Background: Virus capsid assembly and maturation are regulated by viral and host molecular factors.
Results: The puromycin-sensitive aminopeptidase cleaves the capsid protein precursor of a double-stranded RNA virus (IBDV).
Conclusion: Puromycin-sensitive aminopeptidase activity is essential for IBDV replication.
Significance: Understanding crucial host factors for virus assembly is important for selecting efficient heterologous expression systems and for determining potential antiviral targets.
Keywords: Protease, Protein Self-assembly, RNA Silencing, Viral Replication, Virus Assembly, Birnavirus, IBDV, PSA, Capsid Maturation, Macromolecular Assembly
Abstract
In many viruses, a precursor particle, or procapsid, is assembled and undergoes massive chemical and physical modification to produce the infectious capsid. Capsid assembly and maturation are finely tuned processes in which viral and host factors participate. We show that the precursor of the VP2 capsid protein (pVP2) of the infectious bursal disease virus (IBDV), a double-stranded RNA virus, is processed at the C-terminal domain (CTD) by a host protease, the puromycin-sensitive aminopeptidase (PurSA). The pVP2 CTD (71 residues) has an important role in determining the various conformations of VP2 (441 residues) that build the T = 13 complex capsid. pVP2 CTD activity is controlled by co- and posttranslational proteolytic modifications of different targets by the VP4 viral protease and by VP2 itself to yield the mature VP2-441 species. Puromycin-sensitive aminopeptidase is responsible for the peptidase activity that cleaves the Arg-452-Arg-453 bond to generate the intermediate pVP2-452 polypeptide. A pVP2 R453A substitution abrogates PurSA activity. We used a baculovirus-based system to express the IBDV polyprotein in insect cells and found inefficient formation of virus-like particles similar to IBDV virions, which correlates with the absence of puromycin-sensitive aminopeptidase in these cells. Virus-like particle assembly was nonetheless rescued efficiently by coexpression of chicken PurSA or pVP2-452 protein. Silencing or pharmacological inhibition of puromycin-sensitive aminopeptidase activity in cell lines permissive for IBDV replication caused a major blockade in assembly and/or maturation of infectious IBDV particles, as virus yields were reduced markedly. PurSA activity is thus essential for IBDV replication.
Introduction
Viral capsid assembly is a paradigm for analysis of the dynamic processes of macromolecular complexes. Capsid structural units have built-in conformational flexibility, allowing them to acquire transient conformations (1, 2) and, for complex capsids, to maintain slightly distinct contacts among these units (3, 4). For successful production of closed capsids of the correct size, self-assembly of viral components is normally accompanied by mechanisms that control the intrinsic structural polymorphism of capsid proteins. Complex capsid assembly is assisted by specific virus components such as scaffolding and accessory and proteolytic proteins as well as by nucleic acids and/or host cell machinery elements (membrane-specific regions, chaperones, enzyme activity).
Birnavirus capsid assembly is a model for studying the coordination of molecular factors involved in this multistep process. Here we describe a host cell protease essential for capsid assembly of the chicken birnavirus infectious bursal disease virus (IBDV)4.
Birnaviruses are non-enveloped icosahedral viruses with a polyploid bipartite double-stranded (ds) RNA genome (5). Structural units are derived from a 1012-amino acid polyprotein precursor. The polyprotein is co- and posttranslationally self-cleaved to release three polypeptides: pVP2 (the capsid protein precursor), VP4 (the protease), and VP3 (6, 7). pVP2 (512 residues) undergoes a series of much slower VP4-mediated subsequent processing events at three secondary targets (positions 487, 494, and 501) (8). Most resulting pVP2 intermediates are further cleaved by VP2 itself (9) to yield mature VP2 (441 residues) and several C-terminal fragments. The released C-terminal segments remain associated with the capsid (10) and are involved in cell entry by promoting disruption of host cell membranes (11, 12). VP3, the other major structural protein, is multifunctional and interacts with itself (13, 14), pVP2 (15, 16), the RNA polymerase (17–19), and the dsRNA genome (20, 21).
VP2 and a variable amount of pVP2 assemble into 260 trimers, the basic structural units that form a ∼70-nm-diameter capsid on the basis of a triangulation number T = 13 levo lattice. VP2 trimers adopt five distinct conformations and form 12 pentamers and 120 hexamers (22–24). VP2 atomic structure was resolved by x-ray crystallography (23, 25, 26). VP2 monomers encompass three domains termed projection (P), shell (S), and base (B). Domains S and P are β-barrels with a jelly roll topology, whereas the B domain is formed by N- and C-terminal α-helices facing the shell interior.
IBDV assembly has been studied intensively using various heterologous expression systems (10, 19, 27, 28). IBDV polyprotein expression with a recombinant Vaccinia virus in mammalian cells results in correct processing of the polyprotein and assembly of virus-like particles (VLP) similar to authentic IBDV virions. Polyprotein expression in insect cells using a baculovirus-based expression system results in the assembly of T = 1 subviral particles (SVP) built of 20 identical VP2 trimers arranged into pentamers and/or tubular assemblies with pVP2 trimers arranged into hexamers with inefficient assembly of IBDV-like particles.
We established that the pVP2 71-residue C-terminal domain (CTD) is important for VP2 switching among multiple quasi-equivalent conformational states (22). The 443-GFKDIIRAIR-452 amphipathic α-helix (helix α5) is the conformational switch of the VP2 polymorphism (11, 15). Expression of mature VP2 alone results in the spontaneous assembly of T = 1 SVP. pVP2 or intermediate pVP2 variant expression leads to tubular structures. We recently showed that VP3 also participates in the pVP2 conformational polymorphism, acting as a canonical scaffolding protein. VP3 interacts with the basic face of the α5 helix through its C terminus (16, 29).
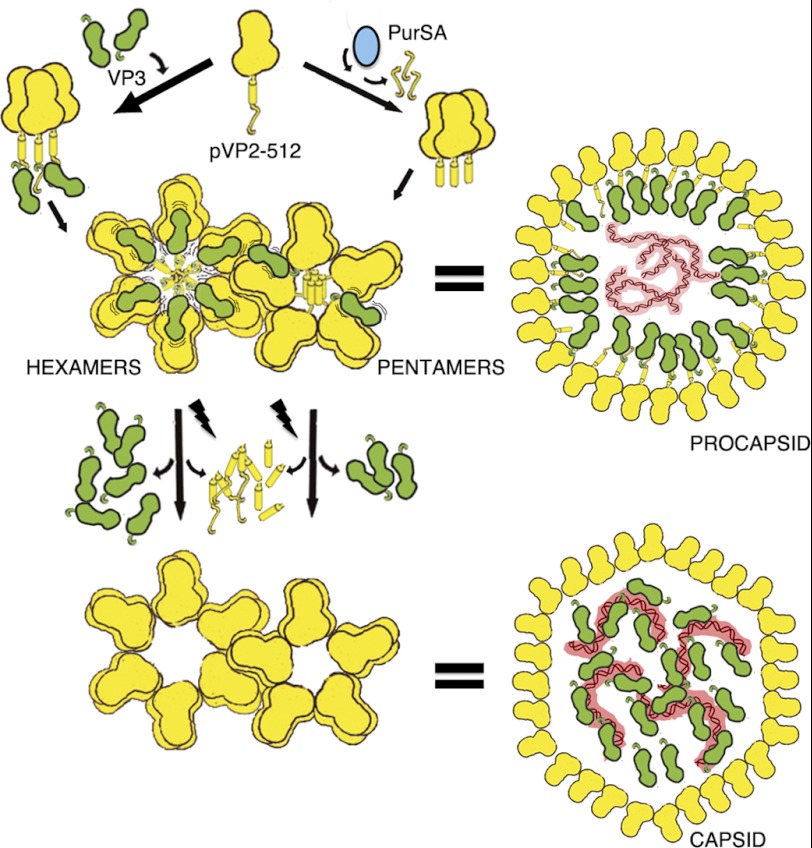
In the IBDV assembly model, a spherical procapsid is initially assembled, as is the case of the fish birnavirus infectious pancreatic necrosis virus (30). According to our hypothesis, the pVP2 coat protein (or its associated intermediates) coassembles with the VP3 scaffold polypeptide to form a double-layered-like particle, except at the pentameric vertices. The VP3:pVP2 interaction directs the assembly of pVP2 trimers into hexamers, thus retarding pVP2 CTD processing. If processing is rapid, the VP3 C-terminal region cannot interact with VP2-441, and a pentamer is formed from VP2 trimers. VP2 and VP3 proteins do not colocalize when coexpressed in insect cells (16).
