a antibodies 0.8% of the load and flow-through were loaded on the gel. D, shown is a schematic representation of structural features of human Mybbp1a and its proteolytic cleavage products p140MBP and p67MBP. Mybbp1a contains several potential leucine charged domain motifs (△), basic amino acid repeats (*), an acidic domain (black box), and two highly conserved regions (CR) 1 and 2 harboring several strictly conserved polar residues (31, 46). Sequence analysis of Mybbp1a (ref AF147709.1) with the Conserved Domain data base search (70) (Version v.2.22) revealed two conserved domains: the DNA polymerase φ domain (DNA pol φ; pfam 04931; E value, 1.54e-138) and the transcription initiation factor IIF α subunit (TFIIF-a; pfam 05793; E value, 6.55e-03). Upon transcription impairment by chemical stress Mybbp1a gets partially proteolyzed to generate the N-terminal fragments p140MBP and p67MBP (24, 44). E, localization Mybbp1a in HeLa cells is shown. 48 h after transfection with human FLAG-hMybbp1a, mouse FLAG- mouse Mybbp1a, and mouse FLAG-p67MBP*NLS (FLAG-mp67MBP*), expression constructs cells were fixed and stained with the antibodies indicated on top. Co-localization is indicated by the orange color. Non-transfected HeLa cells stained with α-Mybbp1a recognizing endogenous Mybbp1a are shown on the right. The positions of the nucleoli are indicated by arrows. F, shown is subnuclear localization of Mybbp1a in relation to nucleolar marker proteins analyzed with super resolution-structured illumination microscopy (71). Shown is co-immunofluorescence staining of HeLa cells with anti-Mybbp1a antibody versus anti-B23 antibody (granular component; top panel), anti-fibrillarin antibody (dense fibrillar component; middle panel), and GFP-RPA43 (fibrillar center, bottom panel). DNA was counterstained with DAPI. Mid-section with conventional optical resolution is shown for comparison (left column). Scale bars, 5 and 0.5 μm (inset).
Whole cell extracts were prepared from MB III and FLAG-RPA116-MB III cells growing in suspension. Immunoaffinity purifications were performed under conditions that preserve complex integrity and generate transcriptionally active RNA Pol I complexes (supplemental Fig. S1). The purified proteins were subjected to protein gel electrophoresis and subsequent mass spectrometric analysis (data not shown). Mybbp1a (accession number NP_058056) was identified as an RNA Pol I interacting protein by MALDI-TOF analysis. The specific association of Mybbp1a and RNA Pol I was verified by co-immunoprecipitation experiments and Western blot analysis (Fig. 1C).
Mybbp1a was originally identified as an interacting partner of the c-myb proto-oncogene product in mouse cell lines. In certain cell lines, a specific portion of mouse Mybbp1a is processed by proteolytic cleavage to generate the N-terminal fragment p67MBP (23). The existence of such post-translational cleavage products has also been confirmed in human cell lines. After exposing HeLa cells to UV light or chemicals that impair transcription, Ishii and co-workers (44) observed the appearance of N-terminal fragments of Mybbp1a with molecular masses of 67 and 140 kDa (p67MBP and p140MBP, respectively) as well as a translocation of the full-length protein and its cleavage products from the nucleolus to the nucleoplasm (Fig. 1D).
Next, we verified the potential Mybbp1a/RNA Pol I association in human HeLa cells by studying protein localization. In agreement with previous observations (44, 45), Mybbp1a preferentially localizes to the nucleolus also in HeLa cells as shown by immunofluorescence experiments that detect the endogenous human protein and transiently transfected mouse and human FLAG-Mybbp1a protein (Fig. 1E). To reveal the subnucleolar distribution of Mybbp1a, the protein was visualized at subdiffraction resolution in parallel with marker proteins of the different functional regions of the nucleolus. The RNA Pol I subunit RPA43, fibrillarin, and B23/nucleophosmin served as markers for the fibrillar center, dense fibrillar component, and granular component, respectively. The pictures reveal that Mybbp1a localizes closer to or overlaps in part with RNA Pol I and fibrillarin but not with B23/nucleophosmin, suggesting a role in transcription and early processing (Fig. 1F). Mybbp1a was shown to shuttle between the nucleus and the cytoplasma, and in agreement with these studies we detected different levels of Mybbp1a in these compartments (24, 44).
Mybbp1a Represses rRNA Gene Transcription
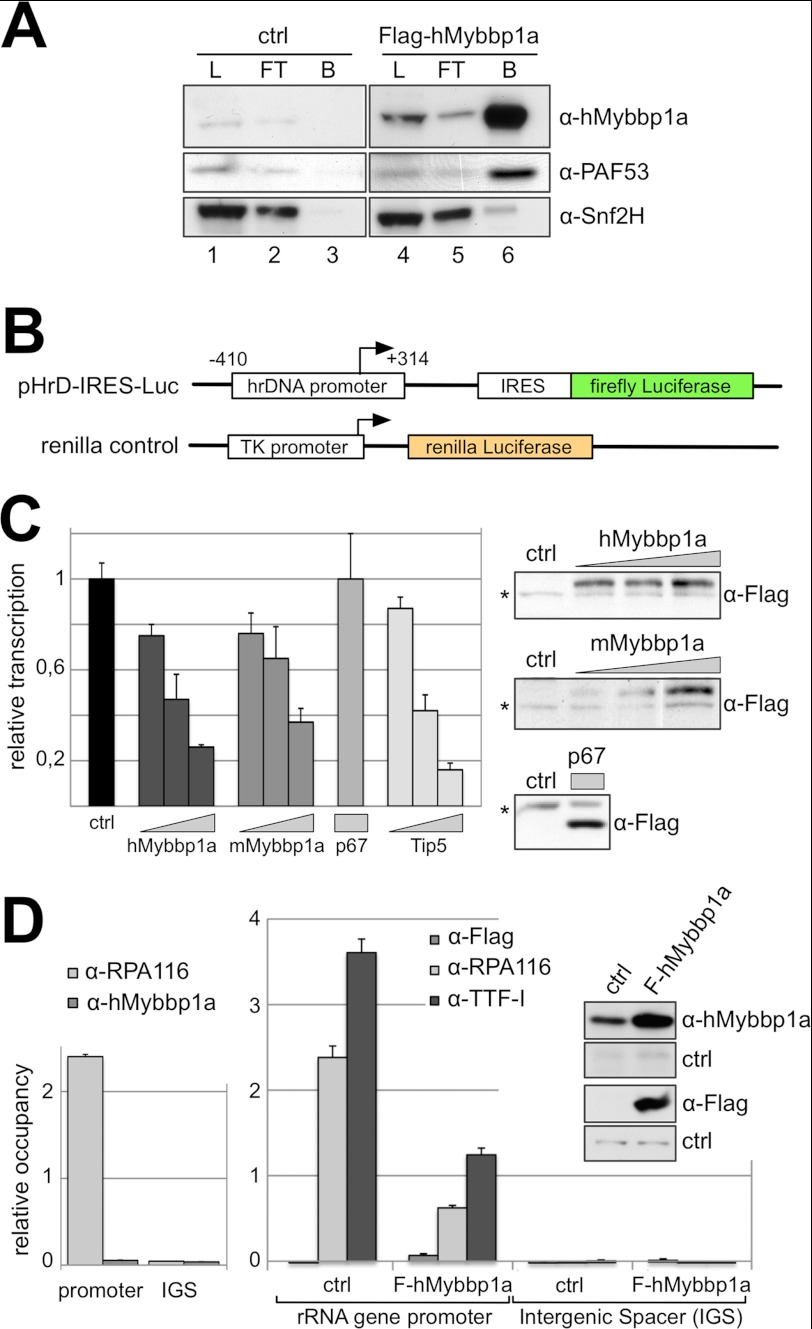
Because of the subnucleolar distribution of Mybbp1a, its interaction with RNA Pol I, and the fact that the protein is widely conserved throughout evolution, orthologs that have been identified in animals, plants, and fungi (23, 31, 46) suggest that Mybbp1a may play a role in the regulation of rRNA synthesis. First, the physical association of Mybbp1a with the RNA Pol I complex was confirmed by co-immunoprecipitation experiments using the transiently transfected FLAG-Mybbp1a construct. Affinity tag purification revealed the specific interaction of Mybbp1a with PAF53, a subunit of the RNA Pol I complex (Fig. 2A). Snf2h has been previously described as an interacting protein of Mybbp1a and was used as a positive control (47). Next we tested Mybbp1a function in the regulation of rRNA gene transcription using a reporter system in HeLa cells. To visualize RNA Pol I activity, we employed the human rRNA minigene construct pHrD-IRES-Luc that harbors the rRNA gene promoter (−410 to +314 bp relative to the transcription start site) followed by an internal ribosome entry site (IRES) and the coding sequence for the firefly luciferase protein (Fig. 2B) (41, 48). The IRES sequence allows translation of the rRNA Pol I transcripts and relative quantification of the minigene RNA levels via the luciferase activity. RNA Pol I transcription efficiency from the minigene construct was normalized to a co-transfected RNA Pol II-dependent construct expressing the Renilla luciferase (Fig. 2B).
