Background: Lysosome membrane permeabilization is required for TRAIL-induced apoptosis in liver cells.
Results: TRAIL induces recruitment of PACS-2 to lysosomes where it interacts with Bim and Bax to permeabilize lysosomes and trigger apoptosis.
Conclusion: The PACS-2, Bim, and Bax-containing PIXosome is a proapoptotic Bcl-2 scaffold.
Significance: Understanding TRAIL-induced lysosome permeabilization may lead to new treatments for liver diseases.
Keywords: Apoptosis, Bax, Lysosomes, Signal Transduction, Trail, Bim, Death Receptor 5, PACS-2
Abstract
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis of liver cancer cell lines requires death receptor-5 (DR5)-dependent permeabilization of lysosomal membranes. Ligated DR5 triggers recruitment of the proapoptotic proteins Bim and Bax to lysosomes, releasing cathepsin B into the cytosol where it mediates mitochondria membrane permeabilization and activation of executioner caspases. Despite the requirement for lysosome membrane permeabilization during TRAIL-induced apoptosis, little is known about the mechanism that controls recruitment of Bim and Bax to lysosomal membranes. Here we report that TRAIL induces recruitment of the multifunctional sorting protein phosphofurin acidic cluster sorting protein-2 (PACS-2) to DR5-positive endosomes in Huh-7 cells where it forms an immunoprecipitatable complex with Bim and Bax on lysosomal membranes. shRNA-targeted knockdown of PACS-2 prevents recruitment of Bim or Bax to lysosomes, blunting the TRAIL-induced lysosome membrane permeabilization. Consistent with the reduced lysosome membrane permeabilization, shRNA knockdown of PACS-2 in Huh-7 cells reduced TRAIL-induced apoptosis and increased clonogenic cell survival. The determination that recombinant PACS-2 bound Bim but not Bax in vitro and that shRNA knockdown of Bim blocked Bax recruitment to lysosomes suggests that TRAIL/DR5 triggers endosomal PACS-2 to recruit Bim and Bax to lysosomes to release cathepsin B and induce apoptosis. Together, these findings provide insight into the lysosomal pathway of apoptosis.
Introduction
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)2 TNFSF10, also referred to as Apo-2 ligand, is a member of the tumor necrosis factor superfamily (1). TRAIL is expressed by immune effector cells (2) and induces target cell apoptosis, especially of malignant cells (1). TRAIL can also exacerbate disease processes such as cholestatic liver injury (3, 4). TRAIL initiates proapoptotic signaling cascades by binding two cognate death receptors termed DR4 (death receptor 4, TNFRSF10A, also referred to as TRAIL receptor 1), and DR5 (death receptor 5, TNFRSF10B, also referred to as TRAIL receptor 2) (1). Following TRAIL-induced receptor aggregation, procaspases 8 and 10 are recruited to the death-inducing signaling complex (5), where they are activated. Cellular demise then occurs by direct activation of downstream executioner caspases or by triggering the mitochondrial pathway of cell death (6).
In malignant hepatobiliary cells and diseased hepatocytes, as well as in other cell types, lysosomal membrane permeabilization (LMP) occurs upstream of mitochondrial dysfunction and is required for efficient cell death (7–13). Indeed, TRAIL-induced release of cathepsin B into the cytosol occurs following bile duct ligation (3), and genetic deletion of cathepsin B reduces liver injury in this model (14). Determining the mechanism that mediates LMP is therefore key to understanding the molecular basis of TRAIL-induced liver injury and development of TRAIL agonists for treatment of hepatobiliary malignancies.
TRAIL-induced LMP in hepatobiliary cells requires activated Bax, which likely forms proteolipid pores in lysosomal membranes that release lumenal proteases into the cytosol to trigger cell death (11, 15). Bax activation requires proapoptotic BH3-domain-only proteins such as Bim and Bid, which act in part by binding and interfering with antiapoptotic Bcl-2 family members and also by directly interacting with Bax (16). Indeed, recent studies suggest that the Bim BH3 domain binds Bax to trigger Bax oligomerization and pore formation (17, 18). In healthy cells, Bim is sequestered on microtubules, whereas inactive, monomeric Bax is cytosolic (19, 20). Despite the important role of Bim- and Bax-mediated LMP in TRAIL-induced apoptosis, little is known about the mechanism that controls their translocation to lysosomes.
The phosphofurin acidic cluster sorting (PACS) proteins are multifunctional homeostatic regulators that integrate secretory pathway traffic and interorganellar communication in healthy cells with key steps in death ligand-induced apoptosis in diseased cells (21, 22). PACS-1 and PACS-2 mediate trafficking of client proteins to the endoplasmic reticulum, the trans-Golgi network, between endosomal compartments, and to the primary cilium (22–34). In response to TRAIL, PACS-2 switches from a secretory pathway sorting protein to a proapoptotic effector that mediates the translocation of Bid to mitochondria to promote mitochondria membrane permeabilization (21). Consistent with these studies, PACS-2 is essential for TRAIL-mediated apoptosis of adenovirus-infected hepatocytes in vivo (21).
The determination that death ligands induce PACS-2 to interact with Bid and mediate mitochondrial outer membrane permeabilization together with its role in endosomal trafficking in healthy cells raised the possibility that TRAIL may also redirect PACS-2 endosomal trafficking to trigger LMP (35, 36). Here we report that TRAIL induces formation of a lysosome-associated complex containing PACS-2, Bim, and Bax, which we have termed the PIXosome. This complex is required for LMP, cathepsin B release, and cell death, providing further insight into the mechanisms of LMP and TRAIL-induced apoptosis.
MATERIALS AND METHODS
Cell Lines and Stable Clones
KMCH cholangiocarcinoma cells and Huh-7 hepatocarcinoma cells were cultured in Dulbecco's modified Eagle's medium containing glucose (25 mm), 100,000 units/liter penicillin, 100 mg/liter streptomycin, and 10% fetal bovine serum. Huh-7 cells were transfected with an siRNA targeting Bim (5′- AACCTCCTTGCATAGTAAGCG) using siPORTtm NeoFXtm transfection reagent (Ambion Inc., Austin, TX) and with a shRNA lentiviral plasmid targeting Bax (5′-AAGACGAACTGGACAGTAACACCTGTCTC) from Open Biosystems (Huntsville, AL). Cells were transfected using FuGENE HD (Roche). To generate clones stably expressing shRNA against PACS-1 or PACS-2, Huh-7 cells were transfected with 1 μg/ml DNA plasmid (PACS-1 shRNA lentiviral plasmid; Open Biosystems, Huntsville, AL; targeting sequence 5′- GTTTCAGATGAGGTGGGCTTT and PACS-2 MISSION shRNA lentiviral plasmid; Sigma Aldrich, St. Louis, MO; targeting sequence 5′-GATTGTAAGAACGACGTCCAT) using Lipofectamine 2000 (Invitrogen). Stably transfected clones were selected in medium containing 2 μg/ml puromycin and screened by PACS-1 or PACS-2 immunoblot analysis. To generate clones stably expressing HA-tagged PACS-1 or PACS-2, Huh-7 cells were transfected with 1 μg/ml HA-PACS-1 or HA-PACS-2 plasmid, which have been described previously (21). Stably transfected clones were selected in medium containing 2 μg/ml puromycin and screened by HA immunoblot analysis. Stably transfected S-peptide tagged Mcl-1 Huh-7 cells were prepared as described previously for KMCH cells (37).
Bax and Cathepsin B Immunofluorescence
For cathepsin B immunofluorescence, cells were grown on glass coverslips and fixed in PBS containing 4% paraformaldehyde and 0.19% picric acid. For Bax and cytochrome c immunofluorescence, cells were fixed in PBS containing 4% paraformaldehyde, 0.1 m PIPES, 1 mm EGTA, and 3 mm MgSO4. Cells were permeabilized with 0.0125% (w/v) CHAPS in PBS at 37 °C for 10 min and blocked for 1 h at room temperature with PBS containing 5% bovine serum albumin, 5% glycerol, and 0.04% sodium azide. After incubation with mouse monoclonal anti-Bax antibody (6A7) and goat polyclonal anti-cathepsin B antibody (C-19, Santa Cruz Biotechnologies, Santa Cruz, CA) at 1:500 in blocking buffer at 4 °C overnight, cells were washed three times with PBS and incubated with Alexa Fluor 488-conjugated donkey anti-goat IgG or goat anti-mouse (Molecular Probes, Eugene, OR) at a dilution of 1:500 in blocking buffer for 1 h at 37 °C. Cells were then washed three times in PBS and three times in water and mounted onto slides using the ProLong antifade kit (Molecular Probes). Cells were imaged by confocal microscopy with excitation and emission wavelengths of 488 nm and 507 nm and counted for their punctate versus diffuse fluorescence.