We speculate that the pentamers are the nucleating centers for capsid assembly and that additional (host) factors are needed to supply pentameric VP2 trimers competent for in vivo capsid assembly. These VP2 nucleating trimers would contain a short C-terminal extension long enough to allow pentamer assembly and transient interaction with some VP3 molecules that act as a liaison with surrounding hexamers (which contain VP3). Our results here demonstrate that these VP2 species are generated by the activity of a host protease, the puromycin-sensitive aminopeptidase (PurSA), which cleaves the pVP2 Arg-452-Arg-453 bond to generate intermediate pVP2-452 polypeptides. Supplying PurSA or pVP2-452 proteins efficiently restores assembly of IBDV-like particles in insect cells.
EXPERIMENTAL PROCEDURES
Cells and Viruses
IBDV infections were carried out with the Soroa isolate (19), adapted to grow in QM7 quail muscle cells and DF1 chicken fibroblasts. Influenza virus A/WSN/33 (H1N1) (WSN) was used as a negative control. Recombinant baculoviruses (rBV) FB/POLY, FB/pVP2-452, and FB/VP2-441 have been described (15, 28). rBV infections were performed in Trichoplusia ni (H5) insect cells (Invitrogen). QM7, DF1, and canine NLB2 cells were cultured in DMEM with 10% FCS. H5 cells were cultured as described (29). rBV were grown and titrated as reported (19, 28).
Construction of Recombinant Baculoviruses
The puromycin-sensitive aminopeptidase protease coding sequence was amplified using primers 5′-GCGCAGATCTCCGGCCAAGC and 5′-GCGCAAGCTTTCAGTTACCT GTGGC using cDNA from DT40 cells as template. BglII-HindIII-digested PCR fragments were cloned into the pFastBac HTb BamH1-HindIII polylinker sites. A mutant version of pVP2 bearing a single mutation, R453A (rBV/Poly-PSA−) was generated by PCR overlap extension. PCR reactions were carried out using the polyprotein gene and cloned in the pVOTE.2/POLY plasmid as the template (19) and the set of mutator primers (5′-CATAATCCGGGCCATAAGGGCGATAGCTGTGCCGGTGGTC and 5′-GACCACCGGCACAGCTATCGCCCTTATGGCCCGGATTATG) in combination with primers 5′-GATGCCATCACAAGCCTCAGC and 5′-CGCAGTCGAGGTTGTGTGCAC. The resulting DNA fragments were digested with SacI and SalI and used to replace the original SacI-SalI fragment in the pFastBac/POLY plasmid. Bacmids derived from the DH10Bac Escherichia coli strain were selected and prepared for Lipofectin transfection according to the protocols of the manufacturer (Invitrogen). Constructs were expressed in H5 insect cells (18).
Generation of Silenced DF1 Cells
The PurSA short hairpin (sh) oligonucleotides were cloned in pRETRO-SUPER (Netherlands Cancer Institute, NKI/AvL), previously digested with BglII and HindIII. The annealed oligonucleotides targeting the cellular protease, 5′GATCCCCGGCAACTTTTGATATTTCATTCAAGAGATGAAATATCAAAAGTTGCCTTTTTGGAAA and 5′-AGCTTTTCCAAAAAGGCAACTTTTGATATTTCATCTCTTGATGAAATATCAAAAGTTGCCGGG for sh1; 5′-GATCCCCAACCTGGTGGAAGTCAAGT TTCAAGAGAACTTGACTTCCACCAGGTTTTTTTGGAAA and 5′-AGCTT TTCCAAAAAAACCTGGTGGAAGTCAAGTTCTCTTGAAACTTGACTTCCACCAGGTTGGG for sh2, were ligated with the vector to yield pRETRO-SUPER-sh1 and pRETRO-SUPER-sh2. The 19-mer PurSA targeting sequence in the oligonucleotide is indicated in boldface. pRETRO-SUPER-sh1, sh2, and an empty plasmid as positive control for non-silencing of the protein were transfected using Lipofectamine (Invitrogen) into the packaging cell line 293T, with pCL-anfo as a helper plasmid. pRETROSUPER expresses a transcript containing the viral packaging signal, the H1-shRNA cassette, and the puromycin resistance gene. At 48 h post-transfection, supernatant was collected, filtered (0.45 μm), and added to DF1 cells. DF1 cells were infected with retrovirus-containing supernatant three times in 24 h, with the addition of polybrene (4 or 8 μg/ml), and silenced cells were selected by adding puromycin (2 μg/ml).
Construction of pcDNA3/pVP2-452
From pFastBac HTb PurSA, PurSA was amplified using primers 5′-GCGCAAGCTTATGTCGTACTACCATCACCATCAC and 5′-GCGCAGATCTTCAGTTACCTGTGGCTGCATTCTTC. HindIII-BglII-digested PCR fragments were cloned into the pcDNA 3 HindIII-BamH1 polylinker sites for protein expression.
IBDV Infection and Titration
For IBDV virion purification, QM7 cultures were infected at a multiplicity of infection (moi) of 1–2 pfu/cell in a minimal volume. After adsorption (1 h), the inocula were removed, and DMEM supplemented with 2% FCS was added. Culture supernatants were collected at 48–72 hpi (hours post-infection), depending on the cytopathic effect. For IBDV stock production, cultures were infected at a moi of 0.05 pfu/cell, and supernatants were collected at 96 hpi. Viral titers were determined by counting lysis plaques.
WSN Titration
NLB2 cell monolayers were infected at 50–60% cell confluence with several virus dilutions. After adsorption, inocula were removed, and DMEM was supplemented with 2% FCS. At 72 hpi, cells were fixed and crystal violet-stained, and lysis plaques were counted.
Purification of IBDV Polyprotein-derived Structures
H5 cells (2–5 × 108 cells) were infected with appropriate rBV (moi, 1–5 pfu/cell). At 48 hpi, cells were harvested, lysed in PES buffer (25 mm piperazine-N,N′-bis (2-ethanesulfonic acid) (pH 6.2), 150 mm NaCl, and 20 mm CaCl2 plus 1% IGEPAL CA-630 (Sigma) and 1% protease inhibitor (Complete Mini, Roche) on ice for 30 min. Purifications were performed as described (22).
SDS-PAGE and Western Blot Analysis
Concentrated sucrose gradient fractions (2–5 μl) were added to Laemmli sample buffer and heated (100 °C, 3 min), followed by electrophoresis in 11% polyacrylamide gels. For Western blot analyses, rabbit anti-VP2 and anti-VP3 sera (19) and an anti-His tag antibody (Sigma) were used.
Electron Microscopy
Samples (2–5 μl) of concentrated sucrose gradient fractions were applied to glow-discharged carbon-coated grids (2 min) and negatively stained with 2% aqueous uranyl acetate. Micrographs (Kodak SO-163) were recorded with a JEOL 1200 EXII electron microscope operating at 100 kV at a nominal magnification of ×40,000.
Immunofluorescence and Confocal Laser Scanning Microscopy Analysis
Silenced DF1 cells seeded onto glass coverslips were infected with IBDV (moi, 1 pfu/cell). At 30 hpi, cells were washed twice with PBS and fixed in methanol-acetone (1:1, 5 min). Coverslips were air-dried, blocked in PBS with 20% FCS, and incubated with rabbit anti-VP2, followed by Alexa Fluor 488 goat anti-rabbit Ig (green). Nuclei were stained with ToPro-3 (Invitrogen). Fluorescent signals were recorded using appropriate filters. Samples were visualized by epifluorescence in a Zeiss Axiovert 200 microscope equipped with a Bio-Rad Radiance 2100 confocal system. Images were captured using LaserSharp software (Bio-Rad).
