Background: Human B cells respond to Toll-like receptor (TLR) 9 stimulation by cytokine production.
Results: A rare, novel TLR9 allele fails to activate NF-κB in HEK293 cells. Heterozygous carrier B cells show defective IL-6 and IL-10 production.
Conclusion: Heterozygosity for this TLR9 allele modifies CpG oligonucleotide responsiveness.
Significance: This is the first analysis of human TLR9 variants in primary cells.
Keywords: Immunodeficiency, Lymphocyte, Pathogen-associated Molecular Pattern (PAMP), Signal Transduction, Toll-like Receptors (TLR), CpG Oligonucleotides
Abstract
Toll-like receptors (TLR) are employed by the innate immune system to detect microbial pathogens based on conserved microbial pathogen molecules. For example, TLR9 is a receptor for CpG-containing microbial DNA, and its activation results in the production of cytokines and type I interferons from human B cells and plasmacytoid dendritic cells, respectively. Both are required for mounting an efficient antibacterial or antiviral immune response. These effects are mimicked by synthetic CpG oligodeoxynucleotides (ODN). Although several hyporesponsive TLR9 variants have been reported, their functional relevance in human primary cells has not been addressed. Here we report a novel TLR9 allele, R892W, which is hyporesponsive to CpG ODN and acts as a dominant-negative in a cellular model system. The R892W variant is characterized by increased MyD88 binding and defective co-localization with CpG ODN. Whereas primary plasmacytoid dendritic cells isolated from a heterozygous R892W carrier responded normally to CpG by interferon-α production, carrier B cells showed impaired IL-6 and IL-10 production. This suggests that heterozygous carriage of a hyporesponsive TLR9 allele is not associated with complete loss of TLR9 function but that TLR9 signals elicited in different cell types are regulated differently in human primary cells.
Introduction
Toll-like receptors (TLRs)8 are pattern recognition receptors able to detect conserved molecular patterns on invading bacteria or viruses, such as lipoproteins, glycolipids, and nucleic acids, from bacteria or viruses (1). The central function of TLRs is the induction of immune responses via intracellular signaling events involving the nuclear factor (NF)-κB, and interferon regulatory factor transcription factors and the subsequent production of proinflammatory cytokines and type I interferons (IFN). Mammalian TLRs are a family of transmembrane glycoproteins consisting of 10 members in humans (1, 2). Whereas TLR1, TLR2, TLR4, TLR5, and TLR6 operate on the plasma membrane, TLR3, TLR7, TLR8, and TLR9 are located in intracellular compartments and are able to detect pathogen-derived nucleic acids (1). TLR9 resides in the endoplasmic reticulum in resting cells, whereas ligand binding occurs in endolysosomes (3, 4). Preformed TLR9 dimers specifically recognize DNA in endosomes via an N-terminal (Lys-51 and Arg-74) and a central binding site (Asp-535 and Tyr-537) within their horseshoe-shaped leucine-rich repeat extracellular domains (ECD) (1, 5–8). Ligand-induced conformational changes relay the signal to the cytoplasmic Toll/Interleukin 1-receptor (TIR) domain (9), which mediates intracellular signaling by homotypic interaction with the adaptor molecule MyD88 (myeloid differentiation primary response gene 88) (5, 10).
In human peripheral blood mononuclear cells (PBMC), TLR9 expression is restricted to plasmacytoid dendritic cells (pDC) and B cells. In B cells, TLR9 activation drives co-stimulatory molecule expression, cellular survival and proliferation, the production of IL-6 and IL-10, terminal differentiation, and Ig secretion (11, 12). In pDC, co-stimulatory molecule expression and type I IFN are activated (10, 11). TLR9 activation has mainly been studied using synthetic CpG motif-containing oligodeoxynucleotides (CpG-ODN).
Extremely rare point mutations or single-nucleotide polymorphisms in TLRs and their adaptors have recently been linked to altered susceptibility to infectious agents (13–16). The profile of currently known genetic variants in TLR9 and other nucleic acid-sensing TLRs has been predicted to associate with severe clinical phenotypes (17). TLR9 alleles have been studied epidemiologically with regard to susceptibility for asthma, human immunodeficiency virus, malaria, and systemic lupus erythematosus and functionally in tissue culture systems, where two TLR9 sequence alleles, rs5743844 and rs41308230, were associated with altered receptor function regarding NF-κB activation and cytokine induction (see Ref. 18 and citations therein).
The extent of variation in human TLR9 is generally poorly defined, and functional consequences on a cellular or immunological level have so far not been assessed sufficiently. This prompted us to search for TLR9 alleles by whole gene sequencing and to characterize the protein variants by in vitro and ex vivo functional analyses. In this process, we identified four novel TLR9 alleles, one of which, R892W, showed a hyporesponsive phenotype in the HEK293T cellular system. In primary cells from heterozygous carriers, TLR9 functionality was generally given but, interestingly, in B cells IL-6 and IL-10 production were abrogated, whereas carrier pDC responded normally with IFN-α production.
EXPERIMENTAL PROCEDURES
Study Subjects
Genomic DNA isolation and sequencing of the TLR9 coding region was performed on genomic DNA isolated from blood samples of patients and healthy blood donors (n = 200) treated at the Department of Medical Oncology, Division of Oncology, University Hospital Center “Zagreb,” Zagreb, and the Department of Transfusion Medicine, Rijeka, Croatia. Allele frequency analysis was conducted on blood samples of 810 healthy donors (Department of Transfusion Medicine, Zagreb and Rijeka, Croatia) and 210 tuberculosis patients (Department of Internal Medicine, Section of Pulmonology, Clinical Hospital Center, University of Rijeka, Rijeka, Croatia) as described (19, 20). All study subjects provided oral and written informed consent. The Medical Research Council ethics committees at Zagreb and Rijeka approved the research. During this discovery phase, there was no particular reason for the choice of patient or healthy donor samples. Information on the health status of the TLR9 R892W allele is found in the supplemental information. Of note, the patient suffered from recurring infections (suppurative throat anginas and tuberculosis of the ear) and breast cancer.
Genomic PCR and DNA Sequencing
Genomic DNA was isolated from whole peripheral blood by standard salting out procedure. PCRs containing 150 ng of genomic DNA were amplified (AmplyTaq Gold; Applied Biosystems) on a DNA Thermal Cycler, Type 2700 (Applied Biosystems), using sequential amplification of various fragments to cover the entire coding region of TLR9 (NM_017442). PCR products were purified by ExoSAP-IT (USB Products, Affymetrix). Direct sequencing was performed using a BigDye terminator cycle sequencing kit, 3.1v (Applied Biosystems), followed by analysis on ABI PRISM 3100 DNA Sequencers (Applied Biosystems). All of the primer sequences are available upon request.
Allele Discrimination PCR
Genotyping of the novel TLR9 sequence alleles, 175delG, and C2674T were genotyped by TaqMan allelic discrimination (Assay-by-Design Assays; Applied Biosystems). PCR was carried out according to the manufacturer's protocol on a CFX96 real time PCR detection system (Bio-Rad) including control samples (sequence-verified heterozygous human DNA, plasmid DNA corresponding to variants of interest generated by site-directed mutagenesis, and a “no template” control).
Plasmids and Mutagenesis
The pCIneoTLR9 plasmid containing human TLR9 cDNA (NT_022517.18/NP_059138.1) was kindly provided by David Segal (NCI, National Institutes of Health). To generate TLR9 C-terminal YFP or protein A fusion constructs LR clonase (Invitrogen), reactions were performed using a Gateway entry clone containing the open TLR9 ORF (ImaGenes) and the appropriate destination plasmids (pT-REx-DEST30-ctProteinA and pdEYFP-N1gen; German Cancer Research Center Genomics core facility). DNA sequencing confirmed identical sequences in all TLR9 expression constructs and the correct presence of point mutations corresponding to the described TLR9 alleles introduced by QuikChange mutagenesis (Stratagene). Mutated TLR9 inserts were always back-cloned into the original plasmid backbone and verified again.