Quantitation of Apoptosis
Recombinant human TRAIL was obtained from R&D Systems (Minneapolis, MN). Staurosporine and 3-methyladenine were obtained from Sigma-Aldrich. Apoptosis was quantified by assessing the characteristic nuclear changes of apoptosis (i.e. chromatin condensation and nuclear fragmentation) after staining with DAPI and by fluorescence microscopy using excitation and emission wavelengths of 380 and 430 nm, respectively. Caspase-3/7 activity in cell cultures was assessed by measuring rhodamine release from the caspase-3/7 substrate rhodamine 110, bis(N-CBZ-L-aspartyl-L-glutamyl-L-valyl-L-aspartic acid amide) (Z-DEVD-R110), using the Apo-ONEtm homogeneous caspase-3/7 kit (Promega, Madison, WI) following the instructions of the supplier.
Multiparameter Fluorescent Confocal Microscopy
Huh-7 cells were loaded with spectrally resolved probes for 30 min at 37 °C to determine the kinetic relationship between LMP, mitochondrial membrane depolarization, and breakdown of the plasma membrane (release of a cytosolic dye). The fluorescent intensity and compartmentalization of the dyes were measured in the same cells sequentially over time. Lysosomal integrity was measured with 500 nm LysoSensor Green DND-189 (Molecular Probes, λex = 443 nm and λem = 505 nm). Mitochondrial membrane potential was measured with 50 nm tetramethylrhodamine ethyl ester perchlorate (TMRE, Sigma Aldrich, λex = 520 nm and λem = 580 nm). Plasma membrane breakdown was measured with 1 μm fura-2 AM (Molecular Probes, λex = 360 nm and λem = 480 nm). Total cellular fluorescence was quantified using Image J (National Institutes of Health) by measuring the mean. Integrated density and mean gray value fluorescent density per cellular area minus background fluorescence.
Clonogenic Assay
Cells were seeded at 100 cells per 35-mm dish in 1 ml of medium. 24 h later the cells were treated with TRAIL as indicated in the figure legend. After removal of TRAIL, cells were then allowed to grow until isolated colonies were visible (∼7 days). The colonies were then washed once with PBS, stained with Coomassie Blue, and counted.
Colocalization of PACS-1-EGFP, PACS-2-EGFP, or Bax with Lysosomes
The PACS-1 and PACS-2 EGFP vectors were described previously (21). The vectors were transiently transfected into Huh-7 cells using FuGENE HD (Roche). For Bax immunofluorescence, PACS-1-EGFP- or PACS-2-EGFP-expressing Huh-7 cells were grown on glass coverslips, treated with TRAIL, and loaded with 100 nm LysoTracker Red (Molecular Probes) for 15 min at 37 °C and then fixed with 4% paraformaldehyde in PBS containing 0.1 m PIPES, 1 mm EGTA, and 3 mm MgSO4. Cells were permeabilized with 0.0125% (w/v) CHAPS in PBS at 37 °C for 10 min and blocked for 1 h at room temperature in PBS containing 5% bovine serum albumin, 5% glycerol, and 0.04% sodium azide. After incubation with mouse monoclonal anti-Bax (6A7) antibody, which only recognizes the active conformation of Bax, at a concentration of 1:100 in blocking buffer at 4 °C overnight, cells were washed three times with PBS and incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG (Molecular Probes) at a dilution of 1:500 in blocking buffer for 1 h at 37 °C. Cells were then washed three times in PBS and three times in water, mounted onto slides using the ProLong antifade kit (Molecular Probes), and imaged by confocal microscopy with excitation and emission wavelengths of 488 nm and 507 nm (Alexa Fluor 488) and 577 nm and 592 nm (LysoTracker Red), respectively. Quantitation of colocalization was accomplished using the LSM 510 imaging software as described previously by us (11). Briefly, random lines one-pixel wide were drawn through the cells, and fluorescence intensity for both the green and red channels was quantified. A ratio of the green to red fluorescence was calculated, yielding a percent pixel overlap. The data are expressed as percent pixel overlap for each experimental condition.
Cell Fractionation and Immunoblot Analysis
Lysosomes were isolated from cell cultures using a commercially available lysosome isolation kit (Sigma Aldrich). Whole cell lysates were obtained by incubating cells for 30 min on ice with lysis buffer (50 mm Tris-HCl (pH 7.4); 1% Nonidet P-40; 0.25% sodium deoxycholate; 150 mm NaCl; 1 mm EDTA; 1 mm PMSF; 1 μg/ml aprotinin, leupeptin, and pepstatin; 1 mm Na3VO4; 1 mm NaF) followed by centrifugation at 14,000 × g for 15 min at 4 °C. Samples were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and blotted with the indicated primary antibodies at a dilution of 1:1000. Horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnologies) were incubated at a dilution of 1:5000. Bound antibodies were visualized using ECL (Amersham Biosciences, Arlington Heights, IL) and Kodak X-OMAT film. Primary antibodies were as follows: mouse monoclonal anti-lysosomal-associated membrane protein 1 (LAMP1) (BD Biosciences); rabbit polyclonal anti-DR5 (ProSci, Poway, CA); rat monoclonal anti-HA (Roche); rabbit polyclonal anti-PACS-2 (Sigma-Aldrich); rabbit polyclonal anti-PACS-2 (31); rabbit polyclonal anti-PACS-1 (Abcam, Cambridge, MA); mouse monoclonal anti-activated BAX (6A7), rabbit polyclonal anti-Bax (N-20), goat polyclonal anti-actin (C-11), and goat polyclonal anti-cytochrome c oxidase II (K-20) (Santa Cruz Biotechnologies); and rabbit polyclonal anti-phospho-Ser-69 Bim (Cell Signaling Technology, Danvers, MA).
DR5 Internalization
DR5 internalization was performed as described previously by us in detail (38). Briefly, Huh-7 wild-type, Huh-7 shPACS-1, or Huh-7 shPACS-2 cells were incubated for 30 min on ice in the presence of preoligomerized FLAG-tagged TRAIL (Alexis, Plymouth Meeting, PA). The cells were then incubated at 37 °C for selected time intervals, acid washed, and fixed. Cells were next incubated with Alexa Fluor 488-conjugated anti-mouse IgG at a concentration of 1:1000 in blocking buffer for 1 h at 37 °C and imaged using a Zeiss LSM 510 confocal microscope. Cells were scored as either positive or negative for acid wash resistant fluorescence.
Immunoprecipitation and Pull-down Assays
For HA-tagged PACS protein immunoprecipitations, Huh-7 cells stably transfected with either HA-PACS-1 or HA-PACS-2 were lysed on ice for 30 min using the lysis buffer described above. Protein concentrations in the whole cell lysates were measured using the Bradford reagent (Sigma-Aldrich). Aliquots of 200 μg of protein were incubated with 40 μl of HA-tagged affinity beads (Sigma-Aldrich) at 4 °C overnight with gentle rotation. For Bax immunoprecipitation, Huh-7 cell lysates were collected as indicated above, and aliquots of 100 μg of protein were incubated with mouse monoclonal anti-Bax (6A7) at 1:100 at 4 °C overnight with rotation. 40 μl of protein A-agarose beads (Millipore, Billerica, MA) were added to the protein-antibody complex and incubated for 2 h at room temperature. For FLAG-TRAIL immunoprecipitation, Huh-7 cells were grown to ∼80% confluence and then placed on ice. The medium was removed and replaced with medium containing FLAG-tagged TRAIL (400 ng/ml), and cells were rapidly placed at 37 °C for the desired time intervals. The cells were then lysed on ice using lysis buffer, and aliquots of 100 μg of protein were incubated with 40 μl of anti-FLAG M2-agarose beads (Sigma-Aldrich) at room temperature for 2 h. For all experiments, the samples were centrifuged at 13,000 × g for 15 min, and the supernatant was removed and saved for analysis. The beads were washed three times with lysis buffer without detergent, and the bound proteins were eluted by adding 30 μl of 2× sample buffer and boiling for 10 min. Samples were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblot analysis was performed as described above. S-peptide-tagged Mcl-1 was affinity-purified from whole cell lysates as described previously in detail (37).
Protein Binding Assays
Plasmids expressing GST, GST-BimEL, GST-Bax (obtained from Hong Tang, Chinese Academy of Science, Beijing), and His6-PACS-2 FBR38–217 (32) were transformed in BL21 Escherichia coli, and cultures were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside (Calbiochem, La Jolla, CA) for 4 h at 37 °C. Bacterial pellets were resuspended in lysis buffer (50 mm Tris (pH 7.6), 1.5 mm EDTA, 100 mm NaCl, 0.5% Triton X-100, 0.1 mm DTT, 10 mm MgCl2) containing protease inhibitors (0.5 mm PMSF and 0.1 μm each of aprotinin, E-64, and leupeptin) and lysed using a French press. Soluble material was collected after a 1-h 25,000 × g spin and subsequently bound to glutathione-Sepharose 4B (GE Healthcare) of nickel-nitrilotriacetic acid (Qiagen, Valencia, CA) for 1 h at 4 °C. For the His6-PACS-2 FBR38–217 interaction with GST-BimEL and GST-Bax, proteins were mixed at a 1:1 ratio for 30 min at 4 °C in binding buffer (20 mm Tris (pH 7.9) containing 150 mm NaCl, 0.1 mm EDTA, and 0.1% Nonidet P-40). After incubation, glutathione-Sepharose 4B was added to the reaction and incubated for an additional 30 min at 4 °C. The resin was subsequently washed three times in binding buffer and resuspended in SDS-PAGE sample buffer.