FIGURE 2.
Mybbp1a represses rRNA gene transcription. A, nuclear extracts of HEK239T cells either transfected with FLAG-Mybbp1a expression plasmid (lanes 4–6) or control DNA (lanes 1–3) were incubated with α-FLAG-agarose. For the detection with α-Mybbp1a and α-Snf2h load (L, 5%), flow-through (FT, 5%), bead-bound (B), and eluted (E) proteins were analyzed by Western blotting. For the detection with α-PAF53, 0.3% of the load and flow-through were loaded on the gel. B, the pHrD-IRES-Luc (41) construct contains the human rDNA (hrDNA) promoter from −410 to + 314 (relative to the transcription start site) followed by an IRES fused to the firefly luciferase coding region. The renilla control construct carrying the renilla luciferase gene under the control of a thymidine kinase promoter (transcribed by Pol II) was used to normalize for transfection efficiency. C, HeLa cells were co-transfected with both luciferase reporter constructs, pHrD-IRES-Luc and renilla control, and increasing amounts (188, 375, 750 ng) of either human Mybbp1a, mouse Mybbp1a, or Tip5 expression plasmids. The mouse p67MBP*NLS (p67) expression plasmid was transfected with an amount of 188 ng. 48 h after transfection, luciferase activity was measured, and firefly luciferase counts (RNA Pol I reporter) were normalized to renilla luciferase counts and compared with the control transfection reaction (ctrl). Protein levels were analyzed by Western blotting with α-FLAG antibody. An unspecific band (*) served as loading control. D, ChIP experiments were performed with chromatin derived from purified nucleoli (left) or whole cell extracts (right) of cells either transfected with human FLAG-Mybbp1a expression plasmid (FLAG-hMybbp1a) or control DNA (ctrl). For immunoprecipitation α-FLAG, α-hMybbp1a, α-RPA116, and α-TTF-I were used as indicated. Purified rabbit immunoglobulin G was used for background estimation. FLAG-hMybbp1a protein levels were analyzed by Western blotting with α-Mybbp1a and α-FLAG antibodies as indicated.
Overexpression of human or mouse Mybbp1a in HeLa cells resulted in a dose-dependent transcriptional repression of the rRNA minigene expression, comparable to the repressive effect of Tip5 (Fig. 2C and supplemental Fig. S2), which is the large subunit of the nucleolar remodeling complex, NoRC, that serves to silence rRNA genes by changing their chromatin structure and initiating heterochromatinization of rDNA (32, 49, 50). In contrast, overexpression of equal amounts of a C-terminal-truncated form of mouse Mybbp1a (mp67MBP*NLS) containing the SV40-T-antigen NLS to target the protein to the nucleus and nucleolus (24) (Fig. 1D) had no effect on RNA Pol I-dependent transcription (Fig. 2C). These results suggest that the C terminus of Mybbp1a is required for efficient transcriptional repression. To obtain more insight into the action of Mybbp1a at rRNA genes, ChIP experiments were performed. Nuclear and nucleolar extracts of control cells and cells overexpressing the FLAG-tagged Mybbp1a were immunoprecipitated with antibodies against the FLAG tag, hMybbp1a, RPA194, RPA116, and transcription termination factor-I (TTF-I; Fig. 2D and supplemental Fig. S3). In these experiments neither the α-FLAG nor the α-Mybbp1a antibodies were able to co-precipitate rDNA fragments, which suggests that Mybbp1a functions at a distance that cannot be captured by formaldehyde cross-link. However, we did observe reduced rDNA binding of RNA Pol I and TTF-I upon overexpression of FLAG-Mybbp1a, correlating with Mybbp1a-dependent Pol I transcription repression. The results suggest that Mybbp1a may repress transcription through its binding to the RNA Pol I complex.