Reagents and Cells
Chemicals and cell culture reagents were from Sigma, unless otherwise stated. HEK293T cells (A. Dalpke, University of Heidelberg, Heidelberg, Germany) were cultured in DMEM with 10% fetal calf serum (PAA), l-glutamine, and penicillin/streptomycin (Invitrogen) at 37 °C and 5% CO2 and were transfected using CaPO4 (18). The phosphorothioate-modified (lowercase) CpG-ODN 2006 (tcgtcgttttgtcgttttgtcgtt) and 2216 (ggGGGACGATCGTCgggggg) were from MWG Biotech. R848 was from Axxora.
Immunoblots and Dual Luciferase Assays
Immunoblots and dual luciferase assays were performed in HEK293T cells as described (18). Rat anti-TLR9 (1:5000; eBioscience) and anti-rat HRP conjugate (1:5000; Cell Signaling), or mouse anti-β-tubulin (1:5000, Sigma) and anti-mouse HRP conjugate (1:5000, Promega) Abs were used. For luciferase assays, the mean values of triplicates (± S.D.) of one of at least three independent experiments are shown. Student's t test were performed by using GraphPad Prism v.4.03. (GraphPad Software). p values of <0.05 and <0.01 were denoted as * and **, respectively.
Co-immunoprecipitation
For TLR9 WT-mutant heterodimerization assays, HEK293 cells were transfected with HA and/or protein A-tagged TLR9 and lysed as described above. Immunoprecipitation was performed from cleared cell lysates with monoclonal rabbit α-protein A antibody (Sigma) and protein A/G beads (Pierce). Immunocomplexes were washed three times with lysis buffer and analyzed by immunoblot with mouse α-HA antibody (Cell Signaling). For MyD88, co-immunoprecipitation HEK293 cells were transiently transfected with TLR9-YFP, TLR9 R892W-YFP, or empty vector plus MyD88-AU1. The cells were stimulated with 3 μm CpG 10104 (5′-TCG TCG TTT CGT CGT TTT GTC GTT-3′; MWG Operon) DNA (Type B) for 1 h and lysed (137 mm NaCl, 20 mm Tris, pH 7.4, 1 mm EDTA, 5% Triton X-100, and protease inhibitor mixture (Roche Applied Science)). In parallel, whole cell lysates were prepared in SDS-PAGE loading dye. TLR9 was immunoprecipitated with anti-GFP (Molecular Probes), which also reacts with YFP. Complexes were purified using protein A/G-agarose (Pierce), washed, and resolved by SDS-PAGE. Once transferred to nitrocellulose, TLR9 and MyD88 were detected by immunoblotting with the respective antibodies (anti-GFP, Clontech; anti-MyD88, Santa Cruz). Densitometric analysis on TLR9 and MyD88 bands in the IP fraction was carried out using ImageJ.
Isolation and Stimulation of Peripheral Blood B Cells and pDC
PBMC were freshly isolated from 50 ml of heparinized peripheral blood by Ficoll-Hypaque (GE Healthcare) density gradient centrifugation (Miltenyi Biotec). pDC were enriched from PBMC by positive selection with anti-BDCA4 microbeads, B cells by anti-CD19 microbeads (Miltenyi Biotec) with a purity of 46.5% (±19.3%) for pDC (anti-CD123-FITC antibody; BD Pharmingen) and 77.4% (± 18.4%) for CD19+ B cells (anti-CD20-FITC antibody; BD Pharmingen), as assessed by flow cytometry. All of the cell stimulations were performed at 37 °C and 5% CO2 in RPMI1640 medium (Invitrogen) with l-glutamine and 10% heat-inactivated human AB serum (Department for Transfusion Medicine, Zagreb, Croatia). BDCA4+ pDC and CD19+ B cells were plated at ∼6 × 104 (pDC) or 1.5 × 105 (B cells), respectively, in 200 μl/well in 96-well plates. For both B cells and pDC, stimulation was carried out for 48 h with CpG-ODN 2006 or CpG-ODN 2216, respectively, or R848.
Flow Cytometry
To analyze surface expression of HLA-DR on stimulated and unstimulated B cells and pDC, the cells were washed in PBS and stained (anti-human HLA-DR-FITC; eBioscience) directly in PBS/1% FCS according to standard procedures. The data were collected on a FACSCalibur flow cytometer and analyzed by FACS Diva software (BD Biosciences).
Cytokine ELISA
Cytokine concentrations were quantified by ELISA in cell-free supernatants, after 48 h of stimulation, using the human IFN-α1 ELISA Ready-SET-Go, human IL-10 ELISA Ready-SET-Go, and human IL-6 ELISA Ready-SET-Go (e-Bioscience) kits. The data were expressed as values ± S.E.
Sequence Alignments and Homology Modeling
The TLR9 (sequence NP_059138) ECD was modeled as described in Ref. 8 and citations therein; the TIR domain was modelled on the TLR1 TIR structure as described in Ref. 21. GROMACS molecular dynamics, quality analysis tools (ANOLEA, VERIFY_3D, and ERRAT), N-glycan analysis (GlyProt server), visualization/analysis tools (SwissPBD Viewer and PyMol), and software for the computation of surface charges were employed as described (Ref. 8 and citations therein).
RESULTS
Identification of Novel Human TLR9 Alleles
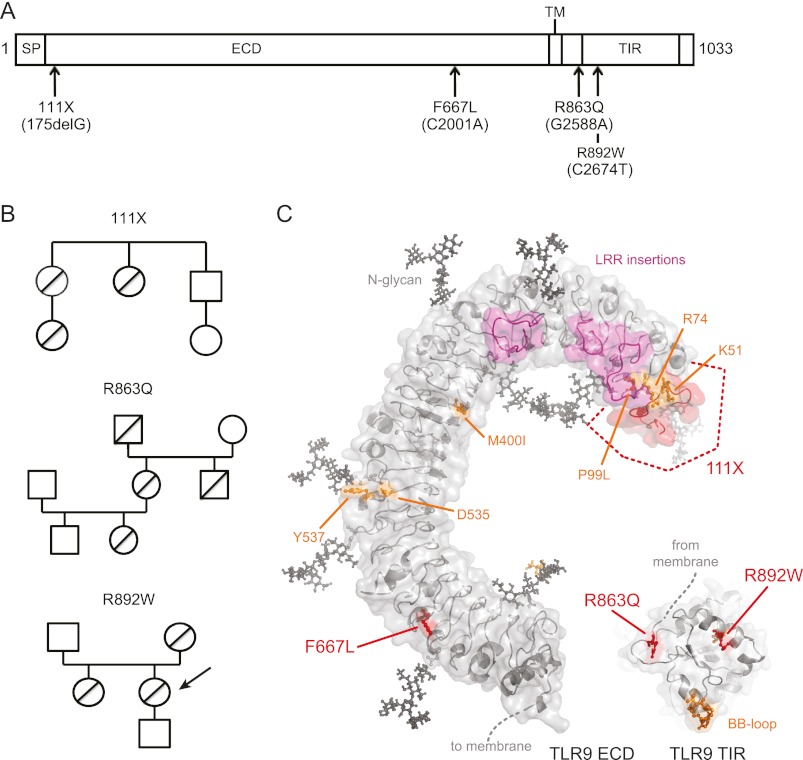
The notion that nonsynonymous single-nucleotide polymorphisms in the human TLR9 gene (GeneID 54106; Chromosome 3p21.3) may impact on the recognition of pathogens or TLR9 therapeutics (11) prompted us to search for naturally occurring mis-sense alleles by sequencing of the TLR9 coding region in genomic DNA from 200 Croatian individuals. Of 11 detected TLR9 alleles (Table 1), we initially focused on the following four nonsynonymous single nucleotide polymorphisms, three of which had not been reported so far: 175delG, C2001 → A, C2588 → T (known), and C2674 → T (Fig. 1A and supplemental Fig. S1). TLR9 175delG, because of a deletion of one nucleotide, is a frameshift mutation creating a truncated 111 amino acid (111X) polypeptide consisting of the first 58 amino acids of TLR9 and 53 amino acids of non-sense sequence. The TLR9 C2001 → A mutation resulted in a Phe to Leu substitution in leucine-rich repeat 22 (F667L). G2588 → A (an Arg → Gln substitution, R863Q) and C2674 → T (an Arg → Trp substitution, R892W) mapped between the transmembrane part and the TIR domain or within the TIR region, respectively (Fig. 1A). Most of the residues affected by variation were highly conserved (supplemental Fig. S2) and, according to generated three-dimensional homology models for the TLR9 ECD and TIR domains, mapped to important functional domains of TLR9 (Fig. 1C). In 111X, most features involved in CpG-ODN sensing/binding (6–8, 18) would be absent because of the frameshift. Phe-667 maps to the ligand-binding glycan-free side of the TLR9 ECD (7, 8, 18). Arg-863 coordinates a loop leading up to the first TIR domain β-strand, and Arg-892 is adjacent to Leu-891, which, when mutated to Asn, completely abrogates TLR9-induced NF-κB signaling (22). These in silico analyses prompted us to determine the functional activity of these variants both in tissue culture systems and immune cells isolated from the original carriers (R892W only, indicated in Fig. 1B). To exclude the possibility that these could be sequencing artifacts, we confirmed by genotyping that heterozygous carriage of 111X, R863Q, and R892W had persisted for at least two generations (Fig. 1B). However, in an additional 1020 Caucasian subjects genotyped for 111X and R892W, the 175delG (111X) allele was detected heterozygously only once, whereas the gene allele 2674C/T (R892W) was absent in all of the additional tested samples. This suggests that the 111X and R892W alleles occur in Caucasians with very low frequency and constitute rare familial mutations.