Statistical Analysis
All data represent at least three independent experiments and are expressed as mean ± S.E. Differences between groups were compared using two-tailed Student's t-tests and one-way analysis of variance with post hoc Dunnett test, and significance was accepted at p < 0.05.
RESULTS
PACS-2 Is Required for TRAIL-induced Lysosome Membrane Permeabilization and Apoptosis in Hepatoma Cells
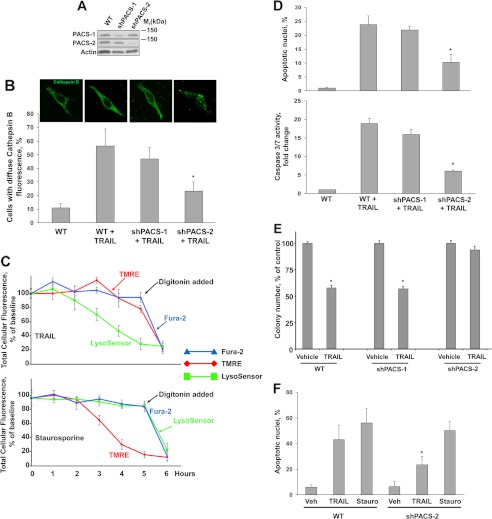
To determine whether one or both PACS proteins were required for TRAIL-induced LMP and apoptosis, we first generated Huh-7 cells stably knocked down for expression of PACS-1 or PACS-2 using shRNA vectors (Fig. 1A). Parallel plates of shPACS-1, shPACS-2 cells, or control Huh-7 cells were treated with TRAIL, and the subcellular distribution of cathepsin B was assessed by immunofluorescence (Fig. 1B). We found that TRAIL induced a marked redistribution of cathepsin B from a punctate lysosomal pattern to a diffuse cytosolic pattern in shPACS-1 and control Huh-7 cells. In contrast, TRAIL failed to induce a redistribution of cathepsin B in shPACS-2 cells.
FIGURE 1.
Knockdown of PACS-2 but not PACS-1 prevents TRAIL-induced cathepsin B release and apoptosis in Huh-7 cells. A, immunoblot analysis showing the effectiveness of shRNA knockdown of PACS-1 or PACS-2 in stably transfected Huh-7 cells. Actin was used as loading control. B, WT and shPACS-1 or shPACS-2 Huh-7 cells were treated with TRAIL (10 ng/ml) for 3 h and then analyzed by immunofluorescence for cathepsin B. In blinded experiments, cells were imaged by confocal microscopy (top panel) and scored for punctate or diffuse appearance of the antigen fluorescence. The results are expressed as percent of cells with diffuse fluorescence over total number of cells per field (bottom panel). C, Wild-type, shPACS-1, or shPACS-2 Huh-7 cells were treated with TRAIL (10 ng/ml) for 3 h and then analyzed by immunofluorescence for cytochrome c (top panel). Release of cytochrome c was quantified as described in B (bottom panel). TMRE, tetramethylrhodamine ethyl ester perchlorate. D, wild-type and shPACS-1 or shPACS-2 Huh-7 cells were treated with TRAIL (10 ng/ml) for 4 h. Apoptosis was quantified using DAPI and fluorescence microscopy (top panel) and by measuring caspase-3/7 activity using a fluorogenic assay (bottom panel). The results are expressed as fold increase of substrate cleavage over the control value, which was arbitrarily set to 1. *, p ≤ 0.01. E, wild-type and shPACS-1 or shPACS-2 Huh-7 cells were treated with TRAIL (10 ng/ml) for 6 h, washed, and cultured for 7 days, and then surviving colonies were detected as described under “Experimental Procedures.” *, p ≤ 0.01. F, wild-type and shPACS-1 or shPACS-2 Huh7 cells were treated with TRAIL (10 ng/ml) or staurosporine (1 μg/ml) for 3 h. Apoptosis was assessed after DAPI staining. *, p ≤ 0.01.
Lysosome membrane permeabilization can occur upstream or downstream of mitochondrial dysfunction (39, 40). If LMP occurs downstream of mitochondrial dysfunction, it likely would be an amplification event in cell death. Under these conditions, inhibiting LMP would delay, but not prevent, cellular demise (41). In contrast, if LMP is upstream of mitochondrial dysfunction, its inhibition should prevent mitochondrial outer membrane permeabilization and cell death. To distinguish between these possibilities, Huh-7 cells were preloaded with fluorescent markers of LMP (LysoSensor), mitochondrial membrane potential (tetramethylrhodamine ethyl ester), and plasma membrane breakdown (Fura-2). The preloaded cells were then treated with TRAIL or vehicle alone, and the subsequent change in fluorescent signal for each probe was monitored as an indication of compartment breakdown (Fig. 1C). We found that treatment of Huh-7 cells with TRAIL induced LMP within 3 h independently of mitochondrial depolarization. The preloaded tetramethylrhodamine ethyl ester and Fura-2 could be released with digitonin, demonstrating that the two probes were localized to releasable membrane compartments. In contrast, treatment of Huh-7 cells with the pan-kinase inhibitor staurosporine, which can directly trigger mitochondrial dysfunction, induced mitochondrial depolarization independently of LMP. Correspondingly, apoptosis was reduced significantly following TRAIL treatment of shPACS-2 cells but not shPACS-1 cells or control Huh-7 cells (Fig. 1D). The resistance of shPACS-2 cells to TRAIL-induced apoptosis suggested that long-term survival of these cells in response to TRAIL would be increased compared with control cells. To test this, we conducted a clonogenic survival assay. The results of the clonogenic assay showed that shRNA knockdown of PACS-2, but not PACS-1, increased survival of Huh-7 cells in response to TRAIL (Fig. 1E). To determine whether PACS-2 was selectively required for the lysosomal pathway of cell death in hepatoma cells, we treated shPACS-2 or wild-type Huh-7 cells with staurosporine, which induces cell death independently of LMP. We found that control Huh-7 and shPACS-2 cells were equally sensitive to staurosporine-induced apoptosis (Fig. 1F). Together, these experiments suggested that TRAIL requires PACS-2 to trigger lysosome membrane permeabilization and cell death of Huh-7 cells.
TRAIL Induces Formation of a PACS-2, Bim, and Bax Multiprotein Complex on Lysosomes
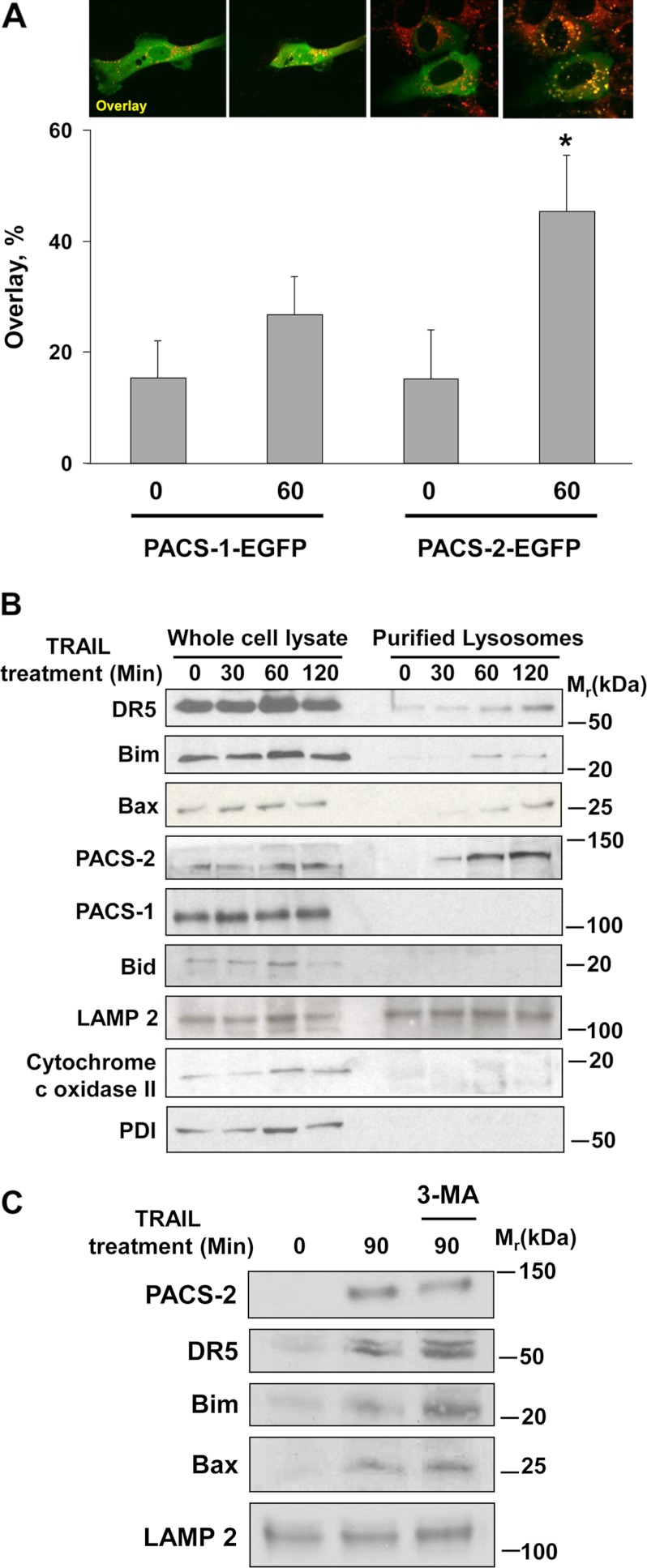
To determine how PACS-2 mediates TRAIL-induced LMP, we first asked whether TRAIL stimulation could promote PACS-2 trafficking to lysosomes. As a control, we monitored trafficking of PACS-1, which is not required for death ligand-induced apoptosis (Fig. 1D and Ref. 36). Huh-7 cells expressing PACS-1-EGFP or PACS-2-EGFP were treated with TRAIL and imaged by confocal microscopy. In agreement with the role of PACS-2 in TRAIL-induced apoptosis, we observed that TRAIL stimulated translocation of PACS-2-EGFP, but not PACS-1-EGFP, to LysoTracker Red-positive lysosomal compartments (Fig. 2A).