Quantitative Real-time PCR Assays
Transcript levels were determined by quantitative real-time PCR using a 7300 real-time PCR system and SYBR Green (both from Applied Biosystems). Reactions were performed in a final volume of 20 μl containing 10 μl 2× Power SYBR Green PCR Master Mix (including AmpliTaq Gold DNA Polymerase-LD, deoxynucleotides, and SYBR Green dye), 250 nm forward and reverse specific primers (PurSA, 5′-TGCCTTAGACAACAGTC ATCC and PurSA c, 5′-TCCGAAAGTCCTCATCACCA) and a 1:10 dilution of cDNA. After enzyme activation (95 °C, 10 min), amplification was carried out in a two-step PCR procedure (40 cycles: 15 s at 95 °C for denaturation and 1 min at 60 °C for annealing/extension). Gene-specific primers that generated a 143-bp amplification product were designed using the Primer Express 2.0 program (Applied Biosystems). Non-template controls were included for each primer pair, and each PCR reaction was carried in triplicate. Data were analyzed using 7300 SDS software 1.3 (Applied Biosystems). Dissociation curves for each amplicon were analyzed to confirm specificity. The dissociation curve was obtained by heating the amplicon from 60 °C to 95 °C. As an internal control, we used an oligonucleotide pair that generates a 154-bp amplification product of Gallus gallus GAPDH (GAPDH, 5′- ATGGTGAAAGTCGGAGTCAACG and GAPDH c, 5′-GACAGTGCCCTTGAAGTGTC).
RESULTS
Coexpression of pVP2-452 and IBDV Polyprotein Rescues VLP Assembly in Insect Cells
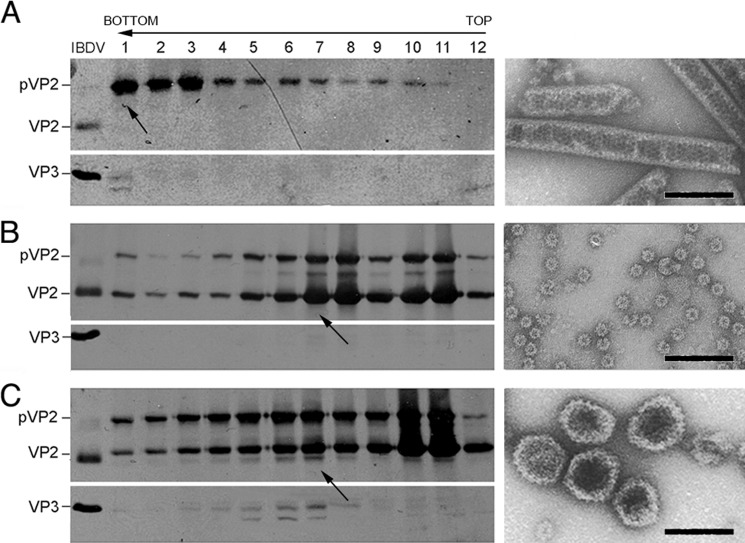
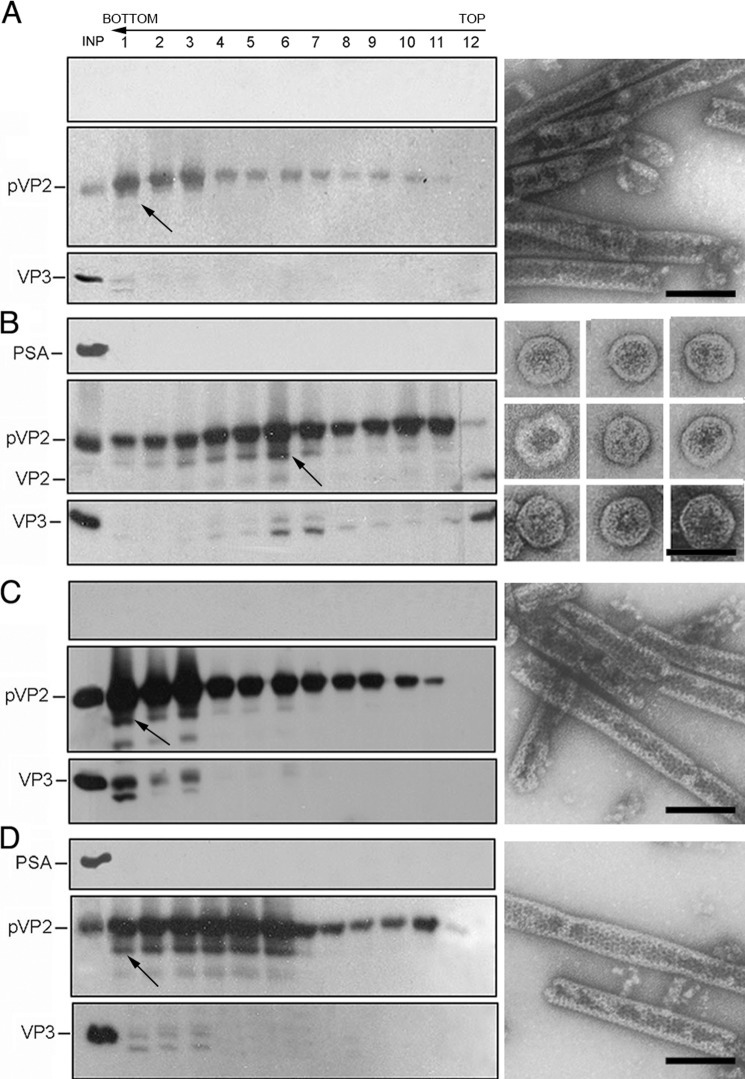
Use of the IBDV polyprotein baculovirus system led to pVP2, VP3, and VP4 accumulation with defective expression of mature VP2. T = 1 SVP, which are composed of VP2 pentamers, are thus not assembled. A schematic representation of VP2 depicting its domains and cleavage sites, including that proposed for PurSA, is shown in Fig. 1. We proposed that T = 13 VLP assembly can be restored by coexpression of shorter pVP2 forms that efficiently assemble into pentamers. We tested this hypothesis using VP2 recombinant forms VP2-441 (lacking the α5 helix) and pVP2-452 (containing the α5 helix). H5 cells were infected with rBV/Poly or coinfected with rBV/Poly and rBV/VP2-441 or rBV/pVP2-452. Cultures were harvested at 48 hpi, extracts were ultracentrifuged on a sucrose gradient, and the resulting fractions were analyzed by SDS-PAGE and Western blot analysis and by negative staining in EM. Extracts from cells expressing rBV/Poly had large pVP2 tubules that migrated mainly to the bottom of the gradient (Fig. 2A). rBV/Poly and rBV/VP2-441 coinfection produced tubular structures (bottom fraction, not shown), T = 1 SVP aggregates (center fractions), and individual T = 1 SVP (top fractions, Fig. 2B). Extracts of cells coinfected with rBV/Poly and rBV/pVP2-452 contained large tubules (bottom), T = 13 VLP with morphology and size similar to IBDV virions (center, Fig. 2C), and T = 1 SVP (top). The middle fractions (fractions 3–8) contained VP3 and a VP2 species in addition to pVP2 and pVP2-452 polypeptides. These results show a biochemical profile similar to bona fide T = 13 IBDV VLP, indicating that pVP2-452 coexpression rescues T = 13 VLP assembly in insect cells expressing the IBDV polyprotein. A similar effect was not observed after coexpression of the fully mature VP2-441 polypeptide.