TABLE 1.
Summary of genotyping results
In the left column, information about known and novel SNPs is summarized (Source: NCBI dbSNP). Genotyping and sequencing results in the initial 200 Caucasian study subjects are also displayed. NC, noncoding; ND, not determined.
| TLR9 variant alleles |
Sequencing/genotyping |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patients |
Controls |
||||||||
| rs number (if available) | Nucleotide change | Amino acid change | Minor allele (frequency) | Allele and number (frequency) | Genotype | Frequency | Allele and number (frequency) | Genotype | Frequency |
| rs352139 | G40A | NC | G (0.49) | A 82/200 (0.41) | GG | 0.36 | A 74/200 (0.37) | GG | 0.37 |
| GA | 0.46 | GA | 0.52 | ||||||
| G 118/200 (0.59) | AA | 0.18 | G 126/200 (0.63) | AA | 0.11 | ||||
| Novel | 175 del G | 111X | del 1/200 (0.005) | del/del | 0 | del 0/200 | del/del | 0 | |
| del/WT | 0.01 | del/WT | 0 | ||||||
| WT 199/200 (0.995) | WT/WT | 0.99 | WT 200/200 | WT/WT | 1 | ||||
| rs35654187 | G1149A | T383T | A (0.026) | A 1/200 (0.005) | GG | 0.99 | A 2/200 (0.01) | GG | 0.98 |
| GA | 0.01 | GA | 0.02 | ||||||
| G 199/200 (0.995) | AA | 0 | G 198/200 (0.99) | AA | 0 | ||||
| rs35342983 | G1527A | S509S | A (0.025) | A 1/200 (0.005) | GG | 0.99 | A 2/200 (0.01) | GG | 0.98 |
| GA | 0.01 | GA | 0.02 | ||||||
| G 199/200 (0.995) | AA | 0 | G 198/200 (0.99) | AA | 0 | ||||
| rs352140 | G1635A | P454P | G (0.52) | A 85/200 (0.43) | GG | 0.2 | A 78/200 (0.39) | GG | 0.13 |
| GA | 0.45 | GA | 0.52 | ||||||
| G 115/200 (0.57) | AA | 0.35 | G 122/200 (0.61) | AA | 0.35 | ||||
| Novel | C2001A | F667L | A 0/200 | CC | 1 | A 1/200 (0.005) | CC | 0.99 | |
| CA | 0 | CA | 0.01 | ||||||
| C 200/200 | AA | 0 | C 199/200 (0.995) | AA | 0 | ||||
| Novel | C2514T | L838L | T 1/200 (0.005) | CC | 0.99 | T 0/200 | CC | 1 | |
| CT | 0.01 | CT | 0 | ||||||
| C 199/200 (0.995) | TT | 0 | C 200/200 | TT | 0 | ||||
| rs5743845 | G2588A | R863Q | A (0.04) | A 1/200 (0.005) | GG | 0.99 | A 2/200 (0.01) | GG | 0.98 |
| GA | 0.01 | GA | 0.02 | ||||||
| G 199/200 (0.995) | AA | 0 | G 198/200 (0.99) | AA | 0 | ||||
| Novel | C2674T | R892W | T 0/200 | CC | 1 | T 1/200 (0.005) | CC | 0.99 | |
| CT | 0 | CT | 0.01 | ||||||
| C 200/200 | TT | 0 | C 198/200 (0.995) | TT | 0 | ||||
| rs445676 | T2940C | Y980Y | ND | C 5/200 (0.03) | TT | 0.95 | C 3/200 (0.02) | TT | 0.97 |
| TC | 0.05 | TC | 0.03 | ||||||
| T 195/200 (0.97) | CC | 0 | T 197/200 (0.98) | CC | 0 | ||||
| rs5743848 | G3117C | NC | ND | C 0/200 | GG | 1 | C 1/200 (0.005) | GG | 0.99 |
| GC | 0 | GC | 0.01 | ||||||
| G 200/200 | CC | 0 | G 199/200 (0.995) | CC | 0 | ||||
FIGURE 1.
Novel familial TLR9 gene variants map to functional domains of TLR9. A, schematic representation of TLR9 with location of variants and functional domains (see text) indicated. SP, signal peptide; TM, transmembrane domain. B, genotyping results of individual family members. Allele carriers are denoted by diagonal lines, females are denoted by circles, and males are denoted by boxes. An arrow points to the R892W carrier whose cells were subsequently analyzed ex vivo. C, computer models of TLR9 ECD and TIR domains. Identified novel TLR9 alleles are in red. Residues previously identified as functionally important are in orange, leucine-rich repeat (LRR) loop insertions are in magenta, and putative N-glycans are in black.