FIGURE 2.
TRAIL induces PACS-2 to interact with DR5, Bim, and Bax on lysosomes. A, Huh-7 cells were transfected with either PACS-1-EGFP or PACS-2-EGFP and loaded with LysoTracker Red. The cells were imaged at 0 and 60 min after TRAIL (10 ng/ml) treatment (top panel). The percent of green fluorescence (EGFP-tagged PACS-1 or PACS-2) colocalizing with red fluorescence (LysoTracker Red) after TRAIL treatment in each experimental condition was quantified (bottom panel). *, p ≤ 0.01. B, Huh-7 cells were treated with TRAIL for the indicated times. Cell lysates and purified lysosomal fractions were obtained and analyzed by immunoblot analysis. LAMP 2, lysosomal marker; cytochrome c oxidase II, mitochondria marker; PDI, protein disulfide isomerase, endoplasmic reticulum marker. C, Huh-7 cells were treated with TRAIL (10 ng/ml) for 90 min in the presence or absence of 3-methyladenine (3-MA) (5 mm). Lysosomes were isolated and analyzed by immunoblot analysis for the indicated proteins.
To determine whether the translocation of PACS-2-EGFP to lysosomes reflected the action of endogenous PACS-2, an independent biochemical assay was conducted. Replicate plates of Huh-7 cells were treated with or without TRAIL. At increasing times following TRAIL addition, cells were harvested, and lysosomes were isolated by subcellular fractionation. Immunoblot analysis showed that TRAIL induced a time-dependent recruitment of PACS-2, but not PACS-1, to lysosomes (Fig. 2B). Interestingly, the lysosomal translocation of endogenous PACS-2 was coupled temporally with the recruitment of DR5, Bax, and Bim to the lysosomal fraction. In contrast, Bid was not translocated to lysosomes, consistent with our prior observations and the role of Bid action in mitochondria membrane permeabilization (Fig. 2B and Refs. 11, 42). To determine whether the TRAIL- and PACS-2-dependent recruitment of DR5, Bim, and Bax to lysosomes reflected an apoptotic or autophagic pathway, the experiment was repeated in the presence of the autophagy inhibitor 3-methyladenine (Fig. 2C). We found that 3-methyladenine failed to block TRAIL-induced recruitment of PACS-2, DR5, Bim, or Bax to lysosomes. Together, these results suggest that TRAIL-induced apoptotic signaling triggers recruitment of PACS-2 and proapoptotic effectors to lysosomes.
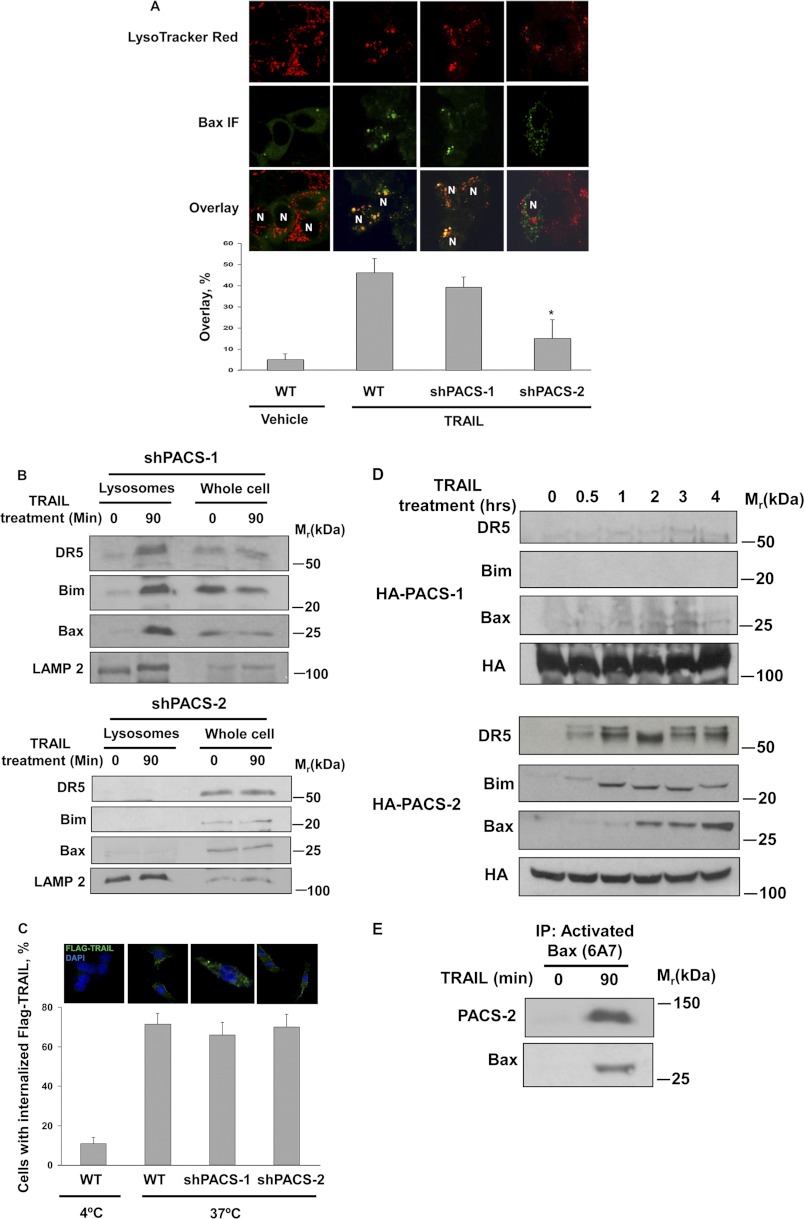
The temporally coupled recruitment of PACS-2, DR5, Bim, and Bax to lysosomes led us to ask whether PACS-2 was required for their translocation. By confocal microscopy we found that TRAIL induced the recruitment of Bax to LysoTracker-positive compartments in control and shPACS-1 cells but not in shPACS-2 cells (Fig. 3A). In agreement with the image analysis, immunoblotting showed that TRAIL induced recruitment of DR5, Bax, and Bim to lysosomes in Huh-7 wild-type and PACS-1 shRNA cells but not in PACS-2 shRNA cells (Figs. 2B and 3B). To determine whether the failure of DR5 to traffic to lysosomes in shPACS-2 cells resulted from a block in TRAIL receptor endocytosis, we monitored the internalization of the FLAG-TRAIL-bound receptor and found that knockdown of PACS-2 had no measurable effect on FLAG-TRAIL:DR5 internalization (Fig. 3C).
FIGURE 3.