Mybbp1a Is Involved in Pre-rRNA Processing
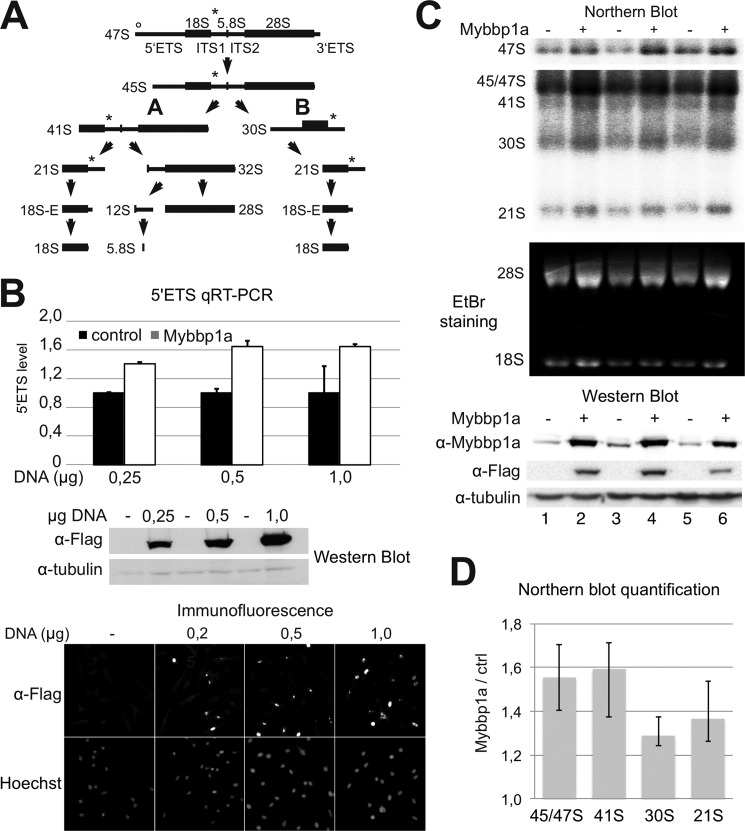
In a recent study, Greenblatt and co-workers (51) analyzed the composition of yeast rRNA-processing complexes and found that Pol5p, the suggested yeast homolog of Mybbp1a, is associated with the UtpA complex. UtpA is the possible equivalent of the t-Utp subcomplex of the small ribosome subunit processome that was identified in an independent study (2). Accordingly, we studied the role of Mybbp1a in pre-rRNA processing, which involves a number of ordered and consecutive endo- and exonucleolytic cleavages of the 47 S precursor to produce the 18 S, 5.8 S, and 28 S rRNAs. A short overview of the major mammalian rRNA processing pathways is depicted in Fig. 3A (for a detailed review, see Ref. 52).
FIGURE 3.
Mybbp1a overexpression perturbs rRNA processing. A, shown is a schematic representation of the rRNA processing in human cells (for more details see Ref. 52). Intermediates and final processing products are shown. ETS, external transcribed spacer; ITS, internal transcribed spacer. A circle and a star indicate the locations of qRT-PCR and Northern probes. B, HeLa cells were transfected with increasing amounts of FLAG-Mybbp1a expression plasmid or control DNA. Steady-state levels of the 5′ETS were quantified in duplicate qRT-PCR measurements and normalized to β-actin mRNA levels. The upper diagram shows the average and S.D. values, and the middle and lower panels illustrate the overexpression of Mybbp1a by Western blot and immunofluorescence, respectively. C, rRNAs were detected on Northern blots after Mybbp1a overexpression in HeLa cells (upper panels). EtBr staining shows the mature 18 S and 28 S rRNA in the middle panel, and a Western blot shows the expression of FLAG-Mybbp1a of three independent experiments in the lower panel. D, quantification of the Northern blot shown in C. rRNA levels were normalized to 18 S rRNA and expressed relative to the control reaction (ctrl). Averages and minimum and maximum values are shown. Two-tailed, paired t tests were performed on the datasets, and they showed that p < 0.05 in all cases (p = 0.02 for 47 S, p = 0.039 for 41 S, p = 0.011 for 30 S, and p = 0.036 for 21 S).
The reporter assay described above allowed the uncoupling of rRNA transcription from processing, whereas the analysis of cellular rRNA levels reflects the sum of these processes. To characterize gain-of-function effects of Mybbp1a, 47 S precursor, intermediate and mature rRNA levels were measured in qRT-PCR and Northern blot experiments (Fig. 3). To our surprise, overexpression of Mybbp1a resulted in elevated steady-state levels of the 5′ETS) fragment of rRNA when measured by qRT-PCR (Fig. 3B).
This result did apparently contradict the reporter assay that monitors only transcription initiation (Fig. 2C). To confirm the qRT-PCR in an independent assay and to rule out the possible accumulation of the 5′ETS fragment after co-transcriptional cleavage, 47 S pre-rRNA levels were measured by the Northern blot assay (Fig. 3C, upper panel). Because the result confirmed the qRT-PCR observations, we hypothesize a dominant negative effect of Mybbp1a on pre-rRNA processing. The difference between both assays is that the reporter assay using the core promoter is only capable of revealing the effects on transcription initiation, whereas qRT-PCR and Northern blot analyze the combined effects of transcription initiation and pre-rRNA processing. An inhibition of rRNA processing steps may result in 47/45 S rRNA accumulation even though rRNA transcription levels are decreased. Thus, we investigated the levels of rRNA processing intermediates by Northern blot analysis of the RNA (Fig. 3C). The quantification of the data (Fig. 3D) approved our hypothesis, demonstrating accumulation of the 47/45 S, 41 S, and to a lower level of 30 S and 21 S rRNA intermediates. Mainly pathway A (Fig. 3A) was affected by overexpression of Mybbp1a; however, also minor effects on the pathway B were observed. Our data suggest that Mybbp1a participates in transcriptional repression and is involved in pre-rRNA processing.
Mybbp1a Associates with the Ribosome Biogenesis Machinery
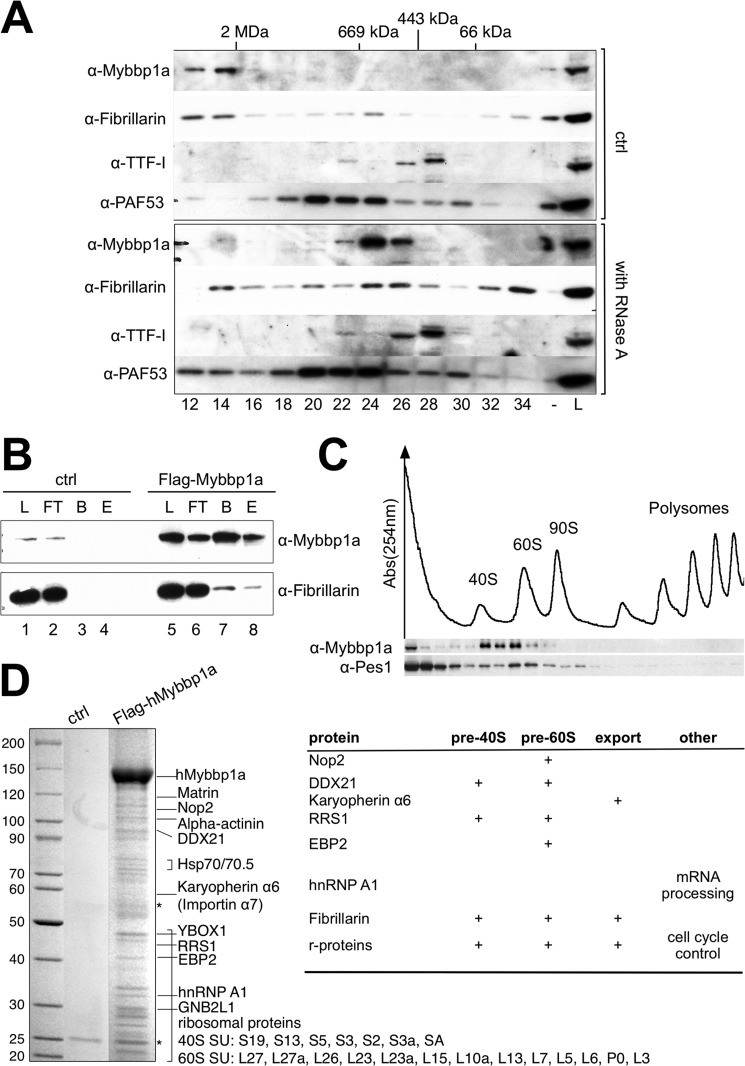
Several large RNA-containing complexes are involved in ribosome synthesis and implement pre-rRNA processing and pre-ribosome assembly in accordance with rDNA transcription (for review, see Ref. 53). To reveal the binding of Mybbp1a to RNA-containing complexes, we first monitored the cellular localization of Mybbp1a after RNase A treatment in HeLa cells (supplemental Fig. S4). Immunodetection of the endogenous protein revealed a relocalization of Mybbp1a from the nucleoli to the nucleoplasm upon RNase A treatment, whereas the Pol I-specific and rDNA binding TTF-I remained nucleolar. Furthermore, control treatment with DNase I, an enzyme that does not hydrolyze RNA, did not change the localization of Mybbp1a. These findings suggest that nucleolar localization of Mybbp1a depends on the interaction with a nucleolar RNA component and confirm very recent data obtained in the human breast cancer cell line MCF-7 (54).