Novel Human TLR9 Alleles Affect NF-κB Activation
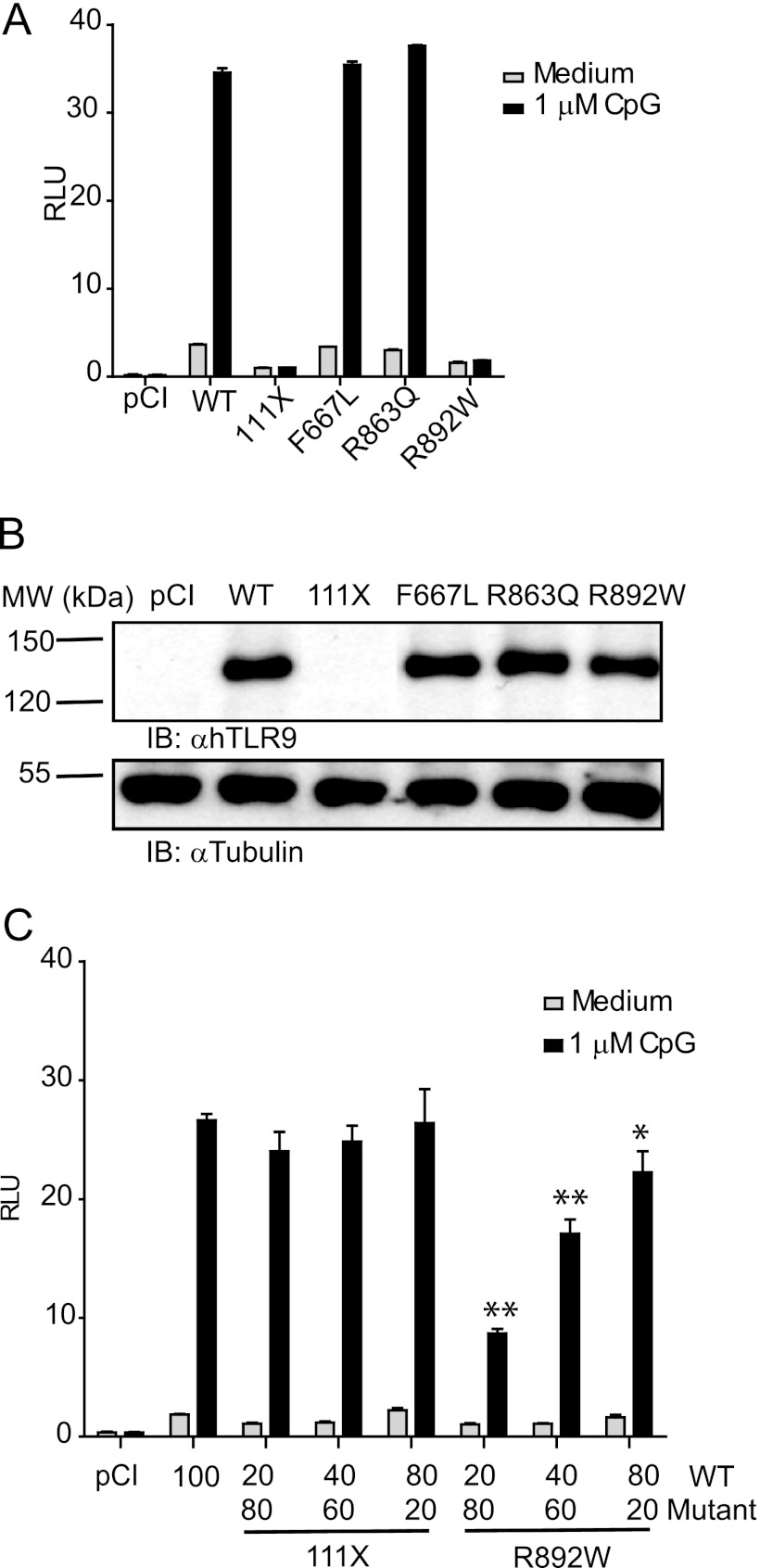
We first sought to determine whether the four nonsynonymous single-nucleotide polymorphisms affect the function of TLR9 in a genetic complementation system in HEK293 cells, which are unresponsive to CpG-ODN prior to TLR9 transfection (18). Point mutations corresponding in sequence to the identified alleles were generated in a human TLR9 expression construct, transfected, and compared with TLR9 WT in NF-κB-dependent dual luciferase assays upon stimulation with CpG-ODN. HEK293T cells expressing F667L and R863Q mutants responded to 1 μm CpG-ODN 2006 similarly to TLR9 WT-expressing cells (Fig. 2A). Conversely, cells transfected with 111X and R892W TLR9 did not show significant NF-κB-dependent activation. Except 111X, which would lack the anti-TLR9 epitope (amino acids 273–288), all of the constructs were detected by anti-TLR9 immunoblot similarly to WT TLR9 (Fig. 2B). 111X, which would be predicted to result in a secreted product based on data regarding other TLR ectodomain truncations (23), did not show a dominant-negative effect when co-transfected with WT TLR9 (Fig. 2C). In contrast, R892W strongly and dose-dependently inhibited TLR9 WT-mediated NF-κB-dependent luciferase production. These data suggest that in a heterozygous carrier for R892W, WT TLR9 function may be compromised.
FIGURE 2.
Two TLR9 variants show reduced NF-κB activation in response to CpG-ODN. A and B, HEK293T cells were transfected with TLR9 WT, 111X, F667L, R863Q, or R892W and assayed for NF-κB activity with (black) or without (gray) CpG-ODN stimulation by dual luciferase assay (A) or immunoblotted for TLR9 and β-tubulin to confirm equivalent expression levels (B). C, HEK293T cells were transfected with the indicated amounts of TLR9 WT (80, 40, or 20 ng) in the presence of TLR9 111X or TLR9 R892W (20, 60, or 80 ng). The cells were CpG-ODN stimulated (black) or unstimulated (gray). pCI, empty vector transfected. p values of < 0.05 and <0.01 were denoted as * and **, respectively. One representative of three independent experiments is shown. In A and C, triplicate points ± S.D. are shown. RLU, relative light unit(s); IB, immunoblot.
Possible Effect of the R892W Mutation on MyD88 Interaction
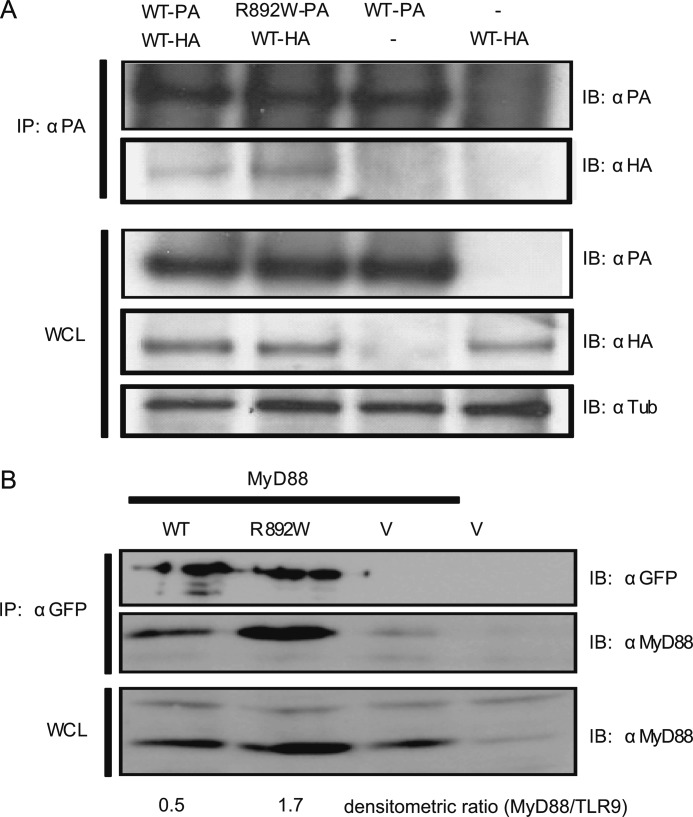
Regarding the molecular cause for the R892W loss of function, we verified correct preformed dimer formation (18) by co-immunoprecipitation in transfected HEK293T cells. As seen in Fig. 3A, R892W was able to interact with WT TLR9, in keeping with the observed dominant-negative effect. Next we asked whether TLR9 R892W associated with the critical adaptor molecule MyD88 because the mutation affects the TLR9 TIR domain. Co-immunoprecipitation analysis in transfected HEK293 cells demonstrated that the mutant retains the ability to bind MyD88 (n = 4). Although in some experiments MyD88 abundance was lower in TLR9 WT than R892W-transfected cell lysates (n = 2), in all four experiments more MyD88 was precipitated by TLR9 R892W compared with TLR9 WT as judged by densitometric analysis (Fig. 3B), suggesting that increased MyD88 affinity may at least partially contribute to the molecular basis for the R892W loss of function (see “Discussion”).
FIGURE 3.
TLR9 R892W co-immunoprecipitates with WT TLR9, binds to MyD88, but fails to reach CpG DNA containing endosomes. A, HEK293T cells were transiently co-transfected with TLR9 WT-protein A (PA) and TLR9 R892W-HA fusion protein expression vectors and lysed after 48 h. Cleared lysates were immunoprecipitated (IP) with α-protein A antibody and immunoblotted (IB) for protein A or HA. Whole cell lysates were immunoblotted for protein A, HA, and tubulin. One of two representative experiments is shown. B, HEK293T cells were transfected with TLR9-YFP (WT), TLR9 R892W-YFP (R892W), or empty pEYFP vector (V) together with MyD88-AU1 (MyD88) as indicated. The lysates were immunoprecipitated for GFP (reacts with YFP) and immunoblotted for GFP and MyD88. Whole cell lysates (WCL) were immunoblotted for MyD88. Note that endogenous MyD88 is detected at low abundance in all lanes. TLR9 and MyD88 bands were subjected to densitometry analysis using ImageJ. One of three independent experiments shown.