TRAIL-induced Bim, Bax, and DR5 translocation to lysosomes is PACS-2 dependent. A, wild-type, shPACS-1, or shPACS-2 Huh-7 cells were treated with TRAIL (10 ng/ml) for 60 min and then analyzed by confocal microscopy for activated Bax (green) and LysoTracker Red. The extent of colocalization of activated Bax and LysoTracker Red in each sample was then quantified (bottom panel). *, p ≤ 0.01. B, shPACS-1 (top panel) or shPACS-2 (bottom panel) cells were treated with TRAIL (10 ng/ml) for 90 min. Lysosomes were isolated and analyzed for the indicated proteins by immunoblot analysis. LAMP 2, lysosomal marker. C, DR5 internalization assay was performed in wild-type, shPACS-1, and shPACS-2 Huh-7 cells as described under “Experimental Procedures.” Top panel, representative confocal microscopy images of internalized FLAG-TRAIL under each condition. Bottom panel, quantification of FLAG-TRAIL-positive staining under each experimental condition. D, Huh-7 HA-PACS-1 or HA-PACS-2 cells lines were treated with TRAIL (10 ng/ml) for the indicated times. HA-PACS-1 and HA-PACS-2 were immunoprecipitated, and associated proteins were identified by immunoblot analysis. E, lysates from Huh-7 cells and Huh-7 cells treated with TRAIL (10 ng/ml) for 90 min cells were used for immunoprecipitation (IP) with the Bax antibody 6A7, which recognizes activated Bax. The immune complexes were then separated by SDS-PAGE electrophoresis and subjected to immunoblot analysis for Bax and associated PACS-2.
The PACS-2-dependent recruitment of apoptotic effectors to lysosomes following TRAIL treatment raised the possibility that this death ligand may trigger PACS-2 to interact with DR5, Bax, or Bim. To test this hypothesis, Huh-7 cells stably transfected with HA-tagged PACS-1 or PACS-2 were treated with TRAIL. At increasing time intervals, PACS-1 or PACS-2 were immunoprecipitated from whole cell lysates, and coprecipitating proteins were detected by immunoblot analysis. We found that TRAIL induced a time-dependent association of DR5, Bim, and Bax with HA-tagged PACS-2 but not PACS-1 (Fig. 3D). In a reciprocal experiment, we treated Huh-7 cells with TRAIL and immunoprecipitated activated Bax using the conformation-specific monoclonal antibody 6A7. Immunoblot analysis showed that TRAIL induced the interaction of activated Bax with PACS-2 (Fig. 3E). Together, these results suggest that PACS-2 is required for mediating endosomal trafficking steps necessary for the TRAIL-induced recruitment of Bax, Bim, and endocytosed DR5 to lysosomes.
PACS-2 Recruitment to Lysosomes Is Bim- and JNK-dependent
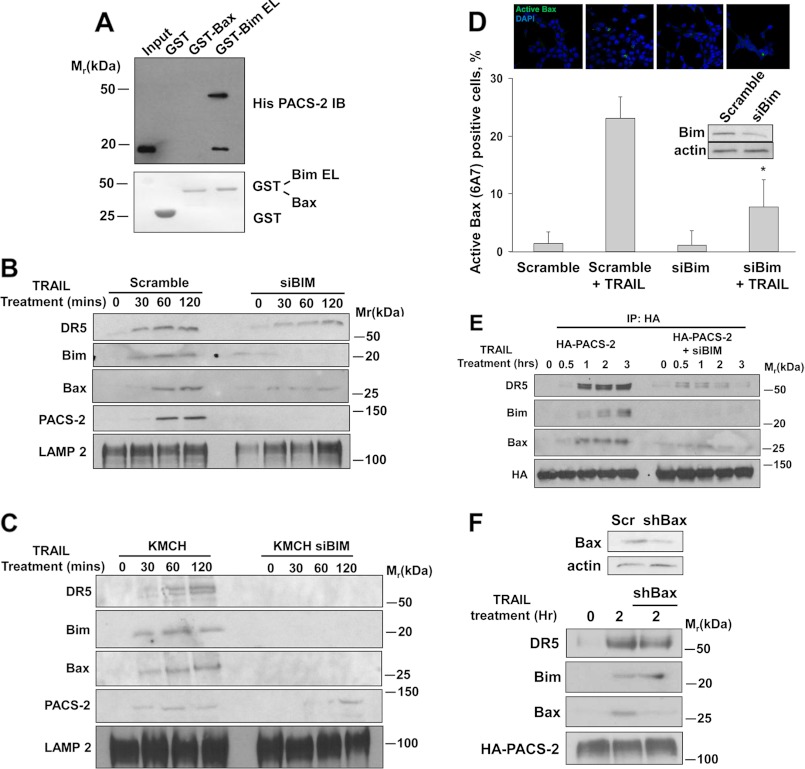
The determination that TRAIL triggers PACS-2 to form an immunoprecipitable complex with Bim and Bax led us to ask whether PACS-2 can bind one or both proapoptotic proteins in vitro. To test this possibility, His6-tagged PACS-238–217, which contains the cargo-binding region (furin binding region), was mixed with GST-tagged Bax, BimEL, or GST alone. Protein complexes were captured with Ni2+-agarose and analyzed by immunoblot analysis. We found that PACS-238–217 bound Bim but not Bax (Fig. 4A). Because Bim binds both PACS-2 and Bax but PACS-2 binds only Bim, these findings suggest that binding of PACS-2 to Bim on lysosome membranes promotes recruitment of Bax.
FIGURE 4.
Bim binds PACS-2 and is required for recruitment of PACS-2 and activated Bax to lysosomes. A, His6-PACS-2FBR38–217 was mixed with GST-Bax, GST-BimEL, or GST alone. GST proteins were captured, and bound His6-PACS-2FBR38–217 was detected by immunoblot analysis. B, Huh-7 cells transfected with scramble siRNA or with a Bim siRNA were treated with TRAIL (10 ng/ml) for the indicated times. Lysosomal fractions were obtained and analyzed by immunoblot analysis. LAMP 2, lysosomal marker. C, KMCH cells and KMCH cells transfected with a Bim siRNA were treated with TRAIL (10 ng/ml) for the indicated times. Lysosomal fractions were obtained and analyzed by immunoblot analysis. D, Huh-7 cells transfected with scramble siRNA or with a Bim siRNA (siBIM) were treated with TRAIL (10 ng/ml) for 3 h, fixed, and analyzed by immunofluorescence for activated Bax (green) and nuclei (DAPI, blue). Bottom panel, quantification of the percent of cells containing activated Bax expressed as percent of total number of cells per field. Inset, efficiency of Bim knockdown was verified by immunoblot analysis. E, Huh-7 HA-PACS-2 transfected with scramble siRNA or with a Bim siRNA were treated with TRAIL for the indicated times. Cell lysates were collected, HA-PACS-2 was immunoprecipitated (IP) from the cell lysates, and associated proteins were identified by immunoblot analysis. F, scramble-transfected HA-PACS-2 Huh-7 cells or HA-PACS-2 cells transfected with a Bax shRNA were treated with TRAIL (10 ng/ml) for 2 h. HA-PACS-2 was immunoprecipitated from the cell lysates, and interacting proteins were detected by immunoblot analysis. Top panel, Bax expression in scramble and Bax shRNA knockdown cells.
Although PACS-2 was required for recruitment of Bim to lysosomes (Fig. 3B), this finding did not preclude the possibility that, conversely, Bim may be required for the TRAIL-induced trafficking of PACS-2 to lysosomes. To test this possibility, we used Bim-specific siRNAs to knock down Bim expression in Huh-7 cells. Next, Bim knockdown or control Huh-7 cells were treated with TRAIL, and the recruitment of PACS-2 and Bax to lysosomes was monitored by immunoblot analysis. We found that siRNA knockdown of Bim blunted the TRAIL-induced trafficking of PACS-2 in Huh-7 cells (Fig. 4B). TRAIL-induced trafficking of PACS-2 was also inhibited in KMCH cells with siRNA knockdown of Bim (Fig. 4C), suggesting that the TRAIL-induced trafficking of PACS-2 to lysosomes occurs in multiple hepatobiliary cancer cell lines. Knockdown of Bim also reduced Bax trafficking to lysosomes, as well as TRAIL-induced Bax activation (Fig. 4, B–D). Likewise, Bim knockdown also reduced the interaction of HA-tagged PACS-2 with Bax as determined by coimmunoprecipitation (Fig. 4E). In contrast, shRNA knockdown of Bax failed to prevent the TRAIL-induced interaction of HA-tagged PACS-2 with DR5 or Bim (Fig. 4F). Collectively, these data suggest that Bim binding to PACS-2 is pivotal for Bax recruitment to this protein complex.