We next investigated whether Mybbp1a is part of an RNA-containing complex, as suggested by the immunofluorescence experiments. Therefore, HeLa nuclear extract was fractionated by gel filtration on a Superose 6 column, and the fractions were assayed for the presence of Mybbp1a (Fig. 4A). Mybbp1a mostly eluted with an apparent molecular mass in the MDa range, co-migrating with the RNA Pol I holoenzyme complex. In agreement with this observation, a recent analysis of the protein composition of transcription factories also revealed the association of Mybbp1a with the RNA Pol I transcription factory (55). Furthermore, the protein co-migrates with fibrillarin, a protein involved in several steps of ribosome biogenesis (56). Mybbp1a, and in part fibrillarin, were shifted to fractions corresponding to lower molecular masses when the extract was incubated with RNase A, confirming that it is part of a large multisubunit RNase-sensitive complex such as the ribosome biosynthesis machinery. To confirm that fibrillarin and Mybbp1a are present in a single complex, we transfected HeLa cells with the human FLAG-Mybbp1a expression construct and performed co-immunoprecipitation experiments. FLAG-Mybbp1a co-precipitated fibrillarin, and fibrillarin also specifically eluted with Mybbp1a from the affinity matrix (Fig. 4B). Our data confirm the proteomics data and identify Mybbp1a as an interactor of fibrillarin (57).
FIGURE 4.
Mybbp1a is part of a RNA-sensitive protein complex. A, RNase A-treated and mock-treated (ctrl) HeLa nuclear extracts were separated on a Superose 6 gel filtration column. Load (9%), and collected fractions (every second fraction from 12 to 34) were analyzed by Western blotting with the indicated antibodies. B, nuclear extracts of HeLa cells either transfected with FLAG-Mybbp1a expression plasmid (lane 5–8) or control DNA (ctrl; lane 1–4) were incubated with α-FLAG-agarose. Load (L; 5%), flow-through (FT; 5%), bead-bound (B), and eluted proteins (E) were analyzed by Western blotting with the indicated antibodies. C, polysome fractionation was performed on a sucrose gradient, and Mybbp1a and Pes1 protein levels of the fractions were detected by immunoblotting. Pre-assembled ribosomal subunit, monosome, and polysome fractions were detected by measuring UV absorbance at 254 nm as indicated. D, nuclear extracts of HeLa cells either transfected with FLAG-Mybbp1a expression plasmid or control DNA (ctrl) were used for immunoprecipitation with α-FLAG-agarose. Eluted protein fractions were separated by SDS-PAGE, silver-stained, and protein bands were cut and subjected to MALDI-TOF mass spectrometry. Proteins identified are indicated, and their participation in ribosome biogenesis is summarized on the right. hnRNP, heterogeneous nuclear ribonucleoprotein.
Next we studied the association of Mybbp1a with the ribosomal biogenesis machinery. We studied its sedimentation behavior on a sucrose gradient with regard to pre-ribosomal particles and polyribosomes. Sucrose gradient centrifugation in combination with UV profiling and Western blotting clearly revealed a co-migration of Mybbp1a with the 40 S and 60 S pre-ribosomal particles, whereas no Mybbp1a was detected in the mRNA-associated polysome fractions (Fig. 4C). Pes1, a factor involved in the processing of the 32 S precursor, showed a preference for the 60 S pre-ribosomal particle as expected. This finding suggests that Mybbp1a is involved in the maturation of the ribosomal subunits, but it does not interact with the mature, translating ribosomal particles.
To reveal additional binding partners of Mybbp1a, the FLAG-tagged protein was transiently transfected into HeLa cells, and associated proteins were purified by affinity chromatography, separated by gel electrophoresis, and analyzed by MALDI-TOF mass spectrometry (Fig. 4D). As suggested by our experiments, Mybbp1a interacts with proteins belonging to the ribosome biogenesis pathway and the small and large ribosomal proteins that are assembled during rRNA transcription. We tested several of those proteins for co-migration with Mybbp1a in the presence or absence of RNA. Interestingly, those proteins co-migrated with Mybbp1a in the large complex and were converted to distinct smaller complexes after RNase A treatment (supplemental Fig. S5).
Mybbp1a Is Essential for Cellular Proliferation
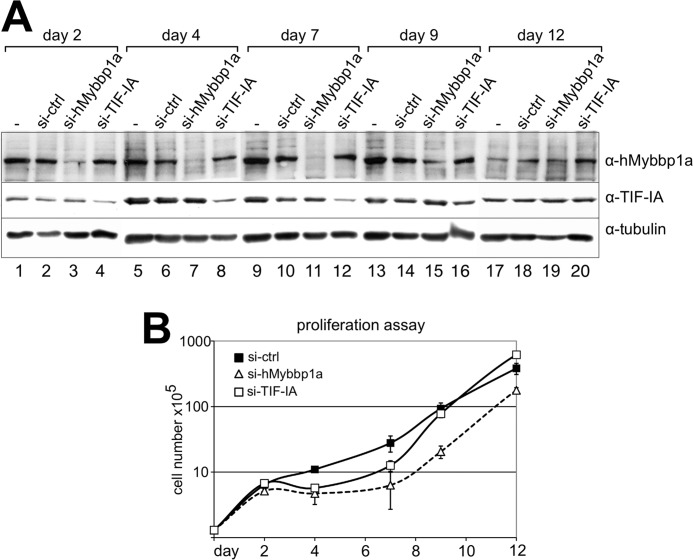
To study the consequence of Mybbp1a-dependent perturbation of ribosome biogenesis on cell proliferation, loss of function experiments were performed using siRNA. Knockdown experiments were performed with GL3- (a control siRNA directed against the luciferase gene), TIF-IA-, and Mybbp1a-specific siRNAs (Fig. 5A and supplemental Fig. S6). Knockdown of Mybbp1a and TIF-IA was monitored at the protein level and followed for up to 12 days (Fig. 5A). siRNA-mediated knockdown of Mybbp1a efficiently reduced protein levels from day 2 through day 7 followed by partial (day 9) and full (day 12) recovery of Mybbp1a protein levels. siRNA-mediated knockdown of TIF-IA was not as efficient as Mybbp1a depletion, but a visible reduction in the protein level was detected from day 2 to day 7.