The TLR9 R892W Modulates the Function in Primary Immune Cells
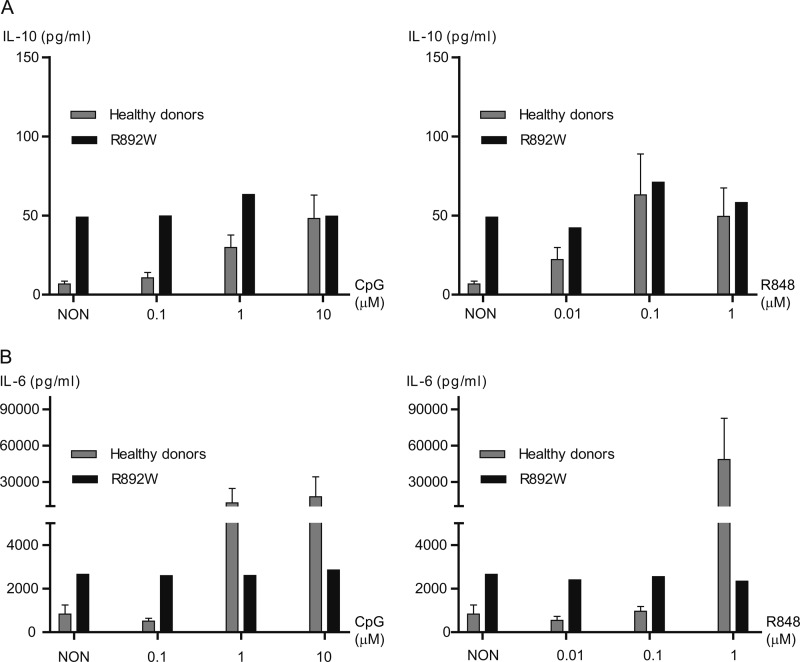
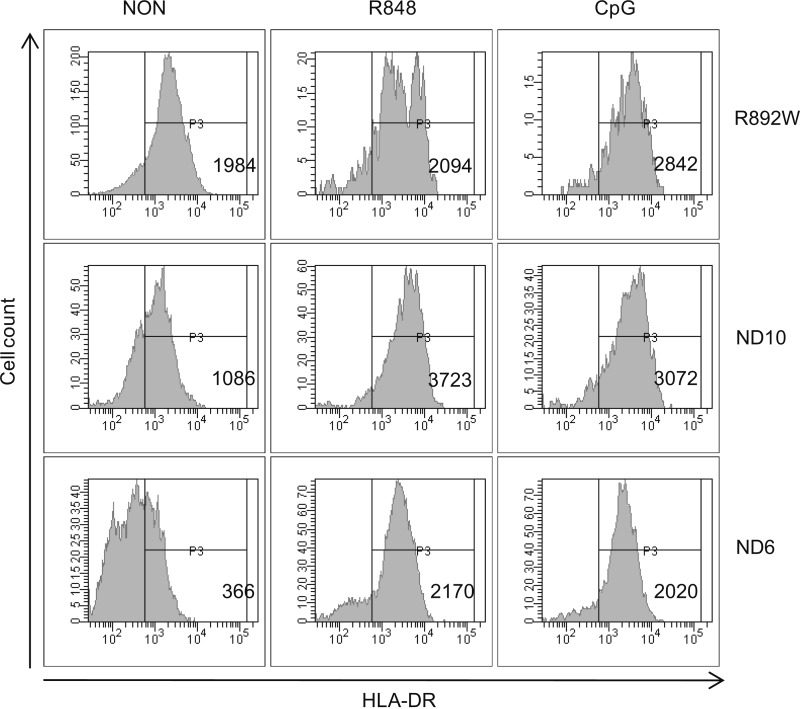
We next sought to investigate the functional role of the TLR9 R892W variant in purified B cells and pDC from an allele carrier. Responses were compared with a panel (n = 11) of healthy control subjects to compare the range of responses because previous studies in human volunteers have shown variability in basal stimulation levels and cellular responses to CpG DNA (24). B cell cultures from healthy donors potently induced IL-10 production in response to 0.1–10 μm CpG-ODN 2006 (Fig. 4A, left panel). Stimulated R892W B cells did not produce IL-10 above a base-line level, which was very elevated compared with the panel of healthy donors and corresponded to levels found only in CpG-stimulated normal donor samples. In response to R848, a human TLR7 agonist sharing downstream signaling components with TLR9 (1), R892W B cells again did not respond above an elevated base-line level (Fig. 4A, right panel). When the same supernatants were analyzed for the presence of IL-6 (Fig. 4B), B cells from the R892W carrier showed no response to CpG-ODN or R848, whereas B cells from normal donors strongly induced IL-6 secretion. Assessing up-regulation of HLA-DR by flow cytometry (Fig. 5) showed that the basal HLA-DR expression levels under nonstimulated conditions varied considerably between samples. Independent of the basal expression levels in normal donors, we observed a marked increase in HLA-DR expression in response to both CpG-ODN and R848 stimulation. HLA-DR expression in R892W B cells was not altered by R848 stimulation. CpG-ODN-dependent HLA-DR expression was lower than the increase observed in normal donors but nevertheless significant (Fig. 5). The reason(s) for the rather high basal cytokine and activation levels in the R892W carrier (especially for IL-10) are difficult to derive from these data and may be altogether unrelated to the presence of the R892W allele or TLR9 function and due to other genetic or environmental factors. However, it can be concluded that in B cells from the R892W allele, donor responses to stimulation with TLR9-specific ligand were diminished when compared with wild-type donors. In the case of IL-6, even high TLR ligand concentrations (10 μm) were not sufficient to reach the cytokine concentrations induced in cells from healthy donors. Overall, we observed significant differences in cytokine production of TLR7/9-stimulated B cells in TLR9 variant R892W donor compared with normal donors.
FIGURE 4.
Modulated cytokine production in TLR9 R892W primary B cells. Secretion of IL-10 (A) and IL-6 (B) measured by ELISA in supernatants (in pg/ml) of CD19-enriched primary B cells stimulated with CpG-ODN 2006 (Type B ODN; 0.1, 1 and 10 μm, left panels) or R848 (0.01, 0.1, and 1 μm, right panels) for 48 h. NON, non-stimulated (media only). One of two representative experiments done in duplicate. The error bars represent the standard error of the mean.
FIGURE 5.
TLR9-mediated HLA-DR up-regulation in R892W B cells. Peripheral blood B cells enriched with anti-CD19 microbeads from the TLR9 R892W carrier (R892W) and from two representative normal healthy donors (ND6 and ND10) were stimulated with or without R848 or CpG 2006 for 48 h. NON, non-stimulated (media only). After harvest, the cells were stained for the expression of surface HLA-DR. The histograms display the mean fluorescence intensities.
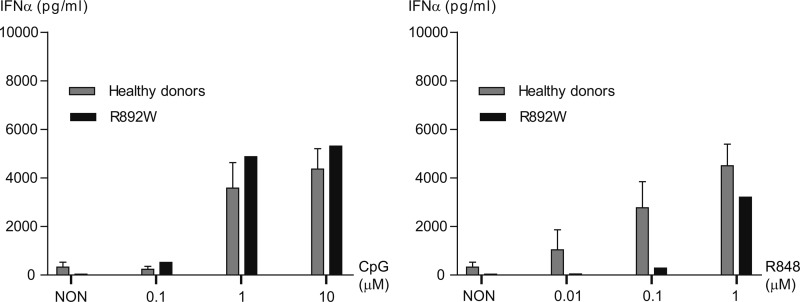
We also analyzed TLR7/9-induced IFNα production, a read-out specific for pDC (1) using anti-BDCA4-enriched PBMC fractions stimulated with CpG-ODN 2216 or R848. CpG-ODN 2216-induced IFNα production induced levels of ∼4000 pg/ml in pDC from healthy donors using 1 and 10 μm CpG-ODN concentrations (Fig. 6, left panel). R892W pDC secreted amounts of IFNα comparable with control donors in response to 1 and 10 μm CpG-ODN. Upon R848 stimulation (Fig. 6, right panel), the levels were reduced at low (0.01 μm) and intermediate (0.1 μm) R848 concentrations but comparable with the control donors at 1 μm. Thus, whereas IL-6 and IL-10 secretion from R892W B cells appeared reduced, B cell HLA-DR expression and pDC IFNα were intact in R892W carrier cells. Our data thus illustrate that heterozygous carriage of R892W loss of function alleles does not result in a general TLR9 insufficiency but may modulate certain TLR9-dependent cellular responses (see “Discussion”).
