Background: The molecular basis for human α-defensin 5 (HD5) binding to non-enveloped viruses is unknown.
Results: Residues critical for virus binding and antiviral activity were identified by mutational analysis.
Conclusion: Multimerization, hydrophobicity, and specific arginine residues dictate HD5 antiviral activity.
Significance: These studies inform the role of enteric α-defensins in immunity against non-enveloped viruses.
Keywords: Adenovirus, Antimicrobial Peptides, Antiviral Agents, Defensins, Innate Immunity, Papillomavirus, HD5
Abstract
Human α-defensins, such as human α-defensin 5 (HD5), block infection of non-enveloped viruses, including human adenoviruses (AdV), papillomaviruses (HPV), and polyomaviruses. Through mutational analysis of HD5, we have identified arginine residues that contribute to antiviral activity against AdV and HPV. Of two arginine residues paired on one face of HD5, Arg-28 is critical for both viruses, while Arg-9 is only important for AdV. Two arginine residues on the opposite face of the molecule (Arg-13 and Arg-32) and unpaired Arg-25 are less important for both. In addition, hydrophobicity at residue 29 is a major determinant of anti-adenoviral activity, and a chemical modification that prevents HD5 self-association was strongly attenuating. Although HD5 binds to the capsid of AdV, the molecular basis for this interaction is undefined. Capsid binding by HD5 is not purely charge-dependent, as substitution of lysine for Arg-9 and Arg-28 was deleterious. Analysis of HD5 analogs that retained varying levels of potency demonstrated that anti-adenoviral activity is directly correlated with HD5 binding to the virus, confirming that the viral capsid rather than the cell is the relevant target. Also, AdV aggregation induced by HD5 binding is not sufficient for neutralization. Rather, these studies confirm that the major mechanism of HD5-mediated neutralization of AdV depends upon specific binding to the viral capsid through interactions mediated in part by critical arginine residues, hydrophobicity at residue 29, and multimerization of HD5, which increases initial binding of virus to the cell but prevents subsequent viral uncoating and genome delivery to the nucleus.
Introduction
Human α-defensins, which include human neutrophil peptides (HNPs)2 1–4 and human α-defensins (HD) 5 and 6, are a class of small antimicrobial peptides with well characterized antibacterial activity against both Gram-positive and Gram-negative organisms (1). More recently, the ability of human α-defensins to neutralize viruses has been described. For enveloped viruses, neutralization has generally been ascribed to disruption of the viral lipid envelope, steric interference with receptor binding, interaction with cellular factors, or less well defined post-entry blocks (2). In most cases, these activities are dependent upon the ability of α-defensins to perturb lipid bilayers or, for certain human α-defensins, to function as lectins. For non-enveloped viruses including human adenoviruses (AdV), papillomaviruses (HPV), and polyomaviruses, the proposed mechanisms of neutralization are distinct. For HPV, the virus appears to uncoat efficiently, but the viral genome fails to escape from the endosome during cell entry (3). For human polyomaviruses such as BK virus, α-defensin binding aggregates virions and blocks receptor binding (4). We have shown that for AdV, sensitivity to α-defensin-mediated neutralization is serotype-dependent (5, 6). α-Defensin binding to neutralized serotypes, such as AdV-5, stabilizes the virus capsid and prevents uncoating of the virus during cell entry, thereby blocking infection (5–7). In each case, the interaction between these non-enveloped viruses and α-defensins cannot be explained by the lipid-interacting or lectin properties of α-defensins. Thus, the molecular basis for α-defensin binding to non-enveloped virus capsids is undefined but likely based on specific protein-protein rather than protein-lipid or protein-carbohydrate interactions.