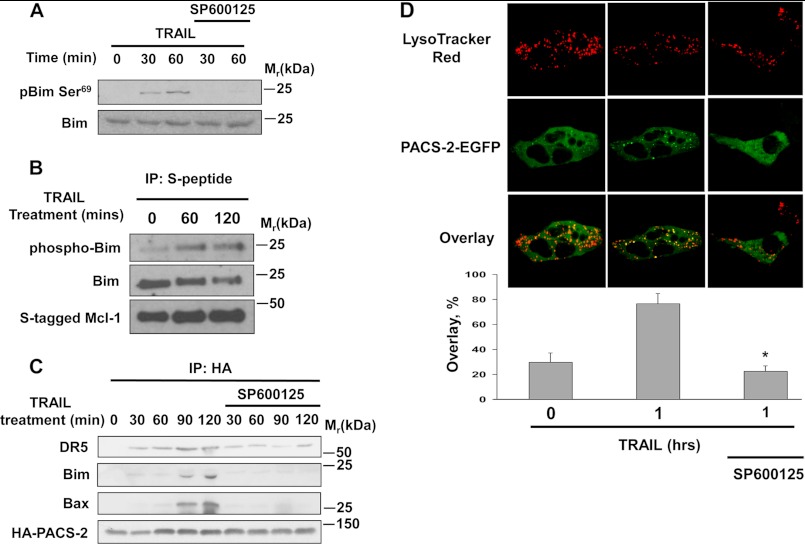
TRAIL-induced lysosomal membrane permeabilization requires JNK activation (11). We therefore asked whether JNK activity was required for the interaction of PACS-2 with Bim. To test this possibility, Huh-7 cells stably expressing HA-PACS-2 were treated with TRAIL in the presence or absence of the JNK inhibitor SP600125, which blocked TRAIL-induced phosphorylation of Bim Ser69, required for release of Bim from the cytoskeleton (Fig. 5A). In a TRAIL-dependent manner, the phosphorylated Bim was found complexed to S-peptide tagged Mcl-1 affinity purified from cell lysates. Phospho Bim, therefore, is capable of binding to Mcl-1, demonstrating its ability to bind to Bcl-2 proteins as a phosphoprotein (Fig. 5B). Correspondingly, co-immunoprecipitation analysis showed that JNK inhibition prevented the TRAIL-induced interaction of HA-PACS-2 with Bim, Bax and DR5 (Fig. 5C). Similarly, immunofluorescence analysis showed SP600125 reduced PACS-2-GFP co-localization with LysoTracker-positive vesicles (Fig. 5D). Together, these data suggest TRAIL stimulates Bim activation via a JNK-mediated pathway, which promotes recruitment of PACS-2 and Bax to form a complex on lysosomes.
FIGURE 5.
JNK activity is required for formation and lysosomal trafficking of PACS-2, DR5, Bim, and Bax. A, Huh-7 cells were treated with TRAIL (10 ng/ml) in the absence or presence of SP600125 (20 μm). Cells were lysed, and phospho-Ser-69 Bim and total Bim were detected by immunoblot analysis. B, Huh-7 cells stably transfected with S-peptide tagged Mcl-1 were treated with TRAIL (10 ng/ml). At the indicated time intervals, S-peptide tagged Mcl-1 was affinity-purified from whole cell lysates, and the associated Bim and phospho Bim were identified by immunoblot analysis. IP, immunoprecipitation. C, Huh-7 cells stably transfected with HA-tagged PACS-2 were treated with TRAIL (10 ng/ml) in the absence or presence of SP600125 (20 μm). At the indicated times, HA-PACS-2 was immunoprecipitated from the cell lysates, and associated proteins were identified by immunoblot analysis. D, Huh-7 cells were transiently transfected with PACS-2-EGFP, pretreated or not pretreated with SP600125 for 1 h, and then treated with TRAIL 10 ng/ml for 1 h. Cells were loaded with LysoTracker Red and imaged by confocal microscopy (top panel). Bottom panel, percent of green fluorescence (PACS-2-EGFP) colocalizing with red fluorescence (LysoTracker Red) was quantified in each experimental condition.
DISCUSSION
Apoptotic pathways are manifested by assembly of protein scaffolds that orchestrate the compartment-specific activation of caspases or Bcl-2 family members. Scaffolds that drive caspase activation have been studied most intensively. For example, the death-inducing signaling complex modulates death ligand-induced activation of caspases 8 or 10 (6), the multimeric apoptotic protease-activating factor-1 (APAF-1) binds cytosolic cytochrome c to activate procaspase 9 (43), the p53-inducible death domain (PIDDosome) regulates caspase 2 activation (44), and the RIP-1-containing ripoptosome modulates caspase 8 activation independently of death ligands (45). Here we identify the PACS-2, Bim, and Bax-containing PIXosome as a multimeric complex required for the compartment-specific activation of Bax. To our knowledge, the PIXosome represents the first identified organellar apoptotic Bcl-2 family scaffold.
Several observations support an important role for the PIXosome in mediating TRAIL-induced apoptosis. PIXosome assembly was TRAIL-dependent and required for apoptosis. Consistent with this finding, PACS-2 knockdown increased clonogenic cell survival in TRAIL- treated Huh-7 cells. The PIXosome was found specifically on lysosomes and required for cathepsin B release, which led to cytochrome c release from mitochondria. The absence of detectable Bid, caspase-8, or RIP-1 (data not shown) suggests that the PIXosome represents a novel lysosome-specific proapoptotic Bcl-2 protein complex that is distinct from mitochondria-associated Bcl-2 complexes or from previously described caspase-activating platforms. The requirement for the PIXosome in mediating TRAIL-induced apoptosis of Huh-7 and KMCH cells supports an important role for this complex in multiple hepatobiliary cell lines.
TRAIL requires Bim and Bax to trigger LMP, but how they are targeted to lysosomes has been unknown (11). Our results suggest that PACS-2 is key. Indeed, genetic studies demonstrated that PACS-2 mediates TRAIL-induced apoptosis in vivo (21). In healthy cells, PACS-2 directs multiple secretory pathway trafficking steps, including the trafficking of endocytosed receptors from early endosomes to the trans-Golgi network (22). TRAIL, however, induces a marked redistribution of PACS-2 to both lysosomes, where it interacts with Bim and Bax, as well as to mitochondria, where it mediates Bid action (Fig. 1C and Ref. 36). Consistent with these findings, PACS-2 knockdown blocked recruitment of Bim and Bax to lysosomes. Moreover, PACS-2 knockdown disrupted the trafficking of ligated DR5 to lysosomes, but not its internalization from the cell surface, suggesting that PACS-2 mediates endocytic trafficking steps in TRAIL-induced cells. Surprisingly, Bim knockdown blocked recruitment of PACS-2 to lysosomes, suggesting that PACS-2 and Bim are mutually dependent for their TRAIL-induced targeting to lysosomes. Together with the determination that Bim binds PACS-2 and Bax but that PACS-2 can only bind Bim, these results suggest that TRAIL-induced binding of Bim to PACS-2 may switch PACS-2 from an early endosome-to-TGN trafficking mediator to a lysosome trafficking protein required for PIXosome assembly and trafficking of endocytosed DR5. It will be important to determine precisely how TRAIL regulates the trafficking itinerary of PACS-2 to assemble Bim and Bax on lysosome membranes.
TRAIL-induced activation of JNK is required for recruitment of Bim and Bax on lysosomal membranes (11). Accordingly, the JNK inhibitor SP600125 blocked TRAIL-induced PIXosome assembly, suggesting that the association of PACS-2, Bim, and Bax was JNK-dependent (Fig. 5). JNK phosphorylates Bim, which releases Bim from dynein light chains (19, 46). Thus, the ability of SP600125 to block Bim phosphorylation suggests how this JNK inhibitor repressed PIXosome assembly (Fig. 5). Consistent with this possibility, PACS-2 bound recombinant Bim in vitro, supporting the possibility that JNK acts upstream of PIXosome assembly (Fig. 4A). However, the mechanism by which JNK activates PIXosome assembly is likely more complex, as the kinase may independently stimulate apoptotic activation of Bim, PACS-2, and Bax. For example, 14-3-3 proteins can sequester Bax, and death ligand-induced JNK phosphorylation of 14-3-3 releases Bax (47). Similarly, Akt phosphorylation of PACS-2 Ser-437 promotes 14-3-3 binding in non-diseased cells. TRAIL triggers release of bound 14-3-3, switching PACS-2 from a homeostatic regulator to a proapoptotic effector (21). Whether JNK activates apoptotic PACS-2 by phosphorylating bound 14-3-3 to activate apoptotic PACS-2 is currently under investigation.
LMP may occur either upstream or downstream of mitochondrial dysfunction during cell death or even represent a postnecrotic phenomenon (39–41). In Huh-7 cells, PACS-2 mediates TRAIL-induced apoptosis by triggering LMP upstream of mitochondrial dysfunction. Despite the fact that TRAIL-induced LMP is upstream of mitochondrial dysfunction in hepatobiliary cells, it can be blocked by antiapoptotic Bcl-2 family members such as Mcl-1 (11). This antiapoptotic protein binds and sequesters Bim, including phospho Bim, as identified in this study. These findings raise the possibility that Mcl-1 sequestration of phospho Bim reduces its availability for binding to PACS-2, which would block formation of the PIXosome and prevent LMP.