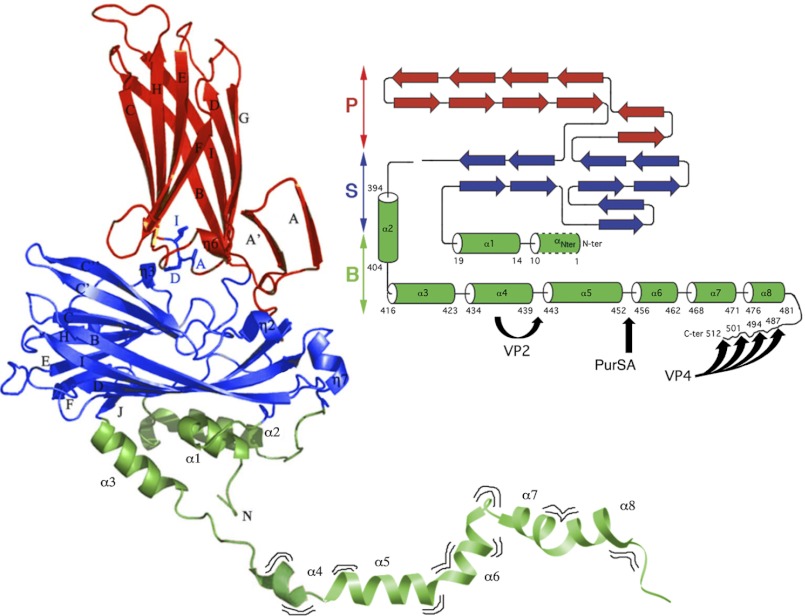
FIGURE 1.
Structure and maturation of pVP2, the capsid protein precursor of IBDV. X-ray structure of VP2 (PDB code 2GSY) manually bound (at residue 441) to part of the pVP2 C terminus determined by NMR analysis (segment 442–488, PDB code 2IMU). Domains S, P and B are colored red, blue, and green, respectively. The VP2 atomic structure extends up to residue 441, including the highly flexible helix α4. The amphipathic α-helix (residues 443–452) is marked as helix α5. The α8 helix ends at residue 481, and the most C-terminal residues assumed display a disordered conformation. Wavy lines indicate that the pVP2 C-terminal extension exhibits conformational flexibility. On the right, a simplified topology schematic (cylinders, α-helices; arrows, β-strands) are shown color-coded by domains. Black arrows indicate processing sites of protease activities performed by VP4, VP2, and PurSA, respectively. VP4 targets: Ala487-Ala488, Ala494-Ala495, Ala501-Ala502 and Ala512-Ala513. VP2 targets: Ala441-Phe442. PurSA targets (this study): Arg452-Arg453.
FIGURE 2.
Coexpression of pVP2-452 and IBDV polyprotein form is necessary for T = 13 VLP assembly in H5 insect cells. A, wild-type rBV/Poly assemblies were purified by ultracentrifugation on sucrose gradients, collected in 12 fractions, concentrated by ultracentrifugation, and analyzed by SDS-PAGE and Western blot analysis using anti-VP2 (left panel, top) and anti-VP3 antibodies (left panel, bottom). The direction of sedimentation was from right to left, with fraction 12 at the gradient top. A representative electron micrograph image is shown of pVP2 tubular structures from lower gradient fractions (arrow). B and C, assemblies from cells coinfected with rBV/Poly and rBV/VP2-441 (B) or rBV/Poly and rBV/pVP2-452 (C) were analyzed as in A. Representative electron micrographs (arrow) of T = 1 SVP from top fractions (B, right panel) and IBDV-like capsids from the center fractions (C, right panel). Bands are indicated for VP2 and VP3 in a purified IBDV sample (left panel). Scale bar = 100 nm.
pVP2-452 Detection in IBDV-infected Cells
The results above suggested a role for pVP2-452 in IBDV assembly. We thus searched for the presence of intermediate pVP2 species in IBDV-infected avian QM7 cells. Cells were harvested at different times post-infection, and extracts were analyzed by SDS-PAGE and Western blot analysis. Anti-VP2 antibody recognized a minor band that migrated between pVP2 and VP2 and whose electrophoretic mobility was similar to that of pVP2-452 (Fig. 3, arrow).
FIGURE 3.
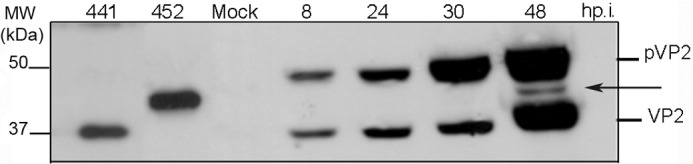
Detection of the intermediate pVP2-452 protein species in IBDV-infected QM7 cells. IBDV-infected QM7 cultures were harvested at 8, 24, 30, and 48 hpi, and the corresponding extracts were analyzed by Western blot analysis. The major bands corresponding to the precursor pVP2 and mature VP2 polypeptides is indicated on the right. The arrow indicates the position of the intermediate pVP2 protein with the same mobility as pVP2-452. Bands corresponding to mature VP2 (441 residues) and pVP2-452 proteins generated by expression of truncated polyprotein genes are indicated (left).
Coexpression of Chicken Puromycin-sensitive Aminopeptidase and IBDV Polyprotein Rescues VLP Assembly in Insect Cells
The results described above highlighted the importance of pVP2-452 for IBDV capsid assembly, although the origin of this protein remained unclear. Two protease activities are associated with VP2 processing, VP4 protease and VP2 itself. As no other viral protease activity has been reported, we hypothesized that a cell protease(s) might cleave the Arg-452-Arg-453 bond to generate pVP2-452. A scan of the Merops protease database (31) yielded a list of suitable candidates, including trypsin 1, cathepsin L, tryptase β, kallikrein 1, thrombin, plasmin, lactoferrin, and PurSA. We excluded those lacking counterparts in chicken cells (trypsin 1, tryptase β, kallikrein 1, and lactoferrin) and sought proteases cleaving at the Arg-Arg site as well as Arg-Lys and Lys-Asn bonds, the homologous sites for the IBDV Arg dipeptide in other birnaviruses (see below). The only protease that met these conditions was PurSA. We used an real-time PCR assay to test for the presence of PurSA mRNA in mammalian and avian cell lines supporting both T = 13 VLP assembly and IBDV replication and found PurSA mRNA expression in all cell lines tested (supplemental Fig. 1). This approach did not allow the detection of PurSA expression in H5 T. ni cells.
Thereafter, chicken PurSAcDNA was generated by real-time PCR and cloned into the pFastBac HTb plasmid vector. We generated an rBV expressing recombinant PurSA fused to an N-terminal His tag, rBV/PurSA. H5 cells were infected with rBV/Poly or coinfected with rBV/Poly and rBV/PurSA, and IBDV-related assemblies were purified from cell extracts. Extracts from rBV/Poly-infected cells showed abundant tubules (Fig. 4A), whereas those from cells coexpressing PurSA contained, in addition to large pVP2 tubules, VLP similar in size and morphology to genuine T = 13 virion capsids (Fig. 4B, center fractions). The protein profile of these pVP2-, VP2-, and VP3-containing fractions resembles that of VLP-containing samples generated in mammalian or avian expression systems (15, 19).
FIGURE 4.
Effect of PurSA on IBDV capsid assembly in H5 insect cells. A, wild-type rBV/Poly assemblies were purified as described in Fig. 2A and analyzed by SDS-PAGE and Western blot analysis using an anti-His tag (left panel, top), anti-VP2 (left panel, center) and anti-VP3 antibodies (left panel, bottom). Direction of sedimentation was from right to left, with fraction 12 at the gradient top. pVP2 tubular structures in the lower fractions (arrow) are shown (right panel). The lane marked INP is a sample loaded from the extract before fractionation. B, assemblies in cells after coinfection of rBV/Poly and rBV/His tag-PurSA (PSA) were analyzed as in A. IBDV-like particles are dominant in the center fractions. A gallery of isolated VLP is shown. C, rBV/PolyR453A assemblies (lower fractions) correspond to tubular structures similar to wild-type rBV/Poly assemblies. D, assemblies from cells coinfected with rBV/PolyR453A and rBV/His tag-PurSA are similar to WT rBV/Poly and rBV/PolyR453A tubular assemblies. Scale bar = 100 nm.