Despite considerable effort toward understanding the structural basis for α-defensin antibacterial activity, comparable studies have not been performed for viruses. Human α-defensins are cationic, amphipathic, and share a common fold with human β-defensins (1). The antibacterial activity of several human α-defensins has been shown to depend on both hydrophobicity and charge, although the relative importance of each of these factors is specific to each α-defensin/bacteria combination (8–12). In addition, there is an exclusive preference in human α-defensins for arginine residues over lysine, and substitution of lysine for arginine is deleterious (10). Another important factor for α-defensin function is the ability to multimerize both in solution and upon ligand binding (8, 12–14). To explore the contribution of these parameters to HD5 antiviral activity, we measured human AdV-5 and HPV16 neutralization by HD5 analogs with amino acid substitutions for arginine residues, variable hydrophobicity at residue 29, and a chemical modification that disrupts dimerization. These studies confirm that the viral capsid rather than a cellular factor is the relevant target for neutralization of infection. They also highlight the specificity of the α-defensin-capsid interaction, which is mediated in part by critical arginine residues on one face of the HD5 molecule and dependent on HD5 multimerization. These insights into HD5 binding to AdV and HPV may be common to other α-defensin-virus interactions and inform the basis for antiviral immunity mediated by these innate immune effectors.
EXPERIMENTAL PROCEDURES
Cells, Viruses, and Peptides
Tissue culture reagents were obtained from Mediatech (Manassas, VA) or Invitrogen. Human A549 and HeLa cells (ATCC) were propagated in DMEM supplemented with 10% fetal bovine serum (Sigma-Aldrich), 4 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.1 mm nonessential amino acids (complete DMEM). Stable 293 cells overexpressing the human β5 integrin subunit (293β5) were a kind gift from Glen Nemerow (The Scripps Research Institute, San Diego, CA) (7). 293TT cells were cultured in complete DMEM supplemented with 0.4 mg/ml hygromycin B and were a kind gift from John Schiller (National Cancer Institute, Bethesda, MD) (15, 16).
The replication-defective human AdV-5 vector used in these studies (AdV5.eGFP) is E1/E3-deleted and contains an enhanced green fluorescent protein (eGFP) reporter gene cassette driven by a CMV promoter. AdV5.eGFP was propagated in 293β5 cells and purified by CsCl gradient centrifugation as described previously (7). Purified AdV5.eGFP was dialyzed against three changes of 150 mm NaCl, 40 mm Tris, 10% glycerol, 2 mm MgCl2, pH 8.1, and stored at −80 °C. For use in experiments, virus aliquots were thawed on ice.
HPV16 pseudoviruses (PsVs) were produced and purified according to established protocols (15, 16). Briefly, plasmids encoding codon-optimized HPV16 L1 and L2 genes (p16L1L2) and a GFP reporter gene (pfwB) were co-transfected using Lipofectamine 2000 (Invitrogen) into 293TT cells. The cells were harvested at 72 h and lysed in Dulbecco's PBS, 9.5 mm MgCl2, 0.5% Triton X-100, 25 mm ammonium sulfate, and 0.1% RNase (Ambion). The lysate was incubated at 37 °C for 20–24 h to allow PsV maturation, and mature PsVs were purified by OptiPrep (Sigma-Aldrich) gradient centrifugation.
Synthetic HD5 was obtained from Peptides Intl., Inc. (Louisville, KY). Alternatively, folded HD5 was generated from a synthesized 80% pure linear peptide (CPC Scientific, Sunnyvale, CA) by thiol disulfide reshuffling overnight at room temperature in the presence of 3 mm reduced and 0.3 mm oxidized glutathione, 2 m guanidine hydrochloride, and 0.25 m sodium bicarbonate, pH 8.3, at a peptide concentration of 0.25 mg/ml, purified to homogeneity by reverse-phase high pressure liquid chromatography, and lyophilized as described previously (17). The synthesis, refolding, purification, and structural validation of the HD5 analogs have been described for R9A, R13A, R25A, R28A, R32A, E21me, and Leu-29 substitutions (12) and R9A/R28A, R9K/R28K, R13A/R32A, and R13K/R32K (9). All α-defensins were quantified by UV absorbance at 280 nm using calculated molar extinction coefficients (18).