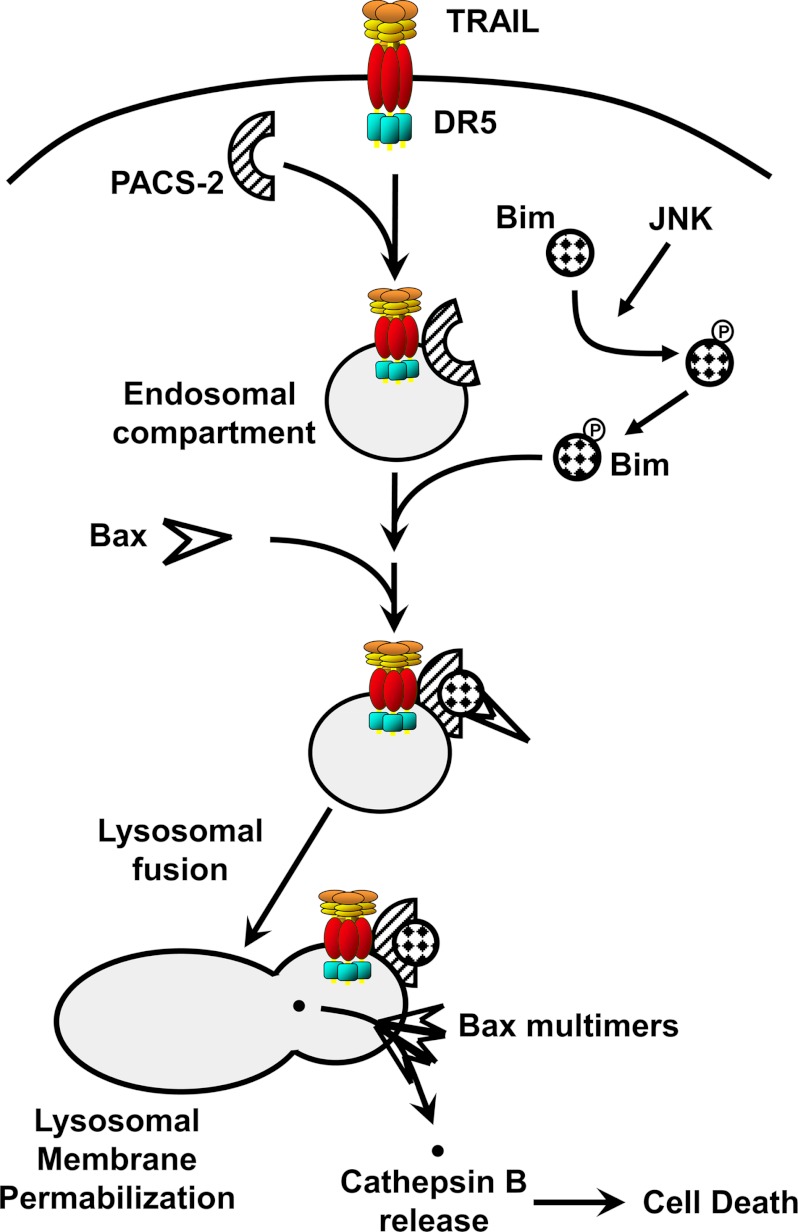
The findings that TRAIL induces PACS-2 to interact with Bim to mediate lysosome permeabilization and with Bid to mediate the subsequent mitochondria permeabilization suggest a broader conceptual framework for the role of PACS-2 in TRAIL action. We suggest that TRAIL induces the JNK-dependent PIXosome formation on lysosomal membranes, triggering LMP (Fig. 6). In parallel, PACS-2 mediates translocation of Bid to mitochondria and Bid cleavage. Because cytosolic cathepsins promote Bid cleavage and mitochondria membrane permeabilization (48–50), our findings suggest that PACS-2 may mediate lysosome-mitochondria interorganellar communication, driving efficient cytochrome c release and activation of executioner caspases. Together, these studies provide insight into the mechanism of TRAIL-induced apoptosis and how inhibition of this pathway may lead to new treatment of cholestatic liver diseases.
FIGURE 6.
Working model for PACS-2-mediated lysosomal membrane permeabilization during TRAIL cytotoxicity. Following binding of TRAIL to DR5, the ligand-receptor complex is internalized, promoting its association with PACS-2 on endosomes. TRAIL also promotes JNK phosphorylation of Bim, liberating it from the cytoskeleton. PACS-2 then binds the cytosolic Bim, which promotes Bax binding to the Bim-PACS-2 complex and Bax activation. After trafficking to lysosomes, Bax inserts into the membrane, homo-oligomerizes, and induces lysosomal membrane permeabilization. Cathepsin B and other lysosomal proteases are released into the cytosol where they contribute to cellular demise.
Acknowledgment
We thank Ms. Courtney Hoover for secretarial services.
This work was supported, in whole or in part, by National Institutes of health Grants DK37274 and CA151564 (to G. T.) and DK 63947 (to G. J. G.). This work was also supported by Canadian Institute of Health Research fellowship HFE-87760, FRQS fellowship 23037 and the Collins Medical Trust (to J. D. D.).
- TRAIL
- tumor necrosis factor-related apoptosis-inducing ligand
- LMP
- lysosomal membrane permeabilization
- PACS
- phosphofurin acidic cluster sorting
- DR5
- death receptor 5
- EGFP
- enhanced green fluorescent protein
- FBR
- furin binding region.
REFERENCES
- 1. Gonzalvez F., Ashkenazi A. (2010) New insights into apoptosis signaling by Apo2L/TRAIL. Oncogene 29, 4752–4765 [DOI] [PubMed] [Google Scholar]
- 2. Johnstone R. W., Frew A. J., Smyth M. J. (2008) The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat. Rev. Cancer 8, 782–798 [DOI] [PubMed] [Google Scholar]
- 3. Kahraman A., Barreyro F. J., Bronk S. F., Werneburg N. W., Mott J. L., Akazawa Y., Masuoka H. C., Howe C. L., Gores G. J. (2008) TRAIL mediates liver injury by the innate immune system in the bile duct-ligated mouse. Hepatology 47, 1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takeda K., Kojima Y., Ikejima K., Harada K., Yamashina S., Okumura K., Aoyama T., Frese S., Ikeda H., Haynes N. M., Cretney E., Yagita H., Sueyoshi N., Sato N., Nakanuma Y., Smyth M. J. (2008) Death receptor 5-mediated apoptosis contributes to cholestatic liver disease. Proc. Natl. Acad. Sci. U.S.A. 105, 10895–10900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang S., El-Deiry W. S. (2003) TRAIL and apoptosis induction by TNF-family death receptors. Oncogene 22, 8628–8633 [DOI] [PubMed] [Google Scholar]
- 6. Guicciardi M. E., Gores G. J. (2009) Life and death by death receptors. FASEB J. 23, 1625–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guicciardi M. E., Deussing J., Miyoshi H., Bronk S. F., Svingen P. A., Peters C., Kaufmann S. H., Gores G. J. (2000) Cathepsin B contributes to TNF-α-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J. Clin. Invest. 106, 1127–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guicciardi M. E., Leist M., Gores G. J. (2004) Lysosomes in cell death. Oncogene 23, 2881–2890 [DOI] [PubMed] [Google Scholar]
- 9. Werneburg N., Guicciardi M. E., Yin X. M., Gores G. J. (2004) TNF-α-mediated lysosomal permeabilization is FAN- and caspase 8/Bid-dependent. Am. J. Physiol. Gastrointest. Liver Physiol. 287, G436–443 [DOI] [PubMed] [Google Scholar]
- 10. Werneburg N. W., Guicciardi M. E., Bronk S. F., Gores G. J. (2002) Tumor necrosis factor-α-associated lysosomal permeabilization is cathepsin B-dependent. Am. J. Physiol. Gastrointest. Liver Physiol. 283, G947–956 [DOI] [PubMed] [Google Scholar]
- 11. Werneburg N. W., Guicciardi M. E., Bronk S. F., Kaufmann S. H., Gores G. J. (2007) Tumor necrosis factor-related apoptosis-inducing ligand activates a lysosomal pathway of apoptosis that is regulated by Bcl-2 proteins. J. Biol. Chem. 282, 28960–28970 [DOI] [PubMed] [Google Scholar]
- 12. Boya P., Andreau K., Poncet D., Zamzami N., Perfettini J. L., Metivier D., Ojcius D. M., Jäättelä M., Kroemer G. (2003) Lysosomal membrane permeabilization induces cell death in a mitochondrion-dependent fashion. J. Exp. Med. 197, 1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boya P., Gonzalez-Polo R. A., Poncet D., Andreau K., Vieira H. L., Roumier T., Perfettini J. L., Kroemer G. (2003) Mitochondrial membrane permeabilization is a critical step of lysosome-initiated apoptosis induced by hydroxychloroquine. Oncogene 22, 3927–3936 [DOI] [PubMed] [Google Scholar]
- 14. Canbay A., Guicciardi M. E., Higuchi H., Feldstein A., Bronk S. F., Rydzewski R., Taniai M., Gores G. J. (2003) Cathepsin B inactivation attenuates hepatic injury and fibrosis during cholestasis. J. Clin. Invest. 112, 152–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johansson A. C., Appelqvist H., Nilsson C., Kågedal K., Roberg K., Ollinger K. (2010) Regulation of apoptosis-associated lysosomal membrane permeabilization. Apoptosis 15, 527–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Strasser A., Cory S., Adams J. M. (2011) Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J. 30, 3667–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gavathiotis E., Reyna D. E., Davis M. L., Bird G. H., Walensky L. D. (2010) BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol. Cell 40, 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gavathiotis E., Suzuki M., Davis M. L., Pitter K., Bird G. H., Katz S. G., Tu H. C., Kim H., Cheng E. H., Tjandra N., Walensky L. D. (2008) BAX activation is initiated at a novel interaction site. Nature 455, 1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Puthalakath H., Huang D. C., O'Reilly L. A., King S. M., Strasser A. (1999) The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol. Cell 3, 287–296 [DOI] [PubMed] [Google Scholar]
- 20. Vogel S., Raulf N., Bregenhorn S., Biniossek M. L., Maurer U., Czabotar P., Borner C. (2012) Cytosolic Bax. Does it require binding proteins to keep its pro-apoptotic activity in check? J. Biol. Chem. 287, 9112–9127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aslan J. E., You H., Williamson D. M., Endig J., Youker R. T., Thomas L., Shu H., Du Y., Milewski R. L., Brush M. H., Possemato A., Sprott K., Fu H., Greis K. D., Runckel D. N., Vogel A., Thomas G. (2009) Akt and 14-3-3 control a PACS-2 homeostatic switch that integrates membrane traffic with TRAIL-induced apoptosis. Mol. Cell 34, 497–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Youker R. T., Shinde U., Day R., Thomas G. (2009) At the crossroads of homoeostasis and disease. Roles of the PACS proteins in membrane traffic and apoptosis. Biochem. J. 421, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bouard D., Sandrin V., Boson B., Nègre D., Thomas G., Granier C., Cosset F. L. (2007) An acidic cluster of the cytoplasmic tail of the RD114 virus glycoprotein controls assembly of retroviral envelopes. Traffic 8, 835–847 [DOI] [PubMed] [Google Scholar]