These findings suggested direct correlation between PurSA expression and the rescue of VLP assembly in insect cells. To confirm this relationship, we coexpressed PurSA with a polyprotein mutant lacking the putative Arg-452-Arg-453 PurSA cleavage site. This Poly-PurSA− contains a single amino acid substitution (Arg-453 to Ala) and was used to generate rBV/Poly-PurSA−. Biochemical and structural analyses showed that, regardless of the presence of PurSA, Poly-PurSA− expression led to the assembly exclusively of large tubules (Fig. 4, C and D) indistinguishable from those generated by rBV/Poly infection (A). These data demonstrate that PurSA promotes assembly of polyprotein-derived VLP in H5 cells, an effect that is abrogated by modifying the putative pVP2 R452-R453 PurSA cleavage site.
Silencing PurSA Expression in DF1 Cells
To study the role of PurSA in the context of IBDV infection, we applied a siRNA approach in DF1 cells. Several retroviruses were generated containing PurSA-derived shRNA susceptible to be targeted by the siRNA-induced silencing complex. An empty retrovirus vector, DF1-sh0, was generated for negative controls. In a set of stably silenced DF1 cells produced with these retroviruses, we used quantitative real-time PCR to analyze the inhibitory effect of the shRNA on PurSA mRNA accumulation. The chicken GAPDH gene was used as an internal control. The inhibitory effect of siRNA on the stably silenced DF1 cells (DF1-sh1 and DF1-sh2) was ∼80% for PurSA, whereas GAPDH remained constant (Fig. 5).
FIGURE 5.

PurSA silencing in DF1 cells. Quantitative real-time PCR was used to quantify the inhibitory effect of shRNA on PurSA (PSA) in silenced cells (DF1-sh1 and DF1-sh2; DF1-sh0, control). cDNA quantification for GAPDH (A) and PurSA (B) is shown for each cell line.
Effect of PurSA Silencing and Inhibition on IBDV Replication
To test the effect of PurSA silencing on IBDV replication, parental DF1-sh0, as well as DF1-sh1 and DF1-sh2 cells were IBDV-infected (moi, 0.05 pfu/cell). Cells were fixed at 30 hpi and analyzed by immunofluorescence with anti-VP2 antibody. Nearly 100% of parental DF1-sh0 cells were infected and showed an intense VP2-specific staining (supplemental Fig. 2A). In contrast, only a small fraction (2 < 5%) of DF1-sh1 and -sh2 cells were stained with the anti- VP2 serum, indicating that PurSA silencing causes a major blockade in IBDV replication. To confirm these results, the three cell lines were infected (moi, 2 pfu/cell) and maintained in the presence or absence of the PurSA inhibitor puromycin (2 μg/ml, Ref. 32). Supernatants from infected cultures were collected at 12 hpi and used for IBDV titration. Virus yields in DF1-sh1 and DF1-sh2 cells were reduced by ∼90% relative to those of parental DF1-sh0 cells (supplemental Fig. 2B). Puromycin treatment reduced virus production in DF1 cells. Noteworthy is that expression of the puromycin resistance gene in DF1-sh0, DF1-sh1, and DF1-sh2 renders them resistant to the inhibition of translation caused by the presence of puromycin in the cell medium. Extracts from infected cells at 48 hpi were analyzed by Western blot analysis with anti-VP2 serum. PurSA silencing resulted in a clear reduction in pVP2 accumulation accompanied by near-complete absence of the mature VP2 species, the marker of pVP2 maturation, thus indicating the blockade of the pVP2 > VP2 maturation step (Fig. 6). This effect was more prominent in the presence of puromycin (Fig. 6, lane 48P).
FIGURE 6.
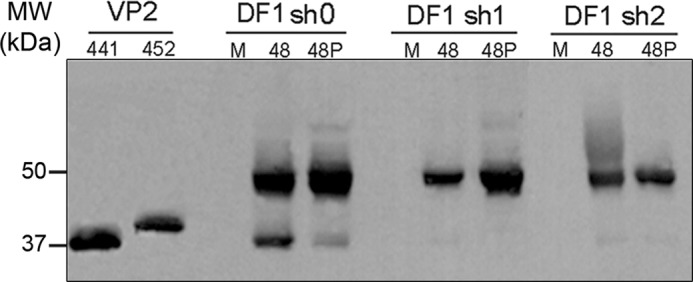
Effect of PurSA silencing on IBDV infection. Western blot analysis of pVP2/VP2 using anti-VP2 antibody in IBDV-infected silenced and control DF1-sh0 cells, alone or with puromycin (P). M, mock. Cultures were harvested at 48 hpi. VP2-441 and pVP2-452 proteins are included as internal markers (left).
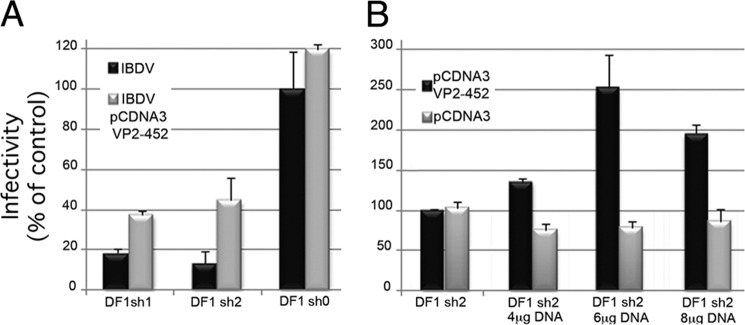
pVP2-452 Expression Counteracts PurSA Silencing Effects on IBDV Replication
The results indicated a relationship between the reduction in PurSA mRNA levels and in IBDV yields. Given its major role in IBDV capsid assembly, pVP2-452 expression should counteract the silencing effect on PurSA. We transfected parental DF1-sh0 as well as DF1-sh1 and DF1-sh2 cells with a plasmid expressing pVP2-452 protein (pcDNA3/pVP2-452) and then infected these cells with IBDV (moi, 2 pfu/cell). Culture media were collected at 12 hpi and used for IBDV titration. pVP2-452 expression resulted in a 2- to 3-fold increase in yields of infectious progeny in both PurSA-silenced cell lines (Fig. 7A). Transfection with the control pcDNA3 plasmid did not affect virus yields (not shown). To assess the specificity of this effect, DF1-sh2 cells were transfected with increasing amounts of pcDNA3/pVP2-452 or control pcDNA3 plasmid and then IBDV-infected (moi, 2 pfu/cell). The effect on IBDV yields was dose-dependent, reaching a maximum (2.5-fold) in cells transfected with 6 μg of plasmid DNA/106 DF1 cells (Fig. 7B).
FIGURE 7.
PurSA silencing of IBDV replication is counteracted by pVP2-452 expression. A, silenced and control cells were transfected with pcDNA3/pVP2-452 plasmid (4 μg/106 cells). After 24 h, transfected (black) and untransfected (gray) cells were infected with IBDV. Thereafter, supernatants were collected at 12 hpi and titrated by serial dilution in QM7 cells. B, DF1-sh2 cells were transfected with pcDNA3/pVP2-452 (black) or empty control pcDNA 3 (gray) plasmids (4, 6, or 8 μg/106 cells). After 24 h, cells were infected with IBDV and virus-titrated as in A.
Pharmacological PurSA Inhibition Abolishes IBDV Replication
We analyzed the effect of pharmacological inhibition of PurSA on IBDV replication. DF1 cells were treated with the non-competitive PurSA inhibitor PAQ-22 (Wako Chemicals GmbH, 10 μm, 12 h). Thereafter, cells were infected with IBDV at 0.5 or 0.05 pfu/cell or influenza virus (strain A/WSN/33) and maintained with PAQ-22. Identical control infections were carried out using untreated cell cultures. When virus-induced cytopathic effect reached 100% in untreated cells, cultures were harvested. Extracts from IBDV-infected cells were used to analyze the accumulation of the structural IBDV VP2 and VP3 polypeptides by Western blot analysis. As shown in Fig. 8A, the presence of PAQ-22 produced an inhibition on the accumulation of both polypeptides that is specially pronounced in infections carried out with the lower moi. Additionally, extracts from cultures infected with IBDV or influenza virus (0.05 pfu/cell) were used to determine the corresponding virus yields. As shown in Fig. 8B, PAQ-22 treatment reduced IBDV yields by nearly two log units (99%) and did not affect influenza virus replication.