FIGURE 5.
siRNA-mediated depletion of Mybbp1a delays cellular proliferation. A, HeLa cells were transfected with siRNA targeting Mybbp1a (si-Mybbp1a.2), TIF-IA, and a control siRNA (si-ctrl). Protein levels were analyzed by Western blotting 2, 4, 7, 9, and 12 days after transfection, as indicated. The α-tubulin staining of the membrane served as loading control. B, cellular proliferation upon siRNA treatment of the cells using si-Mybbp1a (white triangle), si-TIF-IA (white square), or si-ctrl (black square) was followed from days 0 to 12 by counting the number of living cells. Cell numbers are plotted in log 10 scale. The average and S.D. values were calculated from three technical replicates. One of two independent data sets is shown.
To monitor the effect of protein depletion on rRNA levels, we measured first the steady-state levels of the 5′ETS by quantitative real time PCR (supplemental Fig. S6). Total cellular RNA was isolated and reverse-transcribed, and the level of the 5′ETS was quantified and normalized to β-actin mRNA. In agreement with the rRNA minigene expression studies (Fig. 2), depletion of Mybbp1a resulted in an increase in pre-rRNA levels in the cell. As expected, reduced TIF-IA levels resulted in a strong decrease of pre-rRNA synthesis, as this protein is an essential transcription initiation factor (58, 59) (supplemental Fig. S6).
In addition, we observed that cells depleted of either TIF-IA or Mybbp1a behaved similarly and ceased cellular proliferation. BrdU labeling experiments of Mybbp1a-depleted cells showed a reduced number of cells in S-phase (supplemental Fig. S7A). Quantification of cellular proliferation by counting cells after protein knockdown (Fig. 5B) revealed a stronger effect of Mybbp1a on cellular proliferation as compared with TIF-IA. Halted cellular proliferation correlates with the time frame of reduced protein levels (Fig. 5). According to our observation that depletion of Mybbp1a resulted in the accumulation of pre-rRNA, we would have expected increased proliferation rates after Mybbp1a knockdown. These findings appear to be counterintuitive, as high rDNA transcription activity and rRNA levels are the major determinants of cellular growth and proliferation but can be explained by our experimental data on rRNA processing that indicate additional roles for Mybbp1a in growth-related processes other than rDNA transcription. Furthermore, visual inspection of the Mybbp1a knockdown cells revealed an abnormal, flattened, and enlarged morphology (supplemental Fig. S7B).
DISCUSSION
We identified Mybbp1a association with RNA Pol I and demonstrated that Mybbp1a serves both as a negative regulator of rRNA gene transcription and as a functional part of the ribosome biogenesis machinery. The results of the loss-of-function studies together with the gain-of-function experiments suggest that Mybbp1a plays a dual role in rRNA metabolism; first, regulating transcription initiation and, second, being essential for the correct processing of the pre-rRNA. Depletion of the protein leads to a dominant anti-repression effect on rRNA synthesis, whereas the overexpression of Mybbp1a impairs dominantly the rRNA processing. Although ribosome biogenesis is a major determinant of cellular growth and proliferation, additional functions of Mybbp1a in growth-related processes may explain its effects on cellular proliferation. Yet our data showed that Mybbp1a plays a role in the regulation of rRNA transcription and is an essential component of the ribosome biogenesis machinery at the same time, placing the protein at the interface of transcription and processing.
Intriguingly, Mybbp1a was very recently identified to activate the tumor suppressor p53 as a consequence of nucleolar stress (54). Upon inhibition of rRNA synthesis by TIF-IA knockdown or treatment with low doses of actinomycin D, Mybbp1a translocated to the nucleoplasm and increased the acetylation levels of p53. Mybbp1a did stimulate the interaction of p53 with the histone acetyltransferase p300, leading to the p53-dependent gene activation. Taken together, it seems that Mybbp1a plays an important role as a central regulator of proliferation, cell cycle progression, and differentiation in the mammalian system. As shown by our study, Mybbp1a affects simultaneously the Pol II and Pol I transcription systems to achieve its regulatory function.
In budding and bakers' yeast, the protein Pol5p was identified as the potential homolog of Mybbp1a and was shown to bind to the rRNA gene repeat (60, 61). In yeast, Pol5p has been connected with an essential role in rRNA synthesis. We present here substantial evidence for the role of its potential mammalian homologue both in rRNA synthesis and processing. In the absence of Mybbp1a, rRNA synthesis became de-repressed, but cells ceased proliferation at the same time (Fig. 5 and supplemental Fig S6). These results argue that Mybbp1a has a pro-proliferative role, as was observed in yeast. In the case of transcriptional regulation, the further analysis of Mybbp1a interaction with RNA Pol I could provide more mechanistic insights into the function of the protein. Here, the identification and function of the RNA component(s) involved in Mybbp1a-dependent regulation of rRNA synthesis would be particularly interesting. The RNase A sensitivity of the major fraction of RNA Pol I-Mybbp1a-containing complexes (Fig. 4A) suggests that the association of the proteins may take place in a transcriptionally engaged ternary protein-DNA-RNA complex.
We propose that the accumulation of 47 S pre-rRNA upon Mybbp1a depletion is a combined consequence of transcription up-regulation due to de-repression of the rRNA gene and a defect in rRNA processing. A similar effect was suggested for Pwp2 (Utp1), a component of the small ribosome subunit processome (62). Depletion of Pwp2 leads to pre-rRNA accumulation due to stalling rRNA processing. In turn, when cells were overloaded with Mybbp1a, the excess of the protein may perturb dominantly the rRNA processing pathway (Fig. 3). In conclusion, it is likely that the functional role of Mybbp1a involves the coordination of transcription and processing. However, additional experiments have to be performed to unravel the detailed mechanism. These include the dissection of direct and indirect Mybbp1a effects on rDNA transcription by identifying the protein domains and/or RNA that are involved in Mybbp1a-RNA Pol I and Mybbp1a-processome interactions. Functional characterization of specific interaction-defective mutants will give insights into the molecular mechanisms of Mybbp1a-dependent regulation of ribosome biogenesis.
To initially characterize the Mybbp1a mode of action, we have purified Mybbp1a-containing complexes (Fig. 4D). We identified a large variety of ribosomal proteins of both the small and large ribosome subunits as well as of several other factors such as fibrillarin, DDX21, Rrs1, Ebp2, and Nol1 (yeast Nop2) to which roles in rRNA processing and ribosome assembly have been assigned in previous studies (56, 63–66). This result was supported by the observation that Mybbp1a co-migrated with the small and large ribosome subunit pre-ribosomal particles on a polysome sucrose gradient (Fig. 4C). The data show that Mybbp1a is a component of pre-ribosomal complexes and together with the effects in de-regulation pathways A + B (Fig. 3A) in pre-rRNA processing suggests that it is functionally and structurally involved in this process.