Quantification of Virus Infection
Serial dilutions of AdV5.eGFP were used to infect A549 cell monolayers in black wall, clear bottomed 96-well plates (PerkinElmer Life Science). Total monolayer fluorescence was quantified with a Typhoon 9400 variable mode imager (GE Healthcare) 24–30 h post-infection. A virus concentration producing 50–80% maximal signal was chosen for inhibition studies.
To measure their effects on infectivity, increasing concentrations of α-defensins were incubated with purified AdV5.eGFP for 45 min on ice in serum-free DMEM (SFM). The mixture (35 μl/well) was then added to a confluent monolayer of A549 cells in black wall, clear bottomed 96-well plates that had been washed twice in SFM (protocol 1). Alternatively, A549 cell monolayers were incubated with AdV5.eGFP at 4 °C for 45 min, washed twice with SFM to remove unbound virus, and then incubated with increasing concentrations of α-defensins in SFM at 4 °C for 45 min (protocol 2). In both cases, cells were incubated at 37 °C for 2 h with rocking, washed, and then cultured with complete DMEM for 24–30 h. Plates were scanned for eGFP signal as above, and background-subtracted total well fluorescence was quantified using ImageJ software (W. S. Rasband, National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/). Fluorescence values were log-transformed and quantified by linear regression against a standard curve of cells infected with a serial dilution of AdV5.eGFP in the absence of α-defensin using Prism software (version 5.0d, GraphPad Software, Inc., La Jolla, CA). Experiments with HPV PsVs were performed as per protocol 1 above with the following exceptions: 1) infectivity was measured on HeLa cells, 2) the PsVs were incubated at 37 °C for 4 h prior to washing and removal of the inoculum, and 3) GFP was measured 48 h post-infection.
Binding of Virus/α-Defensin to Cells
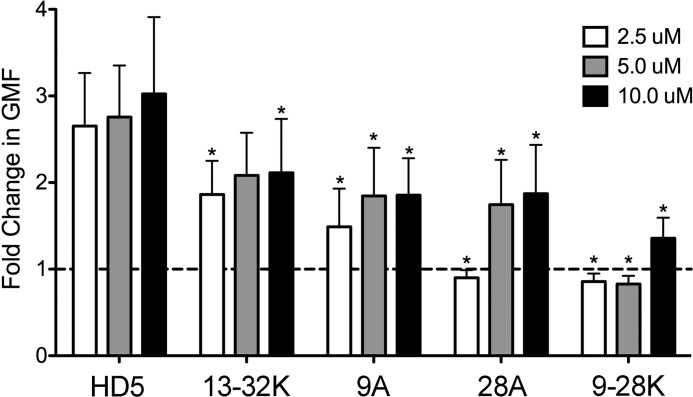
Alexa Fluor 488-labeled AdV5.eGFP (4 × 109 particles/sample) was prepared as described and incubated with or without 5, 10, or 20 μm of the α-defensins indicated in Fig. 2 for 45 min on ice in 50 μl of PBS (6). In parallel, 1.2 × 105 A549 cells/sample were incubated on ice in PBS containing 0.2% sodium azide to inhibit endocytosis. The virus/defensin mixtures were combined with the cells (final volume, 100 μl/sample), incubated on ice for 45 min, washed twice with cold PBS, fixed with 1% paraformaldehyde, and analyzed by flow cytometry for Alexa Fluor 488 signal (10,000 events). The fold increase in the geometric mean fluorescence of samples containing α-defensins was calculated relative to the geometric mean fluorescence of samples containing virus alone.
FIGURE 2.
α-Defensin interaction increases AdV5.eGFP binding to cells. Data are the fold change in the geometric mean fluorescence (GMF) of cells incubated with fluorescently labeled AdV5.eGFP in the presence of the indicated concentrations of HD5 and HD5 analogs relative to cells incubated with virus in the absence of α-defensin and are the means of three independent experiments ± S.D. HD5 analogs are labeled as in Fig. 1. *, p < 0.05.