FIGURE 8.
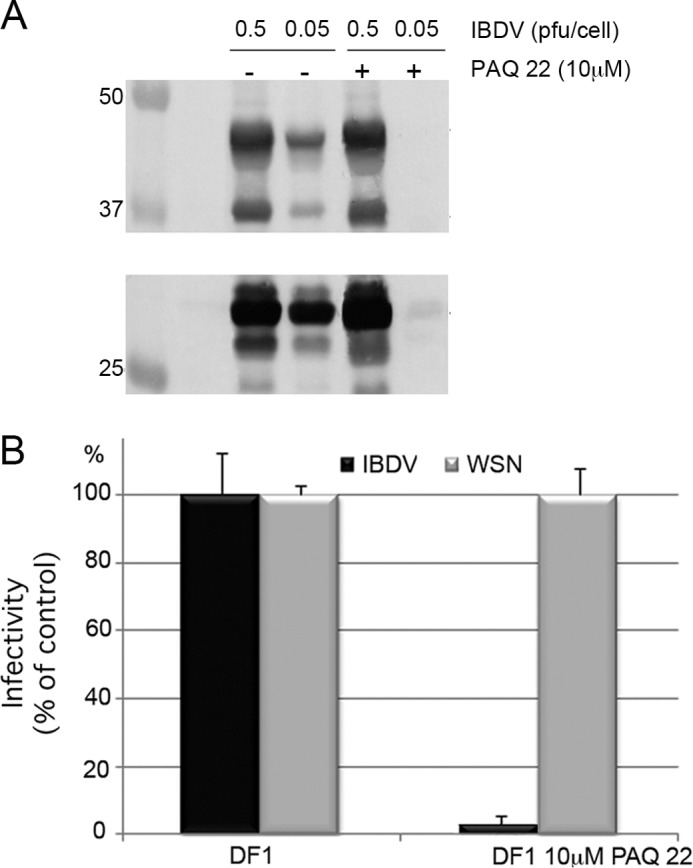
Pharmacological inhibition of PurSA abolishes IBDV replication. A, Western blot analysis of pVP2/VP2 and VP3 using anti-VP2 and anti-VP3 antibodies in DF1 cells, alone or in the presence of 10 μm PAQ-22, infected with two different dilutions of IBDV, 0.5 and 0.05 pfu/cell. B, DF1 cells were cultured alone (gray bars) or in the presence of 10 μm PAQ-22 (black bars) for 12 h and then infected with IBDV or influenza virus (strain A/WSN/33). When virus-induced cytopathic effect reached 100% in untreated cells, cells were harvested, and virus titers were determined in supernatant by serial dilution.
DISCUSSION
Capsid assembly and maturation constitutes an excellent system for the study of mechanisms that control dynamics of multiprotein-nucleic acid complexes (1, 33). The capsid structural subunits of many simple viruses self-assemble directly into an infectious particle that contains a copy of the genome. This process is occasionally accompanied by postassembly maturation. For complex viruses, structural subunits first assemble into a metastable intermediate, the procapsid, which usually requires multiple copies of a scaffolding protein (the morphogenetic factor). After completion of procapsid assembly, the viral genome can be packaged, as is the case for many bacteriophages and some dsDNA viruses, or can nucleate capsid subunits in conjunction with the scaffolding proteins. After cooperative conformational changes, which are irreversible as they are generated by proteases (triggering factor), the procapsids mature into more stable particles (infectious virions).
Expression of IBDV polyprotein via recombinant Vaccinia virus or rBV allows analysis of the assembly of complex capsids that have hundreds of protein subunits (34). Although IBDV polyprotein expression in mammalian cells using recombinant Vaccinia virus supports VLP formation similar to IBDV virions (19, 22), expression using rBV in insect cells leads to aberrant, heterogeneous assemblies with defective VLP production (27, 28). Insect cells might therefore have cellular factor(s) or an environment that interfere with VLP assembly or simply lack a necessary molecular factor. Fusion of a heterologous protein (such as GFP) to the IBDV polyprotein C terminus in rBV yields VLP indistinguishable from those of IBDV virions; i.e. VLP assembly is promoted much more efficiently in a non-natural context (27). Despite these drawbacks, however, recombinant Vaccinia virus and rBV systems provide considerable information in this system.
The pVP2 capsid protein precursor is released from the viral polyprotein by the embedded VP4 viral protease and is further processed at the CTD (also VP4-mediated) to yield shorter pVP2 intermediates. Many positive-sense single-stranded RNA viruses follow this strategy, which is on the basis of the translation of large polyproteins that are self-processed by viral proteases (35). IBDV virions contain mainly mature VP2, as a result of Asp-431-mediated endopeptidase activity that cleaves the 441A-F442 scissile bond from pVP2 intermediates (9). The participation of these two viral proteases in capsid maturation is nonetheless insufficient to explain capsid assembly, and a host-dependent factor was thus considered.
The ubiquitous 100-kDa Zn2+ metallopeptidase PurSA was described in chicken bursal lymphocytes, the IBDV cell target (36), suggesting the importance of this protease in IBDV assembly. PurSA orthologs were not detected in insect cells, probably reflecting a low abundance of its mRNA or the non-specificity of the probes for the PCR assay. IBDV polyprotein expression in yeast also leads to defective VLP assembly (37). Several lines of evidence indicate that PurSA is involved in IBDV capsid assembly. pVP2-452 protein, the product of PurSA activity, rescues assembly of T = 13 VLP of correct size and morphology, as does direct PurSA expression (although at lower efficiency). Moreover, the R453A mutation of the PurSA target scissile bond directs assembly of IBDV structural proteins into tubular structures. Finally, PurSA silencing or pharmacological PurSA inhibition causes a major blockade in IBDV replication, probably by reducing pVP2-452 protein available during virus assembly. This effect is reversed when pVP2-452 is supplied in trans.
Our data indicate that assembly of a birnavirus procapsid is mediated at least by two morphogenetic factors, the VP3 scaffolding protein and the PurSA cellular protease. Although electrostatic interactions between the pVP2 443GFKDIIRAIR452 segment (15) and the VP3 acid C-terminal end lead to hexamer assembly (16), PurSA generates pVP2-452 pentamers. We postulate that, mediated by VP3, these pentamers can interact with neighboring hexamers in the growing procapsid. Only a fraction of newly synthesized pVP2 is cleaved by PurSA, leading to pVP2-452 with a short C-terminal extension able to interact with VP3. Studies by several groups indicated that pentamer assembly is limited by the length of pVP2 C-terminal extensions, as a large CTD cause steric hindrance (12, 15, 38). Limited pVP2 cleavage to yield pVP2-452 is therefore strategic for regulating the assembly of the final product (T = 13 capsids) and, thus, avoiding the assembly of aberrant structures. Processing of pVP2 intermediates into mature VP2 would be completed only when the procapsid is correctly assembled (27). Our T = 13 capsid assembly scheme (Fig. 9) implies that hexamer and pentamer assembly pathways operate separately in the viral factories but are closely coordinated. These results also suggest that the VP3 scaffolding protein promotes pVP2 intermediate interactions through at least two binding modes. Most VP3 interactions with hexameric pVP2 are moderately stable, and a minor VP3 subset interacts more weakly with pentameric pVP2-452. This model is supported by a structural analysis showing that the amphipathic α-helix has two conformations, a five-helix bundle in the pentamers or an open, star-like conformation in hexamers (38). pVP2-452 behaves like a classical assembly pathway intermediate. It does not accumulate and is found at low concentrations relative to a high background of starting material (pVP2 oligomers) and final product (virus particles formed by VP2 oligomers). Mutation R453A confirmed our initial assumption, as the PurSA role is blocked and pVP2 accumulates.
FIGURE 9.