Recently, Yamauchi and co-workers (44) have shown that Mybbp1a is processed upon ribosomal stress induction. Treatment of cells with actinomycin D, cisplatin, or UV, all of which inhibit ribosome biogenesis, lead to proteolytic cleavage of Mybbp1a into C-terminal-truncated p140MBP and p67MBP proteins. Proteolysis of Mybbp1a results in its translocation to the nucleoplasm (24, 44). The full-length Mybbp1a and the processed forms were found in distinct complexes, both of which contained nucleophosmin and nucleolin, and the large complex containing the full-length protein also contained nucleostemin and ribosomal proteins (44). We speculate that the Mybbp1a-containing ribosome assembly/processing machinery identified in this study is likely to be equivalent to the large complex purified by Yamauchi and co-workers (44). Indeed, a recent characterization of Mybbp1a-containing complexes in MCF-7 cells by Yanagisawa and co-workers (54) also revealed some of the factors identified in this study, such as EBP2 and Nol1, confirming our results.
Interestingly, depletion of Rrs1 or Ebp2, factors that co-purified with Mybbp1a, were also shown to affect rRNA processing, mirroring the effects observed for Mybbp1a (63, 64). Rrs1 is a ribosome assembly factor that recruits the 5 S rRNA and the ribosomal proteins rpL5 and rpL11 into nascent ribosomes (67). Moreover, Rrs1 seems to be involved in rDNA transcription regulation, as a yeast rrs1 mutant has been shown to reduce transcriptional repression of the rRNA gene (63). Both Rrs1 and Ebp2 have recently been connected to a role in the progression of mitosis as well. Mybbp1a is phosphorylated by the mitotic kinase Aurora B, and it has been revealed that depletion of Mybbp1a or Rrs1 leads to a mitotic delay and abnormalities in spindle organization (45, 68). Additionally, the involvement of Mybbp1a, rpL5, and rpL11 was shown in p53 acetylation and accumulation, which is dependent on the release of Mybbp1a from nucleolar RNA upon stress (54). This study strongly suggests a role for Mybbp1a in rRNA metabolism, based on its binding to nucleolar RNA.
We have shown that Mybbp1a is required for cellular proliferation, an effect that may relate either to its role in pre-rRNA processing or in the regulation of extra-nucleolar interaction partners. Our experiments cannot discriminate between these scenarios. Mybbp1a-depleted cells exhibit a flattened and enlarged morphology that is clearly visible on day 4 after siRNA-mediated knockdown (supplemental Fig. S7B). This phenotype mirrors the “flat cell phenotype” of retinoblastoma (Rb)-negative SAOS-2 sarcoma cells when overexpression of Rb leads to a cell cycle arrest in late G1 phase (69). Thus, it would be interesting to address the possible role of Mybbp1a in coordinating ribosome biogenesis and cell cycle progression in future studies.
As transcription of rRNA genes and subsequent pre-RNA processing and ribosome assembly processes are the major energy-consuming processes in the cell, the rate of biogenesis is tightly linked to cellular proliferation. We suggest that in proliferating cells, Mybbp1a is mainly associated with the pre-ribosomal complexes where it acts as a scaffold for rRNA processing and assembly factors and is functionally required to drive efficient ribosome biogenesis. Reduced levels of ribosome biogenesis upon stress signals and/or reduced demands of ribosomes potentially result in the disassembly of pre-ribosomal particles and a translocation of Mybbp1a to the nucleoplasm. Although nucleolar Mybbp1a would repress RNA Pol I transcription, the nuclear proteins would modulate the activity of transcription regulators, such as c-myb, peroxisome proliferator-activated receptor-γ coactivator 1α, NF-κB, or p53, to cease cell cycle progression, proliferation, and energy production.
Supplementary Material
Acknowledgments
We thank R. Strohner, A. Brehm, A. Hochheimer, and T. Straub for stimulating and helpful discussions and A. Maiser for technical assistance. We are very thankful to K. Dachauer for technical help. We thank I. Grummt for the generous gifts of antibodies and cDNA construct and T. Misteli for the GFP-RPA43 plasmid.
This work was supported by Deutsche Forschungsgemeinschaft grants (to G. L., D. E., A. I., and H. L.) and the Bayerisches Genomforschungsnetzwerk (BayGene) (to G. L.).

This article contains supplemental Figs. S1–S7.
- pre-rRNA
- rRNA precursor
- PIC
- pre-initiation complex
- 5′ETS
- 5′ external transcribed spacer
- Mybbp1a
- Myb-binding protein 1a
- rDNA
- rRNA gene
- RNA Pol I
- RNA polymerase I
- TTF-I
- transcription termination factor-I
- Utps
- U three protein
- t-Utps
- transcriptional-Utps
- ctrl
- control
- IRES
- internal ribosome entry site
- NLS
- nuclear localization signal
- qPCR
- quantitative real time PCR.
REFERENCES
- 1. Warner J. R. (1999) The economics of ribosome biosynthesis in yeast. TIBS 24, 437–440 [DOI] [PubMed] [Google Scholar]
- 2. Gallagher J. E., Dunbar D. A., Granneman S., Mitchell B. M., Osheim Y., Beyer A. L., Baserga S. J. (2004) RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18, 2506–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schneider D. A., Michel A., Sikes M. L., Vu L., Dodd J. A., Salgia S., Osheim Y. N., Beyer A. L., Nomura M. (2007) Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol. Cell 26, 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grummt I., Pikaard C. S. (2003) Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 4, 641–649 [DOI] [PubMed] [Google Scholar]
- 5. Németh A., Guibert S., Tiwari V. K., Ohlsson R., Längst G. (2008) Epigenetic regulation of TTF-I-mediated promoter-terminator interactions of rRNA genes. EMBO J. 27, 1255–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Németh A., Längst G. (2008) Chromatin organization of active ribosomal RNA genes. Epigenetics 3, 243–245 [DOI] [PubMed] [Google Scholar]
- 7. Németh A., Längst G. (2011) Genome organization in and around the nucleolus. Trends Genet. 27, 149–156 [DOI] [PubMed] [Google Scholar]
- 8. Mayer C., Zhao J., Yuan X., Grummt I. (2004) mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 18, 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hannan K. M., Brandenburger Y., Jenkins A., Sharkey K., Cavanaugh A., Rothblum L., Moss T., Poortinga G., McArthur G. A., Pearson R. B., Hannan R. D. (2003) mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell. Biol. 23, 8862–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao J., Yuan X., Frödin M., Grummt I. (2003) ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell 11, 405–413 [DOI] [PubMed] [Google Scholar]
- 11. Stefanovsky V. Y., Pelletier G., Hannan R., Gagnon-Kugler T., Rothblum L. I., Moss T. (2001) An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 8, 1063–1073 [DOI] [PubMed] [Google Scholar]