FIGURE 6.
Normal IFNα production from TLR9 R892W primary pDC. Concentration of IFNα measured by ELISA in supernatants from nonstimulated or stimulated BDCA-4-enriched pDC. The cells were enriched from PBMC (see “Experimental Procedures”) and stimulated with a range of either CpG-ODN 2216 (Type A ODN; 0.1, 1, and 10 μm, left panel) or R848 (0.01, 0.1, and 1 μm, right panel) for 48 h. NON, non-stimulated (media only). Duplicates ± standard error of the mean are shown.
DISCUSSION
The aim of this present work was to functionally analyze rare TLR9 mutations in both cellular model systems and primary B lymphocytes and pDC. Such studies conducted for naturally occurring variants in other human TLR pathway genes have recently provided important insights into the molecular biology of these proteins (18, 25, 26) and their role in the pathogenesis of different diseases (14, 27).
The naturally occurring variants 111X and R892W described here indeed provide new insights into the molecular biology of TLR9. The 111X dysfunctional phenotype is probably related to the secretion of this polypeptide and/or the disruption of the N-terminal CpG-ODN binding site (Ref. 7 and Fig. 1). 111X did not affect TLR9 WT signaling in a dominant-negative way (Fig. 2C), conversely to a recently identified truncated and secreted TLR9 ECD form (28). One explanation for this discrepancy may be that the secreted 111X product, which is considerably shorter than the one previously observed (28), is unable to re-enter endosomal compartments where WT TLR9 resides and/or the inability of the 111X polypeptide to engage CpG-ODN and WT TLR9 in a receptor-ligand complex. Further molecular characterization of the 111X polypeptide may shed further light on TLR9 determinants of CpG recognition or receptor complex assembly.
Arg-892 is conserved in all known TLR9 sequences (supplemental Fig. S2) and across TIR domains (not shown). The residue maps to a protein region sensitive to mutagenesis (22) that harbors a targeting motif (amino acids 888–902) (29). The observed dominant-negative effect over WT TLR9 and its ability to associate with TLR9 WT under unstimulated conditions (Figs. 2C and 3A, respectively) argue that R892W is initially targeted to the same subcellular compartment as WT, presumably the endoplasmic reticulum, where dysfunctional dimers would be formed. According to preliminary data investigating TLR9 WT and R892W co-localization with labeled CpG-ODN (not shown), there is no statistically significant difference between TLR9 WT and R892W co-localization with labeled CpG-ODN in HEK293 cells, indicating that the molecular cause of the R892W loss of function lies elsewhere. Homology modeling predicts the R892W mutation to destabilize hydrogen bonds with Tyr-888 and Glu-896 (Fig. 1A and supplemental Fig. S3), leading to changes in surface charge and hydrophilicity (supplemental Fig. S3). Apparently, TLR9 homotypic TLR9 dimer formation was not affected (Fig. 3A) but rather heterotypic interaction with adaptors, for example MyD88. Our co-immunoprecipitation analysis suggests that the R892W mutation does not decrease this interaction but rather hints to a stronger association of the R892W mutant with MyD88 (Fig. 3B) that will need to be confirmed using, for example, biochemical methods in the future. Interestingly, a truncated TLR9 form that is defective in signaling on its own also strongly precipitated MyD88 (30), and for RIG-I signaling it was recently shown that an increased association observed for the variant RIG-I S183I with the adaptor Cardif also precluded downstream signaling (31). This suggests that dynamic adaptor association and dissociation may be required for optimum signaling in several PRR pathways and that an increased MyD88 interaction of R892W may contribute to blocking NF-κB activation. It cannot be ruled out that the additional association of TLR9 with protein kinase D1 (32), AP-3 (33, 34), or specific post-translational modification as reported for several other receptor TIR domains (35, 36) could also contribute to the hyporesponsive effect of R892W. The R892W mutant and the TLR9 TIR domain three-dimensional model reported here may serve as useful tools to more comprehensively characterize the structure-function relationships of the TLR9 TIR domain, which remain poorly understood.
Polymorphisms in TLR9 have been predicted to significantly contribute to disease in humans (17), but the immunological consequences of TLR9 variation so far have not been addressed. We present here a first analysis in primary cells of a novel but very rare TLR9 allele, R892W, which, similarly to rs5743844 (P99L), a TLR9 allele reported earlier (18), we found to be associated with complete loss of function effect in a genetic TLR9 complementation system in tissue culture. The range of effects of TLR9 stimulation in terms of IL-10, IL-6, and IFNα secretion or HLA-DR up-regulation in B cells and pDC from the allele carrier and control donors varied considerably, highlighting the complexity of the human experimental system. The comparison between allele carriers and control donors is informative in this regard, but comparisons of TLR9 allele carriers with multiple relatives of the same carriers would have been ideal for reducing the influence of genetic factors apart from TLR9 but unfortunately difficult to accomplish. Nevertheless, at this point we can conclude that the outcome of heterozygous TLR9 R892W carriage is not a general loss of responsiveness to CpG-ODN. For example, R892W B cells did not produce additional IL-10 or IL-6 (Fig. 4), but HLA-DR was moderately up-regulated (Fig. 5), and in R892W pDC robust IFNα production could be observed at high CpG-ODN concentrations (Fig. 5). On the other hand, our data from the HEK293 system and primary B cells indicate that heterozygote carriage may nevertheless have an impact on TLR9-mediated B cell-derived IL-10 and IL-6, whose production depends on NF-κB activation. Conversely, pDC responded with normal IFNα production, which, in pDC, is interferon regulatory factor-dependent and requires TLR9 signaling from early Transferrin receptor+ endosomes, whereas NF-κB-linked TLR9 signaling (e.g., required for the production of TNFα or cellular maturation) is thought to be initiated from endolysosomes in human pDC (37, 44). It was reported before that different TLR9 effector outcomes are differentially regulated, for example MCP-1 and IL-6 (38). Even across different dendritic cells, the spatiotemporal parameters for CpG-dependent responses differ, and it is unknown from which endosomal compartment TLR9 signaling is initiated in B cells (39). Further cell biological studies beyond the scope of this manuscript would be required to elucidate the precise molecular nature of the signaling defect observed for R892W. In the context of the proposed mechanistic interplay of TLR7 and TLR9 (40, 41), it is noteworthy that the effects of R848 stimulation mirror those observed for CpG-ODN stimulation in some (IL-10, IL-6, IFNα) but not all (HLA-DR) experiments. In interaction and stimulation studies in HEK293 cells, the presence of TLR9 was reported to reduce TLR7 responsiveness to R848 (40) and in a murine model of autoimmunity, (homozygous) TLR9 deficiency resulted in an exacerbated, TLR7-driven autoimmune phenotype (41), probably by influencing UNC-93B-mediated trafficking of the latter receptor (34). It is therefore conceivable that TLR9 R892W may directly or indirectly (via UNC-93B) also affect TLR7 in primary human cells. Future work on the interaction of TLR7 and TLR9 is, however, needed, to better understand and verify these findings in human primary cells.
Given the overall moderate heterozygous effects of R892W in immune cells observed here, one might conclude that genetic variability in TLR9 is relatively well tolerated or compensated for. On the other hand, the low genetic variability of TLR9 as evidenced by our sequencing approach may be an indication that heterozygous carriage of dysfunctional TLR9 alleles may still have adverse effects on a clinical level that were not captured here and that might entail a strong negative selective pressure. In this context, the medical history of the TLR9 allele carrier is noteworthy because it contains malignancy and recurring infections (suppurative throat anginas and tuberculosis of the ear; see the supplemental information) but not autoimmunity. At this point, we can only speculate whether TLR9 (partial) dysfunction may have contributed to the manifestation or progression of these diseases. Data on rare dysfunctional alleles in MyD88, interleukin-1 receptor-associated kinase 4, or UNC93B suggest that TLR-related gene defects only cause severe immunodeficiencies in homozygous individuals whose heterozygous siblings are not severely immunocompromised (42, 43). Our data confirm this notion for TLR9.