Confocal Microscopy
Alexa Fluor 555-labeled AdV5.eGFP was prepared as described (6), and 3.0 × 109 particles/sample in ice-cold SFM were centrifuged at 5000 × g for 10 min at 4 °C to remove aggregates and incubated with subconfluent monolayers of A549 cells grown on glass coverslips. Samples were treated or not with 10 μm α-defensins as described in protocol 2 above and fixed with 4% paraformaldehyde for 15 min at room temperature either immediately before (0 min) or after incubating for 45 min at 37 °C. Samples were quench/permeabilized with 20 mm glycine, 0.5% Triton X-100 for 15 min at room temperature and stained sequentially with anti-lamin B1 rabbit polyclonal antibody (BD Biosciences) and Alexa Fluor 488-conjugated anti-rabbit polyclonal antibody (Invitrogen). Samples were mounted with ProLong gold (Invitrogen), and image z-series were obtained with a Zeiss 510 Meta laser scanning confocal microscope (Carl Zeiss, Inc., Thornwood, NY).
ImageJ was used for image analysis. Images in the z-series for each sample above and below the plane of the nucleus were discarded, and a maximum intensity z-profile of the remaining images was obtained. Thresholds for Alexa Fluor 488 and 555 signals above background compared with uninfected samples were determined. The same thresholds were used for all samples obtained during each microscopy session. Cell borders were delineated from bright field images. The outside border of the nucleus was delineated using the Alexa Fluor 488 threshold. The integrated density of the Alexa Fluor 555 signal in the nucleus was divided by that in the whole cell to obtain the percent nuclear localization of the virus for each cell. A total of 40–80 cells were analyzed for each condition.
Direct Measurement of α-Defensin Binding to Virus
Samples containing 10 μg of purified AdV5.eGFP and 10 μm α-defensins in a final volume of 80 μl of 150 mm NaCl were incubated for 1 h on ice. Virus was separated from unbound α-defensin by ultracentrifugation (209,000 × g for 1.5 h at 4 °C) on a discontinuous gradient of 300 μl of 30% Nycodenz (Sigma-Aldrich) and 200 μl of 80% Nycodenz in 20 mm Tris, pH 7.4. The visible virus band was collected and separated by acid-urea polyacrylamide gel electrophoresis. Gels were fixed for 1 h in 28% MeOH, 5% formaldehyde and stained with the manufacturer's rapid protocol using SYPRO-Ruby Protein Stain (Invitrogen). Gels were imaged on a Typhoon 9400 imager, and virus bands were quantified using ImageJ software. The fraction of input α-defensin that was bound to virus was determined for each HD5 analog, normalized to a viral protein band to account for virus recovery from the gradient, and expressed relative to the amount of bound, normalized wild type HD5.
Dynamic Light Scattering and Zeta Potential Analysis
α-Defensins were serially diluted in 10 mm Tris, 150 mm NaCl, pH 7.5, and mixed with 1.3 × 1010 particles/ml of virus in a final volume of 50 μl (dynamic light scattering) or 750 μl (zeta potential) using disposable cuvettes (Malvern Instruments, Westborough, MA). Control samples of AdV5.eGFP or α-defensin only were diluted in the same buffer. Samples were incubated for 1 h on ice, stored briefly at room temperature, and then equilibrated for 3 min at 37 °C prior to analysis. The polydispersity index and Z-average particle size were obtained by cumulant analysis, and the zeta potential was obtained using phase analysis light scattering (M3-PALS) with a Malvern Zetasizer Nano ZS and manufacturer's software (Malvern Instruments).
Statistical Analysis
Experiments were analyzed using Prism (version 5.0d). For Figs. 1 and 2 and Figs. 4–6, data were analyzed by two-way analysis of variance with Bonferroni post-tests to compare each mutant with wild type HD5 at each concentration. For Fig. 3, data were log transformed and analyzed by one-way analysis of variance with Dunnett post-tests to compare each mutant to the no HD5 control separately for each time point. For all tests, p < 0.05 was considered significant.
FIGURE 1.