- 24. Crump C. M., Hung C. H., Thomas L., Wan L., Thomas G. (2003) Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J. Virol. 77, 11105–11113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Crump C. M., Xiang Y., Thomas L., Gu F., Austin C., Tooze S. A., Thomas G. (2001) PACS-1 binding to adaptors is required for acidic cluster motif-mediated protein traffic. EMBO J. 20, 2191–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wan L., Molloy S. S., Thomas L., Liu G., Xiang Y., Rybak S. L., Thomas G. (1998) PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell 94, 205–216 [DOI] [PubMed] [Google Scholar]
- 27. Molloy S. S., Thomas L., Kamibayashi C., Mumby M. C., Thomas G. (1998) Regulation of endosome sorting by a specific PP2A isoform. J. Cell Biol. 142, 1399–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J., Wang J., Meyers K. R., Enns C. A. (2009) Transferrin-directed internalization and cycling of transferrin receptor 2. Traffic 10, 1488–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schermer B., Höpker K., Omran H., Ghenoiu C., Fliegauf M., Fekete A., Horvath J., Köttgen M., Hackl M., Zschiedrich S., Huber T. B., Kramer-Zucker A., Zentgraf H., Blaukat A., Walz G., Benzing T. (2005) Phosphorylation by casein kinase 2 induces PACS-1 binding of nephrocystin and targeting to cilia. EMBO J. 24, 4415–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jenkins P. M., Zhang L., Thomas G., Martens J. R. (2009) PACS-1 mediates phosphorylation-dependent ciliary trafficking of the cyclic-nucleotide-gated channel in olfactory sensory neurons. J. Neurosci. 29, 10541–10551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Köttgen M., Benzing T., Simmen T., Tauber R., Buchholz B., Feliciangeli S., Huber T. B., Schermer B., Kramer-Zucker A., Höpker K., Simmen K. C., Tschucke C. C., Sandford R., Kim E., Thomas G., Walz G. (2005) Trafficking of TRPP2 by PACS proteins represents a novel mechanism of ion channel regulation. EMBO J. 24, 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Atkins K. M., Thomas L., Youker R. T., Harriff M. J., Pissani F., You H., Thomas G. (2008) HIV-1 Nef binds PACS-2 to assemble a multikinase cascade that triggers major histocompatibility complex class I (MHC-I) down-regulation. Analysis using short interfering RNA and knockout mice. J. Biol. Chem. 283, 11772–11784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Myhill N., Lynes E. M., Nanji J. A., Blagoveshchenskaya A. D., Fei H., Carmine Simmen K., Cooper T. J., Thomas G., Simmen T. (2008) The subcellular distribution of calnexin is mediated by PACS-2. Mol. Biol. Cell 19, 2777–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dikeakos J. D., Thomas L., Kwon G., Elferich J., Shinde U., Thomas G. (2012) An interdomain binding site on HIV-1 Nef interacts with PACS-1 and PACS-2 on endosomes to down-regulate MHC-1. Mol. Biol. Cell 23, 2184–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aslan J. E., Thomas G. (2009) Death by committee. Organellar trafficking and communication in apoptosis. Traffic 10, 1390–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simmen T., Aslan J. E., Blagoveshchenskaya A. D., Thomas L., Wan L., Xiang Y., Feliciangeli S. F., Hung C. H., Crump C. M., Thomas G. (2005) PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 24, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kobayashi S., Lee S. H., Meng X. W., Mott J. L., Bronk S. F., Werneburg N. W., Craig R. W., Kaufmann S. H., Gores G. J. (2007) Serine 64 phosphorylation enhances the antiapoptotic function of Mcl-1. J. Biol. Chem. 282, 18407–18417 [DOI] [PubMed] [Google Scholar]
- 38. Akazawa Y., Mott J. L., Bronk S. F., Werneburg N. W., Kahraman A., Guicciardi M. E., Meng X. W., Kohno S., Shah V. H., Kaufmann S. H., McNiven M. A., Gores G. J. (2009) Death receptor 5 internalization is required for lysosomal permeabilization by TRAIL in malignant liver cell lines. Gastroenterology 136, 2365–2376.e1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guicciardi M. E., Bronk S. F., Werneburg N. W., Yin X. M., Gores G. J. (2005) Bid is upstream of lysosome-mediated caspase 2 activation in tumor necrosis factor α-induced hepatocyte apoptosis. Gastroenterology 129, 269–284 [DOI] [PubMed] [Google Scholar]
- 40. Vanden Berghe T., Vanlangenakker N., Parthoens E., Deckers W., Devos M., Festjens N., Guerin C. J., Brunk U. T., Declercq W., Vandenabeele P. (2010) Necroptosis, necrosis and secondary necrosis converge on similar cellular disintegration features. Cell Death Differ. 17, 922–930 [DOI] [PubMed] [Google Scholar]
- 41. Oberle C., Huai J., Reinheckel T., Tacke M., Rassner M., Ekert P. G., Buellesbach J., Borner C. (2010) Lysosomal membrane permeabilization and cathepsin release is a Bax/Bak-dependent, amplifying event of apoptosis in fibroblasts and monocytes. Cell Death Differ. 17, 1167–1178 [DOI] [PubMed] [Google Scholar]
- 42. Tait S. W., Green D. R. (2010) Mitochondria and cell death. Outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11, 621–632 [DOI] [PubMed] [Google Scholar]
- 43. Taylor R. C., Cullen S. P., Martin S. J. (2008) Apoptosis. Controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 9, 231–241 [DOI] [PubMed] [Google Scholar]
- 44. Tinel A., Tschopp J. (2004) The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304, 843–846 [DOI] [PubMed] [Google Scholar]
- 45. Tenev T., Bianchi K., Darding M., Broemer M., Langlais C., Wallberg F., Zachariou A., Lopez J., MacFarlane M., Cain K., Meier P. (2011) The ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol. Cell 43, 432–448 [DOI] [PubMed] [Google Scholar]
- 46. Lei K., Davis R. J. (2003) JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc. Natl. Acad. Sci. U.S.A. 100, 2432–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tsuruta F., Sunayama J., Mori Y., Hattori S., Shimizu S., Tsujimoto Y., Yoshioka K., Masuyama N., Gotoh Y. (2004) JNK promotes Bax translocation to mitochondria through phosphorylation of 14-3-3 proteins. EMBO J. 23, 1889–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stoka V., Turk B., Schendel S. L., Kim T. H., Cirman T., Snipas S. J., Ellerby L. M., Bredesen D., Freeze H., Abrahamson M., Bromme D., Krajewski S., Reed J. C., Yin X. M., Turk V., Salvesen G. S. (2001) Lysosomal protease pathways to apoptosis. Cleavage of bid, not pro-caspases, is the most likely route. J. Biol. Chem. 276, 3149–3157 [DOI] [PubMed] [Google Scholar]
- 49. Repnik U., Stoka V., Turk V., Turk B. (2012) Lysosomes and lysosomal cathepsins in cell death. Biochim. Biophys. Acta 1824, 22–33 [DOI] [PubMed] [Google Scholar]
- 50. Youle R. J., Strasser A. (2008) The BCL-2 protein family. Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]