- 12. Dragon F., Gallagher J. E., Compagnone-Post P. A., Mitchell B. M., Porwancher K. A., Wehner K. A., Wormsley S., Settlage R. E., Shabanowitz J., Osheim Y., Beyer A. L., Hunt D. F., Baserga S. J. (2002) A large nucleolar U3 ribonucleoprotein required for 18 S ribosomal RNA biogenesis. Nature 417, 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Osheim Y. N., French S. L., Keck K. M., Champion E. A., Spasov K., Dragon F., Baserga S. J., Beyer A. L. (2004) Pre-18 S ribosomal RNA is structurally compacted into the SSU processome before being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell 16, 943–954 [DOI] [PubMed] [Google Scholar]
- 14. Mougey E. B., O'Reilly M., Osheim Y., Miller O. L., Jr., Beyer A., Sollner-Webb B. (1993) The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 7, 1609–1619 [DOI] [PubMed] [Google Scholar]
- 15. Kos M., Tollervey D. (2010) Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell 37, 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prieto J. L., McStay B. (2007) Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 21, 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stefanovsky V., Langlois F., Gagnon-Kugler T., Rothblum L. I., Moss T. (2006) Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell 21, 629–639 [DOI] [PubMed] [Google Scholar]
- 18. Schneider D. A., French S. L., Osheim Y. N., Bailey A. O., Vu L., Dodd J., Yates J. R., Beyer A. L., Nomura M. (2006) RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc. Natl. Acad. Sci. U.S.A. 103, 12707–12712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grummt I. (2003) Life on a planet of its own. Regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17, 1691–1702 [DOI] [PubMed] [Google Scholar]
- 20. Milkereit P., Gadal O., Podtelejnikov A., Trumtel S., Gas N., Petfalski E., Tollervey D., Mann M., Hurt E., Tschochner H. (2001) Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105, 499–509 [DOI] [PubMed] [Google Scholar]
- 21. Ferreira-Cerca S., Pöll G., Kühn H., Neueder A., Jakob S., Tschochner H., Milkereit P. (2007) Analysis of the in vivo assembly pathway of eukaryotic 40 S ribosomal proteins. Mol. Cell 28, 446–457 [DOI] [PubMed] [Google Scholar]
- 22. Holmström T. H., Mialon A., Kallio M., Nymalm Y., Mannermaa L., Holm T., Johansson H., Black E., Gillespie D., Salminen T. A., Langel U., Valdez B. C., Westermarck J. (2008) c-Jun supports ribosomal RNA processing and nucleolar localization of RNA helicase DDX21. J. Biol. Chem. 283, 7046–7053 [DOI] [PubMed] [Google Scholar]
- 23. Tavner F. J., Simpson R., Tashiro S., Favier D., Jenkins N. A., Gilbert D. J., Copeland N. G., Macmillan E. M., Lutwyche J., Keough R. A., Ishii S., Gonda T. J. (1998) Molecular cloning reveals that the p160 Myb-binding protein is a novel, predominantly nucleolar protein that may play a role in transactivation by Myb. Mol. Cell. Biol. 18, 989–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keough R. A., Macmillan E. M., Lutwyche J. K., Gardner J. M., Tavner F. J., Jans D. A., Henderson B. R., Gonda T. J. (2003) Myb-binding protein 1a is a nucleocytoplasmic shuttling protein that utilizes CRM1-dependent and independent nuclear export pathways. Exp. Cell Res. 289, 108–123 [DOI] [PubMed] [Google Scholar]
- 25. Fan M., Rhee J., St-Pierre J., Handschin C., Puigserver P., Lin J., Jäeger S., Erdjument-Bromage H., Tempst P., Spiegelman B. M. (2004) Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1α. Modulation by p38 MAPK. Genes Dev. 18, 278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Díaz V. M., Mori S., Longobardi E., Menendez G., Ferrai C., Keough R. A., Bachi A., Blasi F. (2007) p160 Myb-binding protein interacts with Prep1 and inhibits its transcriptional activity. Mol. Cell. Biol. 27, 7981–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Owen H. R., Elser M., Cheung E., Gersbach M., Kraus W. L., Hottiger M. O. (2007) MYBBP1a is a novel repressor of NF-κB. J. Mol. Biol. 366, 725–736 [DOI] [PubMed] [Google Scholar]
- 28. Jones L. C., Okino S. T., Gonda T. J., Whitlock J. P., Jr. (2002) Myb-binding protein 1a augments AhR-dependent gene expression. J. Biol. Chem. 277, 22515–22519 [DOI] [PubMed] [Google Scholar]
- 29. Hara Y., Onishi Y., Oishi K., Miyazaki K., Fukamizu A., Ishida N. (2009) Molecular characterization of Mybbp1a as a co-repressor on the Period2 promoter. Nucleic Acids Res. 37, 1115–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seither P., Grummt I. (1996) Molecular cloning of RPA2, the gene encoding the second largest subunit of mouse RNA polymerase I. Genomics 37, 135–139 [DOI] [PubMed] [Google Scholar]
- 31. Keough R., Woollatt E., Crawford J., Sutherland G. R., Plummer S., Casey G., Gonda T. J. (1999) Molecular cloning and chromosomal mapping of the human homologue of MYB binding protein (P160) 1A (MYBBP1A) to 17p13.3. Genomics 62, 483–489 [DOI] [PubMed] [Google Scholar]
- 32. Strohner R., Nemeth A., Jansa P., Hofmann-Rohrer U., Santoro R., Längst G., Grummt I. (2001) NoRC, a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 20, 4892–4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. (1980) DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc. Natl. Acad. Sci. U.S.A. 77, 3855–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seither P., Zatsepina O., Hoffmann M., Grummt I. (1997) Constitutive and strong association of PAF53 with RNA polymerase I. Chromosoma 106, 216–225 [DOI] [PubMed] [Google Scholar]
- 35. Bodem J., Dobreva G., Hoffmann-Rohrer U., Iben S., Zentgraf H., Delius H., Vingron M., Grummt I. (2000) TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 1, 171–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hölzel M., Rohrmoser M., Schlee M., Grimm T., Harasim T., Malamoussi A., Gruber-Eber A., Kremmer E., Hiddemann W., Bornkamm G. W., Eick D. (2005) Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J. Cell Biol. 170, 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Evers R., Smid A., Rudloff U., Lottspeich F., Grummt I. (1995) Different domains of the murine RNA polymerase I-specific termination factor mTTF-I serve distinct functions in transcription termination. EMBO J. 14, 1248–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Christensen M. E., Banker N. (1992) Mapping of monoclonal antibody epitopes in the nucleolar protein fibrillarin (B-36) of Physarum polycephalum. Cell Biol. Int. Rep. 16, 1119–1131 [DOI] [PubMed] [Google Scholar]
- 39. Wu H., Ceccarelli D. F., Frappier L. (2000) The DNA segregation mechanism of Epstein-Barr virus nuclear antigen 1. EMBO Rep. 1, 140–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Swiercz R., Person M. D., Bedford M. T. (2005) Ribosomal protein S2 is a substrate for mammalian PRMT3 (Protein Arginine Methyltransferase 3). Biochem. J. 386, 85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ghoshal K., Majumder S., Datta J., Motiwala T., Bai S., Sharma S. M., Frankel W., Jacob S. T. (2004) Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J. Biol. Chem. 279, 6783–6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Sullivan A. C., Sullivan G. J., McStay B. (2002) UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 22, 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dundr M., Hoffmann-Rohrer U., Hu Q., Grummt I., Rothblum L. I., Phair R. D., Misteli T. (2002) A kinetic framework for a mammalian RNA polymerase in vivo. Science 298, 1623–1626 [DOI] [PubMed] [Google Scholar]