IBDV capsid assembly model. The IBDV capsid protein is synthesized as a precursor, pVP2 (512 residues). The removable C-terminal extension contains the molecular switch, the 443-GFKDIIRAIR-452 amphipathic α-helix (helix α5), and is represented as a cylinder followed by a random coil. A rapid processing of the pVP2 C-terminal extension by PurSA (blue) determines that the pentamer assembly is favored from pVP2-452 trimers. Amphipathic α-helices associate forming a bundle, and a pentamer is formed with bent contacts between VP2 trimers (for simplicity, only a monomer is shown). The PurSA-mediated processing affects a small fraction of the newly synthesized pVP2. Most pVP2 molecules, protected from proteolytic activities through their interaction with VP3 (green), assemble hexamers formed by pVP2 trimers with flat contacts. The pVP2-VP3 interaction is mediated by the VP2 helix α5 and the VP3 C terminus, respectively. pVP2-452 (but not pVP2-441) is able to establish weak interactions with VP3, which can interact with other VP3 molecules from surrounding hexamers in the growing procapsid. Wavy lines indicate the dynamism of the pVP2/VP3 interaction. Autoprocessing of the pVP2 intermediates is a late step in virus assembly that takes place once the procapsid is completely built. VP3 is a multifunctional polypeptide that, in addition to its ability to interact with pVP2 during capsid assembly via an as yet uncharacterized mechanism, is found associated to the dsRNA genome (red) in mature virions.
Host protease dependence is well established for enveloped viruses. Many viral envelope glycoproteins are synthesized as inactive precursors and require maturation by host cell proteases (39). These proteases, such as endoplasmic reticulum signal peptidases (40) and furin family proteases (41), are normally responsible for trimming cell proteins. It remains to be established whether there are PurSA-related cell proteases that provide pVP2-452 competent for pentamer assembly in birnaviruses other than IBDV. The pVP2 molecular switch, organized as an amphipathic helix, is essential for hexamer assembly and is a conserved feature among viruses of several birnavirus genera. The conservation of the 452RR453 scissile bond in all viruses of this group would suggest that PurSA or other equivalent host proteases are involved in pentamer assembly. Alignment of various pVP2 CTD sequences supports a ubiquitous mechanism for pentamer assembly in birnaviruses. The IBDV 452RR453 scissile bond (or analogous bonds) is conserved or has only conservative substitutions in distinct IBDV strains and related genera, including 428RK429 in blotched snakehead virus (42), 453RK454 in infectious pancreatic necrosis virus (43), 462KK463 in Tellina virus 1 (44), 442KN443 in Drosophila X virus (45), and 442KK443 in Espirito Santo virus (46).
Some features of the IBDV T = 13 capsid assembly remain to be elucidated. Mutations in the VP3 Arg-rich region, which precedes the acidic C-terminal residues, show that this basic segment has an additional role during capsid assembly (29). Another factor involved in the sophisticated sequence of interactions between VP3 and the (p)VP2 CTD is the pVP2 segment 501–512 (12), which is as important as the 443-452 segment that bears the VP2 molecular switch. Additional studies are needed, first to define the role of these VP3 and pVP2 segments and, second, to define the order of the multiple molecular events that have been characterized in studies by different groups.
In summary, our results indicate that IBDV capsid assembly and maturation proceed through defined proteolytic steps controlled by viral and host proteases in addition to the participation of a scaffolding protein. This sequential mode of action by several proteases provides an effective regulatory mechanism for the structural polymorphism of the capsid protein by generating structural subunits (the VP2 protein) with C-terminal extensions of different lengths.
Acknowledgments
We thank Dr. A. Maraver for advice on the pRETRO-SUPER plasmid and silencing, Dr. E. Yangüez-Cano for A/WSN/33 flu virus and NLB2 cells, and C. Mark for editorial assistance.
This work was supported by Spanish Ministry of Science and Innovation Grants BIO2009-12443 and AGL2011-24758 (to J. F. R.) and BIO2008-02361 and BFU2011-25902 (to J. R. C.). This work was also supported by a fellowship from the Spanish Ministry of Education and Science (MEC) with support from the CSIC Residencia de Estudiantes and the Gobierno de Aragón (to N. I.).

This article contains supplemental Figs. 1 and 2.
- IBDV
- infectious bursal disease virus
- ds
- double-stranded
- P
- projection domain
- S
- shell domain
- B
- base domain
- VLP
- virus-like particle
- SVP
- subviral particle
- PurSA
- puromycin-sensitive aminopeptidase
- moi
- multiplicity of infection
- pfu
- plaque-forming unit(s)
- hpi
- hours post-infection
- rBV
- recombinant baculovirus
- CTD
- C-terminal domain.
REFERENCES
- 1. Steven A. C., Heymann J. B., Cheng N., Trus B. L., Conway J. F. (2005) Virus maturation. Dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr. Opin. Struct. Biol. 15, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dokland T. (2000) Freedom and restraint. Themes in virus capsid assembly. Structure Fold. Des. 8, R157-R162 [DOI] [PubMed] [Google Scholar]
- 3. Gertsman I., Gan L., Guttman M., Lee K., Speir J. A., Duda R. L., Hendrix R. W., Komives E. A., Johnson J. E. (2009) An unexpected twist in viral capsid maturation. Nature 458, 646–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cardone G., Purdy J. G., Cheng N., Craven R. C., Steven A. C. (2009) Visualization of a missing link in retrovirus capsid assembly. Nature 457, 694–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luque D., Rivas G., Alfonso C., Carrascosa J. L., Rodríguez J. F., Castón J. R. (2009) Infectious bursal disease virus is an icosahedral polyploid dsRNA virus. Proc. Natl. Acad. Sci. U.S.A. 106, 2148–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Birghan C., Mundt E., Gorbalenya A. E. (2000) A non-canonical lon proteinase lacking the ATPase domain employs the Ser-Lys catalytic dyad to exercise broad control over the life cycle of a double-stranded RNA virus. EMBO J. 19, 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feldman A. R., Lee J., Delmas B., Paetzel M. (2006) Crystal structure of a novel viral protease with a serine/lysine catalytic dyad mechanism. J. Mol. Biol. 358, 1378–1389 [DOI] [PubMed] [Google Scholar]
- 8. Sánchez A. B., Rodriguez J. F. (1999) Proteolytic processing in infectious bursal disease virus. Identification of the polyprotein cleavage sites by site-directed mutagenesis. Virology 262, 190–199 [DOI] [PubMed] [Google Scholar]
- 9. Irigoyen N., Garriga D., Navarro A., Verdaguer N., Rodríguez J. F., Castón J. R. (2009) Autoproteolytic activity derived from the infectious bursal disease virus capsid protein. J. Biol. Chem. 284, 8064–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Da Costa B., Chevalier C., Henry C., Huet J. C., Petit S., Lepault J., Boot H., Delmas B. (2002) The capsid of infectious bursal disease virus contains several small peptides arising from the maturation process of pVP2. J. Virol. 76, 2393–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galloux M., Libersou S., Morellet N., Bouaziz S., Da Costa B., Ouldali M., Lepault J., Delmas B. (2007) Infectious bursal disease virus, a non-enveloped virus, possesses a capsid-associated peptide that deforms and perforates biological membranes. J. Biol. Chem. 282, 20774–20784 [DOI] [PubMed] [Google Scholar]
- 12. Chevalier C., Galloux M., Pous J., Henry C., Denis J., Da Costa B., Navaza J., Lepault J., Delmas B. (2005) Structural peptides of a nonenveloped virus are involved in assembly and membrane translocation. J. Virol. 79, 12253–12263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maraver A., Oña A., Abaitua F., González D., Clemente R., Ruiz-Díaz J. A., Castón J. R., Pazos F., Rodriguez J. F. (2003) The oligomerization domain of VP3, the scaffolding protein of infectious bursal disease virus, plays a critical role in capsid assembly. J. Virol. 77, 6438–6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Casañas A., Navarro A., Ferrer-Orta C., González D., Rodríguez J. F., Verdaguer N. (2008) Structural insights into the multifunctional protein VP3 of birnaviruses. Structure 16, 29–37 [DOI] [PubMed] [Google Scholar]