In conclusion, this present study to our knowledge is the first to functionally analyze TLR9 alleles in human primary cells. The obtained data clearly illustrate that the presence of the dysfunctional TLR9 allele R892W in heterozygous carriers does not result in a general loss of function in TLR9-dependent effector function but may selectively affect B lymphocyte-derived cytokine production. It remains to be investigated how TLR9 alleles influence the clinical course of natural infections or complex diseases in which TLR9 has been implicated rather than CpG-ODN treatment of isolated immune cells. Additionally, future studies may be able to address whether homozygous carriage of alleles like R892W is associated with immunodeficiency. The data presented here warrant such further studies into the significance of TLR9 genetic variation on a molecular, immunological, clinical, and epidemiological level.
Supplementary Material
Acknowledgments
We thank all our colleagues for helpful discussions and Julie George, Hui Wang, and Jody L. Cameron for technical advice on co-immunoprecipitations. We thank Dr. Melita Balija (Department for Transfusion Medicine, Petrova 3, Zagreb, Croatia), who assisted in the collection of blood samples.
This work was supported by Grants from the Croatian Ministry of Science, Education and Sports, the German Research Foundation, and the German Cancer Research Center.

This article contains supplemental Figs. S1–S3.
- ECD
- extracellular domain
- NF
- nuclear factor
- ODN
- oligodeoxynucleotide
- PBMC
- peripheral blood mononuclear cells
- pDC
- plasmacytoid dendritic cell(s)
- TIR
- Toll/interleukin-1 receptor
- TLR
- Toll-like receptor.
REFERENCES
- 1. Kawai T., Akira S. (2008) Toll-like receptor and RIG-I-like receptor signaling. Ann. N.Y. Acad. Sci. 1143, 1–20 [DOI] [PubMed] [Google Scholar]
- 2. Dembic Z. (2005) The Function of Toll-like Receptors: An Immunologic Perspective (Rich T., ed) pp. 18–55, Landes Bioscience Eurekah.com and Kluwer Academic/Plenum, Georgetown, TX [Google Scholar]
- 3. Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K., Akira S. (2000) A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 [DOI] [PubMed] [Google Scholar]
- 4. Chockalingam A., Brooks J. C., Cameron J. L., Blum L. K., Leifer C. A. (2009) TLR9 traffics through the Golgi complex to localize to endolysosomes and respond to CpG DNA. Immunol. Cell Biol. 87, 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gay N. J., Gangloff M., Weber A. N. (2006) Toll-like receptors as molecular switches. Nat. Rev. Immunol. 6, 693–698 [DOI] [PubMed] [Google Scholar]
- 6. Rutz M., Metzger J., Gellert T., Luppa P., Lipford G. B., Wagner H., Bauer S. (2004) Toll-like receptor 9 binds single-stranded CpG-DNA in a sequence- and pH-dependent manner. Eur. J. Immunol. 34, 2541–2550 [DOI] [PubMed] [Google Scholar]
- 7. Peter M. E., Kubarenko A. V., Weber A. N., Dalpke A. H. (2009) Identification of an N-terminal recognition site in TLR9 that contributes to CpG-DNA-mediated receptor activation. J. Immunol. 182, 7690–7697 [DOI] [PubMed] [Google Scholar]
- 8. Kubarenko A. V., Ranjan S., Colak E., George J., Frank M., Weber A. N. (2010) Comprehensive modeling and functional analysis of Toll-like receptor ligand-recognition domains. Protein Sci. 19, 558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Latz E., Verma A., Visintin A., Gong M., Sirois C. M., Klein D. C., Monks B. G., McKnight C. J., Lamphier M. S., Duprex W. P., Espevik T., Golenbock D. T. (2007) Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat. Immunol. 8, 772–779 [DOI] [PubMed] [Google Scholar]
- 10. Watters T. M., Kenny E. F., O'Neill L. A. (2007) Structure, function and regulation of the Toll/IL-1 receptor adaptor proteins. Immunol. Cell Biol. 85, 411–419 [DOI] [PubMed] [Google Scholar]
- 11. Krieg A. M. (2006) Therapeutic potential of Toll-like receptor 9 activation. Nat. Rev. Drug Discov. 5, 471–484 [DOI] [PubMed] [Google Scholar]
- 12. Bekeredjian-Ding I., Jego G. (2009) Toll-like receptors. Sentries in the B-cell response. Immunology 128, 311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang S. Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P., Segal D., Sancho-Shimizu V., Lorenzo L., Puel A., Picard C., Chapgier A., Plancoulaine S., Titeux M., Cognet C., von Bernuth H., Ku C. L., Casrouge A., Zhang X. X., Barreiro L., Leonard J., Hamilton C., Lebon P., Héron B., Vallée L., Quintana-Murci L., Hovnanian A., Rozenberg F., Vivier E., Geissmann F., Tardieu M., Abel L., Casanova J. L. (2007) TLR3 deficiency in patients with herpes simplex encephalitis. Science 317, 1522–1527 [DOI] [PubMed] [Google Scholar]
- 14. von Bernuth H., Picard C., Jin Z., Pankla R., Xiao H., Ku C. L., Chrabieh M., Mustapha I. B., Ghandil P., Camcioglu Y., Vasconcelos J., Sirvent N., Guedes M., Vitor A. B., Herrero-Mata M. J., Aróstegui J. I., Rodrigo C., Alsina L., Ruiz-Ortiz E., Juan M., Fortuny C., Yagüe J., Antón J., Pascal M., Chang H. H., Janniere L., Rose Y., Garty B. Z., Chapel H., Issekutz A., Maródi L., Rodriguez-Gallego C., Banchereau J., Abel L., Li X., Chaussabel D., Puel A., Casanova J. L. (2008) Pyogenic bacterial infections in humans with MyD88 deficiency. Science 321, 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Misch E. A., Hawn T. R. (2008) Toll-like receptor polymorphisms and susceptibility to human disease. Clin. Sci. 114, 347–360 [DOI] [PubMed] [Google Scholar]
- 16. El-Omar E. M., Ng M. T., Hold G. L. (2008) Polymorphisms in Toll-like receptor genes and risk of cancer. Oncogene 27, 244–252 [DOI] [PubMed] [Google Scholar]
- 17. Barreiro L. B., Ben-Ali M., Quach H., Laval G., Patin E., Pickrell J. K., Bouchier C., Tichit M., Neyrolles O., Gicquel B., Kidd J. R., Kidd K. K., Alcaïs A., Ragimbeau J., Pellegrini S., Abel L., Casanova J. L., Quintana-Murci L. (2009) Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 5, e1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kubarenko A. V., Ranjan S., Rautanen A., Mills T. C., Wong S., Vannberg F., Neumaier M., Bekeredjian-Ding I., Hill A. V., Ahmad-Nejad P., Weber A. N. (2010) A naturally occurring variant in human TLR9, P99L, is associated with loss of CpG oligonucleotide responsiveness. J. Biol. Chem. 285, 36486–36494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fraser D. A., Bulat-Kardum L., Knezevic J., Babarovic P., Matakovic-Mileusnic N., Dellacasagrande J., Matanic D., Pavelic J., Beg-Zec Z., Dembic Z. (2003) Interferon-γ receptor-1 gene polymorphism in tuberculosis patients from Croatia. Scand. J. Immunol. 57, 480–484 [DOI] [PubMed] [Google Scholar]
- 20. Bulat-Kardum L., Etokebe G. E., Knezevic J., Balen S., Matakovic-Mileusnic N., Zaputovic L., Pavelic J., Beg-Zec Z., Dembic Z. (2006) Interferon-γ receptor-1 gene promoter polymorphisms (G-611A; T-56C) and susceptibility to tuberculosis. Scand. J. Immunol. 63, 142–150 [DOI] [PubMed] [Google Scholar]
- 21. Xu Y., Tao X., Shen B., Horng T., Medzhitov R., Manley J. L., Tong L. (2000) Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature 408, 111–115 [DOI] [PubMed] [Google Scholar]