- 44. Yamauchi T., Keough R. A., Gonda T. J., Ishii S. (2008) Ribosomal stress induces processing of Mybbp1a and its translocation from the nucleolus to the nucleoplasm. Genes Cells 13, 27–39 [DOI] [PubMed] [Google Scholar]
- 45. Perrera C., Colombo R., Valsasina B., Carpinelli P., Troiani S., Modugno M., Gianellini L., Cappella P., Isacchi A., Moll J., Rusconi L. (2010) Identification of Myb-binding protein 1A (MYBBP1A) as a novel substrate for aurora B kinase. J. Biol. Chem. 285, 11775–11785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang W., Rogozin I. B., Koonin E. V. (2003) Yeast POL5 is an evolutionarily conserved regulator of rDNA transcription unrelated to any known DNA polymerases. Cell Cycle 2, 120–122 [DOI] [PubMed] [Google Scholar]
- 47. Cavellán E., Asp P., Percipalle P., Farrants A. K. (2006) The WSTF-SNF2h chromatin remodeling complex interacts with several nuclear proteins in transcription. J. Biol. Chem. 281, 16264–16271 [DOI] [PubMed] [Google Scholar]
- 48. Németh A., Strohner R., Grummt I., Längst G. (2004) The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Res. 32, 4091–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Santoro R., Li J., Grummt I. (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32, 393–396 [DOI] [PubMed] [Google Scholar]
- 50. Li J., Längst G., Grummt I. (2006) NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 25, 5735–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krogan N. J., Peng W. T., Cagney G., Robinson M. D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D. P., Beattie B. K., Lalev A., Zhang W., Davierwala A. P., Mnaimneh S., Starostine A., Tikuisis A. P., Grigull J., Datta N., Bray J. E., Hughes T. R., Emili A., Greenblatt J. F. (2004) High definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13, 225–239 [DOI] [PubMed] [Google Scholar]
- 52. Henras A. K., Soudet J., Gérus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. (2008) The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65, 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Granneman S., Baserga S. J. (2005) Cross-talk in gene expression. Coupling and co-regulation of rDNA transcription, pre-ribosome assembly, and pre-rRNA processing. Curr. Opin. Cell Biol. 17, 281–286 [DOI] [PubMed] [Google Scholar]
- 54. Kuroda T., Murayama A., Katagiri N., Ohta Y. M., Fujita E., Masumoto H., Ema M., Takahashi S., Kimura K., Yanagisawa J. (2011) RNA content in the nucleolus alters p53 acetylation via MYBBP1A. EMBO J. 30, 1054–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Melnik S., Deng B., Papantonis A., Baboo S., Carr I. M., Cook P. R. (2011) The proteomes of transcription factories containing RNA polymerases I, II, or III. Nat. Methods 8, 963–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E. C. (1993) Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 72, 443–457 [DOI] [PubMed] [Google Scholar]
- 57. Yanagida M., Hayano T., Yamauchi Y., Shinkawa T., Natsume T., Isobe T., Takahashi N. (2004) Human fibrillarin forms a subcomplex with splicing factor 2-associated p32, protein arginine methyltransferases, and tubulins α3 and β1 that is independent of its association with preribosomal ribonucleoprotein complexes. J. Biol. Chem. 279, 1607–1614 [DOI] [PubMed] [Google Scholar]
- 58. Buttgereit D., Pflugfelder G., Grummt I. (1985) Growth-dependent regulation of rRNA synthesis is mediated by a transcription initiation factor (TIF-IA). Nucleic Acids Res. 13, 8165–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schnapp A., Pfleiderer C., Rosenbauer H., Grummt I. (1990) A growth-dependent transcription initiation factor (TIF-IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. EMBO J. 9, 2857–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nadeem F. K., Blair D., McInerny C. J. (2006) Pol5p, a novel binding partner to Cdc10p in fission yeast involved in rRNA production. Mol. Genet. Genomics 276, 391–401 [DOI] [PubMed] [Google Scholar]
- 61. Shimizu K., Kawasaki Y., Hiraga S., Tawaramoto M., Nakashima N., Sugino A. (2002) The fifth essential DNA polymerase phi in Saccharomyces cerevisiae is localized to the nucleolus and plays an important role in synthesis of rRNA. Proc. Natl. Acad. Sci. U.S.A. 99, 9133–9138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bernstein K. A., Bleichert F., Bean J. M., Cross F. R., Baserga S. J. (2007) Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Mol. Biol. Cell 18, 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsuno A., Miyoshi K., Tsujii R., Miyakawa T., Mizuta K. (2000) RRS1, a conserved essential gene, encodes a novel regulatory protein required for ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 2066–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tsujii R., Miyoshi K., Tsuno A., Matsui Y., Toh-e A., Miyakawa T., Mizuta K. (2000) Ebp2p, yeast homologue of a human protein that interacts with Epstein-Barr virus nuclear antigen 1, is required for pre-rRNA processing and ribosomal subunit assembly. Genes Cells 5, 543–553 [DOI] [PubMed] [Google Scholar]
- 65. Hong B., Brockenbrough J. S., Wu P., Aris J. P. (1997) Nop2p is required for pre-rRNA processing and 60 S ribosome subunit synthesis in yeast. Mol. Cell. Biol. 17, 378–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Henning D., So R. B., Jin R., Lau L. F., Valdez B. C. (2003) Silencing of RNA helicase II/Guα inhibits mammalian ribosomal RNA production. J. Biol. Chem. 278, 52307–52314 [DOI] [PubMed] [Google Scholar]
- 67. Zhang J., Harnpicharnchai P., Jakovljevic J., Tang L., Guo Y., Oeffinger M., Rout M. P., Hiley S. L., Hughes T., Woolford J. L., Jr. (2007) Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 21, 2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gambe A. E., Matsunaga S., Takata H., Ono-Maniwa R., Baba A., Uchiyama S., Fukui K. (2009) A nucleolar protein RRS1 contributes to chromosome congression. FEBS Lett. 583, 1951–1956 [DOI] [PubMed] [Google Scholar]
- 69. Hinds P. W., Mittnacht S., Dulic V., Arnold A., Reed S. I., Weinberg R. A. (1992) Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell 70, 993–1006 [DOI] [PubMed] [Google Scholar]
- 70. Marchler-Bauer A., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., He S., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Liebert C. A., Liu C., Lu F., Lu S., Marchler G. H., Mullokandov M., Song J. S., Tasneem A., Thanki N., Yamashita R. A., Zhang D., Zhang N., Bryant S. H. (2009) CDD. Specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37, D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schermelleh L., Carlton P. M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M. C., Agard D. A., Gustafsson M. G., Leonhardt H., Sedat J. W. (2008) Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320, 1332–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.