- 15. Saugar I., Luque D., Oña A., Rodríguez J. F., Carrascosa J. L., Trus B. L., Castón J. R. (2005) Structural polymorphism of the major capsid protein of a double-stranded RNA virus. An amphipathic α helix as a molecular switch. Structure 13, 1007–1017 [DOI] [PubMed] [Google Scholar]
- 16. Oña A., Luque D., Abaitua F., Maraver A., Castón J. R., Rodríguez J. F. (2004) The C-terminal domain of the pVP2 precursor is essential for the interaction between VP2 and VP3, the capsid polypeptides of infectious bursal disease virus. Virology 322, 135–142 [DOI] [PubMed] [Google Scholar]
- 17. Garriga D., Navarro A., Querol-Audí J., Abaitua F., Rodríguez J. F., Verdaguer N. (2007) Activation mechanism of a noncanonical RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 104, 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maraver A., Clemente R., Rodríguez J. F., Lombardo E. (2003) Identification and molecular characterization of the RNA polymerase-binding motif of infectious bursal disease virus inner capsid protein VP3. J. Virol. 77, 2459–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lombardo E., Maraver A., Castón J. R., Rivera J., Fernández-Arias A., Serrano A., Carrascosa J. L., Rodriguez J. F. (1999) VP1, the putative RNA-dependent RNA polymerase of infectious bursal disease virus, forms complexes with the capsid protein VP3, leading to efficient encapsidation into virus-like particles. J. Virol. 73, 6973–6983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hjalmarsson A., Carlemalm E., Everitt E. (1999) Infectious pancreatic necrosis virus. Identification of a VP3-containing ribonucleoprotein core structure and evidence for O-linked glycosylation of the capsid protein VP2. J. Virol. 73, 3484–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luque D., Saugar I., Rejas M. T., Carrascosa J. L., Rodríguez J. F., Castón J. R. (2009) Infectious bursal disease virus. Ribonucleoprotein complexes of a double-stranded RNA virus. J. Mol. Biol. 386, 891–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Castón J. R., Martínez-Torrecuadrada J. L., Maraver A., Lombardo E., Rodríguez J. F., Casal J. I., Carrascosa J. L. (2001) C terminus of infectious bursal disease virus major capsid protein VP2 is involved in definition of the T number for capsid assembly. J. Virol. 75, 10815–10828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coulibaly F., Chevalier C., Gutsche I., Pous J., Navaza J., Bressanelli S., Delmas B., Rey F. A. (2005) The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120, 761–772 [DOI] [PubMed] [Google Scholar]
- 24. Böttcher B., Kiselev N. A., Stel'Mashchuk V. Y., Perevozchikova N. A., Borisov A. V., Crowther R. A. (1997) Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J. Virol. 71, 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee C. C., Ko T. P., Chou C. C., Yoshimura M., Doong S. R., Wang M. Y., Wang A. H. (2006) Crystal structure of infectious bursal disease virus VP2 subviral particle at 2.6A resolution. Implications in virion assembly and immunogenicity. J. Struct. Biol. 155, 74–86 [DOI] [PubMed] [Google Scholar]
- 26. Garriga D., Querol-Audí J., Abaitua F., Saugar I., Pous J., Verdaguer N., Castón J. R., Rodriguez J. F. (2006) The 2.6-Ångstrom structure of infectious bursal disease virus-derived T=1 particles reveals new stabilizing elements of the virus capsid. J. Virol. 80, 6895–6905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chevalier C., Lepault J., Erk I., Da Costa B., Delmas B. (2002) The maturation process of pVP2 requires assembly of infectious bursal disease virus capsids. J. Virol. 76, 2384–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martinez-Torrecuadrada J. L., Castón J. R., Castro M., Carrascosa J. L., Rodriguez J. F., Casal J. I. (2000) Different architectures in the assembly of infectious bursal disease virus capsid proteins expressed in insect cells. Virology 278, 322–331 [DOI] [PubMed] [Google Scholar]
- 29. Saugar I., Irigoyen N., Luque D., Carrascosa J. L., Rodríguez J. F., Castón J. R. (2010) Electrostatic interactions between capsid and scaffolding proteins mediate the structural polymorphism of a double-stranded RNA virus. J. Biol. Chem. 285, 3643–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Villanueva R. A., Galaz J. L., Valdés J. A., Jashés M. M., Sandino A. M. (2004) Genome assembly and particle maturation of the birnavirus infectious pancreatic necrosis virus. J. Virol. 78, 13829–13838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rawlings N. D., Morton F. R., Kok C. Y., Kong J., Barrett A. J. (2008) MEROPS. The peptidase database. Nucleic Acids Res. 36, D320–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fidler I. J., Gersten D. M., Hart I. R. (1978) The biology of cancer invasion and metastasis. Adv. Cancer Res. 28, 149–250 [DOI] [PubMed] [Google Scholar]
- 33. Johnson J. E. (2010) Virus particle maturation. Insights into elegantly programmed nanomachines. Curr. Opin. Struct. Biol. 20, 210–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Castón J. R., Rodríguez J. F., Casrrascosa J. L. (2008) in Segmented Double-stranded RNA Viruses. Structure and Molecular Biology (Patton J. T., ed.) pp. 133–144, Caister Academic Press, Norfolk [Google Scholar]
- 35. Lloyd R. E. (2006) Translational control by viral proteinases. Virus Res. 119, 76–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Caldwell R. B., Kierzek A. M., Arakawa H., Bezzubov Y., Zaim J., Fiedler P., Kutter S., Blagodatski A., Kostovska D., Koter M., Plachy J., Carninci P., Hayashizaki Y., Buerstedde J. M. (2005) Full-length cDNAs from chicken bursal lymphocytes to facilitate gene function analysis. Genome Biol. 6, R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vakharia V. N. (1997) Biotech. Annu. Rev. 3, 151–168 [Google Scholar]
- 38. Luque D., Saugar I., Rodríguez J. F., Verdaguer N., Garriga D., Martín C. S., Velázquez-Muriel J. A., Trus B. L., Carrascosa J. L., Castón J. R. (2007) Infectious bursal disease virus capsid assembly and maturation by structural rearrangements of a transient molecular switch. J. Virol. 81, 6869–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moulard M., Decroly E. (2000) Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta 1469, 121–132 [DOI] [PubMed] [Google Scholar]
- 40. Carrère-Kremer S., Montpellier C., Lorenzo L., Brulin B., Cocquerel L., Belouzard S., Penin F., Dubuisson J. (2004) Regulation of hepatitis C virus polyprotein processing by signal peptidase involves structural determinants at the p7 sequence junctions. J. Biol. Chem. 279, 41384–41392 [DOI] [PubMed] [Google Scholar]
- 41. Fischl W., Elshuber S., Schrauf S., Mandl C. W. (2008) Changing the protease specificity for activation of a flavivirus, tick-borne encephalitis virus. J. Virol. 82, 8272–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Da Costa B., Soignier S., Chevalier C., Henry C., Thory C., Huet J. C., Delmas B. (2003) Blotched snakehead virus is a new aquatic birnavirus that is slightly more related to avibirnavirus than to aquabirnavirus. J. Virol. 77, 719–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Galloux M., Chevalier C., Henry C., Huet J. C., Costa B. D., Delmas B. (2004) Peptides resulting from the pVP2 C-terminal processing are present in infectious pancreatic necrosis virus particles. J. Gen. Virol. 85, 2231–2236 [DOI] [PubMed] [Google Scholar]
- 44. Nobiron I., Galloux M., Henry C., Torhy C., Boudinot P., Lejal N., Da Costa B., Delmas B. (2008) Genome and polypeptides characterization of Tellina virus 1 reveals a fifth genetic cluster in the Birnaviridae family. Virology 371, 350–361 [DOI] [PubMed] [Google Scholar]
- 45. Chung H. K., Kordyban S., Cameron L., Dobos P. (1996) Sequence analysis of the bicistronic Drosophila X virus genome segment A and its encoded polypeptides. Virology 225, 359–368 [DOI] [PubMed] [Google Scholar]
- 46. Vancini R., Paredes A., Ribeiro M., Blackburn K., Ferreira D., Kononchik J. P., Jr., Hernandez R., Brown D. (2012) Espirito Santo virus. A new birnavirus that replicates in insect cells. J. Virol. 86, 2390–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]