- 22. Jiang Z., Georgel P., Li C., Choe J., Crozat K., Rutschmann S., Du X., Bigby T., Mudd S., Sovath S., Wilson I. A., Olson A., Beutler B. (2006) Details of Toll-like receptor:adapter interaction revealed by germ-line mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 10961–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weber A. N., Morse M. A., Gay N. J. (2004) Four N-linked glycosylation sites in human toll-like receptor 2 cooperate to direct efficient biosynthesis and secretion. J. Biol. Chem. 279, 34589–34594 [DOI] [PubMed] [Google Scholar]
- 24. Leifer C. A., Verthelyi D., Klinman D. M. (2003) Heterogeneity in the human response to immunostimulatory CpG oligodeoxynucleotides. J. Immunother. 26, 313–319 [DOI] [PubMed] [Google Scholar]
- 25. George J., Kubarenko A. V., Rautanen A., Mills T. C., Colak E., Kempf T., Hill A. V., Nieters A., Weber A. N. (2010) MyD88 adaptor-like D96N is a naturally occurring loss-of-function variant of TIRAP. J. Immunol. 184, 3025–3032 [DOI] [PubMed] [Google Scholar]
- 26. George J., Motshwene P. G., Wang H., Kubarenko A. V., Rautanen A., Mills T. C., Hill A. V., Gay N. J., Weber A. N. (2011) Two human MYD88 variants, S34Y and R98C, interfere with MyD88-IRAK4-myddosome assembly. J. Biol. Chem. 286, 1341–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Picard C., von Bernuth H., Ghandil P., Chrabieh M., Levy O., Arkwright P. D., McDonald D., Geha R. S., Takada H., Krause J. C., Creech C. B., Ku C. L., Ehl S., Maródi L., Al-Muhsen S., Al-Hajjar S., Al-Ghonaium A., Day-Good N. K., Holland S. M., Gallin J. I., Chapel H., Speert D. P., Rodriguez-Gallego C., Colino E., Garty B. Z., Roifman C., Hara T., Yoshikawa H., Nonoyama S., Domachowske J., Issekutz A. C., Tang M., Smart J., Zitnik S. E., Hoarau C., Kumararatne D. S., Thrasher A. J., Davies E. G., Bethune C., Sirvent N., de Ricaud D., Camcioglu Y., Vasconcelos J., Guedes M., Vitor A. B., Rodrigo C., Almazán F., Méndez M., Aróstegui J. I., Alsina L., Fortuny C., Reichenbach J., Verbsky J. W., Bossuyt X., Doffinger R., Abel L., Puel A., Casanova J. L. (2010) Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine 89, 403–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chockalingam A., Cameron J. L., Brooks J. C., Leifer C. A. (2011) Negative regulation of signaling by a soluble form of toll-like receptor 9. Eur. J. Immunol. 41, 2176–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leifer C. A., Brooks J. C., Hoelzer K., Lopez J., Kennedy M. N., Mazzoni A., Segal D. M. (2006) Cytoplasmic targeting motifs control localization of toll-like receptor 9. J. Biol. Chem. 281, 35585–35592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ewald S. E., Lee B. L., Lau L., Wickliffe K. E., Shi G. P., Chapman H. A., Barton G. M. (2008) The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature 456, 658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pothlichet J., Burtey A., Kubarenko A. V., Caignard G., Solhonne B., Tangy F., Ben-Ali M., Quintana-Murci L., Heinzmann A., Chiche J. D., Vidalain P. O., Weber A. N., Chignard M., Si-Tahar M. (2009) Study of human RIG-I polymorphisms identifies two variants with an opposite impact on the antiviral immune response. PLoS One 4, e7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park J. E., Kim Y. I., Yi A. K. (2008) Protein kinase D1. A new component in TLR9 signaling. J. Immunol. 181, 2044–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blasius A. L., Arnold C. N., Georgel P., Rutschmann S., Xia Y., Lin P., Ross C., Li X., Smart N. G., Beutler B. (2010) Slc15a4, AP-3, and Hermansky-Pudlak syndrome proteins are required for Toll-like receptor signaling in plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 107, 19973–19978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sasai M., Linehan M. M., Iwasaki A. (2010) Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science 329, 1530–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ivison S. M., Khan M. A., Graham N. R., Bernales C. Q., Kaleem A., Tirling C. O., Cherkasov A., Steiner T. S. (2007) A phosphorylation site in the Toll-like receptor 5 TIR domain is required for inflammatory signalling in response to flagellin. Biochem. Biophys. Res. Commun. 352, 936–941 [DOI] [PubMed] [Google Scholar]
- 36. Rajagopal R., Waller A. S., Mendoza J. D., Wightman P. D. (2008) The covalent modification and regulation of TLR8 in HEK-293 cells stimulated with imidazoquinoline agonists. Biochem. J. 409, 275–287 [DOI] [PubMed] [Google Scholar]
- 37. Guiducci C., Ott G., Chan J. H., Damon E., Calacsan C., Matray T., Lee K. D., Coffman R. L., Barrat F. J. (2006) Properties regulating the nature of the plasmacytoid dendritic cell response to Toll-like receptor 9 activation. J. Exp. Med. 203, 1999–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baumann C. L., Aspalter I. M., Sharif O., Pichlmair A., Blüml S., Grebien F., Bruckner M., Pasierbek P., Aumayr K., Planyavsky M., Bennett K. L., Colinge J., Knapp S., Superti-Furga G. (2010) CD14 is a coreceptor of Toll-like receptors 7 and 9. J. Exp. Med. 207, 2689–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Honda K., Ohba Y., Yanai H., Negishi H., Mizutani T., Takaoka A., Taya C., Taniguchi T. (2005) Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature 434, 1035–1040 [DOI] [PubMed] [Google Scholar]
- 40. Wang J., Shao Y., Bennett T. A., Shankar R. A., Wightman P. D., Reddy L. G. (2006) The functional effects of physical interactions among Toll-like receptors 7, 8, and 9. J. Biol. Chem. 281, 37427–37434 [DOI] [PubMed] [Google Scholar]
- 41. Nickerson K. M., Christensen S. R., Shupe J., Kashgarian M., Kim D., Elkon K., Shlomchik M. J. (2010) TLR9 regulates TLR7- and MyD88-dependent autoantibody production and disease in a murine model of lupus. J. Immunol. 184, 1840–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Picard C., Puel A., Bonnet M., Ku C. L., Bustamante J., Yang K., Soudais C., Dupuis S., Feinberg J., Fieschi C., Elbim C., Hitchcock R., Lammas D., Davies G., Al-Ghonaium A., Al-Rayes H., Al-Jumaah S., Al-Hajjar S., Al-Mohsen I. Z., Frayha H. H., Rucker R., Hawn T. R., Aderem A., Tufenkeji H., Haraguchi S., Day N. K., Good R. A., Gougerot-Pocidalo M. A., Ozinsky A., Casanova J. L. (2003) Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science 299, 2076–2079 [DOI] [PubMed] [Google Scholar]
- 43. Casrouge A., Zhang S. Y., Eidenschenk C., Jouanguy E., Puel A., Yang K., Alcais A., Picard C., Mahfoufi N., Nicolas N., Lorenzo L., Plancoulaine S., Sénéchal B., Geissmann F., Tabeta K., Hoebe K., Du X., Miller R. L., Héron B., Mignot C., de Villemeur T. B., Lebon P., Dulac O., Rozenberg F., Beutler B., Tardieu M., Abel L., Casanova J. L. (2006) Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 314, 308–312 [DOI] [PubMed] [Google Scholar]
- 44. Chockalingam A., Rose W. A., Hasan M., Ju C.-H., Leifer C. A. (2012) Cutting edge: a TLR9 cytoplasmic tyrosine motif is selectively required for proinflammatory cytokine production. J. Immunol. 188, 527–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.