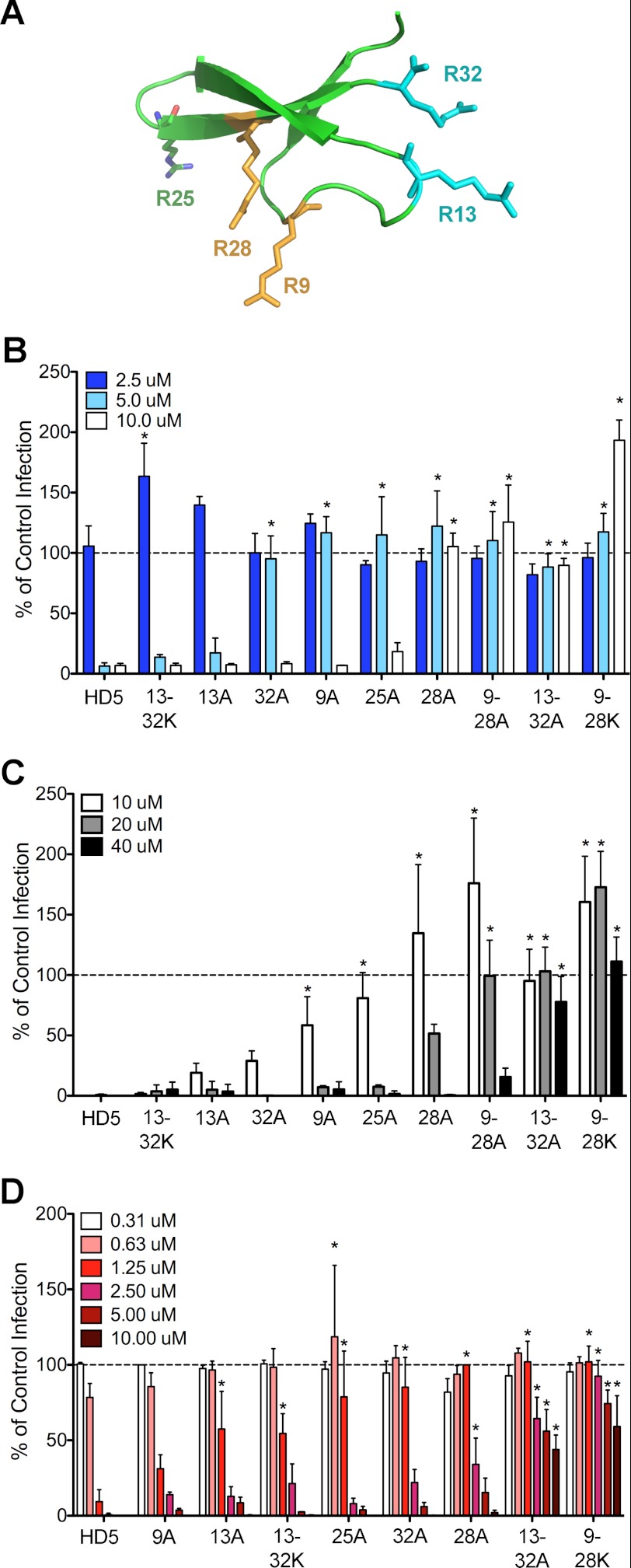
Antiviral activity of HD5 and HD5 analogs against AdV5.eGFP and HPV16. A, ribbon representation of HD5 (Protein Data Bank code 1ZMP). Arginine residues are numbered, depicted as ball and stick models, and color-coded by pairs. B, infection of A549 cells by AdV5.eGFP pre-bound to the indicated concentrations of α-defensins (protocol 1) is expressed relative to control cells infected in the absence of α-defensin (100%, dashed line). Substitutions in the HD5 analogs are labeled by the position of the arginine residue and the single letter code for the substituted amino acid. Double mutants are indicated by a dash. Data are the means of at least three independent experiments ± S.D. *, p < 0.05. C, AdV5.eGFP infection upon prebinding of virus to A549 cells prior to addition of the indicated concentrations of α-defensins (protocol 2) was quantified as described in B. D, infection of HeLa cells by HPV16 PsVs encapsidating a GFP reporter plasmid prebound to the indicated concentrations of α-defensins was quantified as described in B.
FIGURE 4.
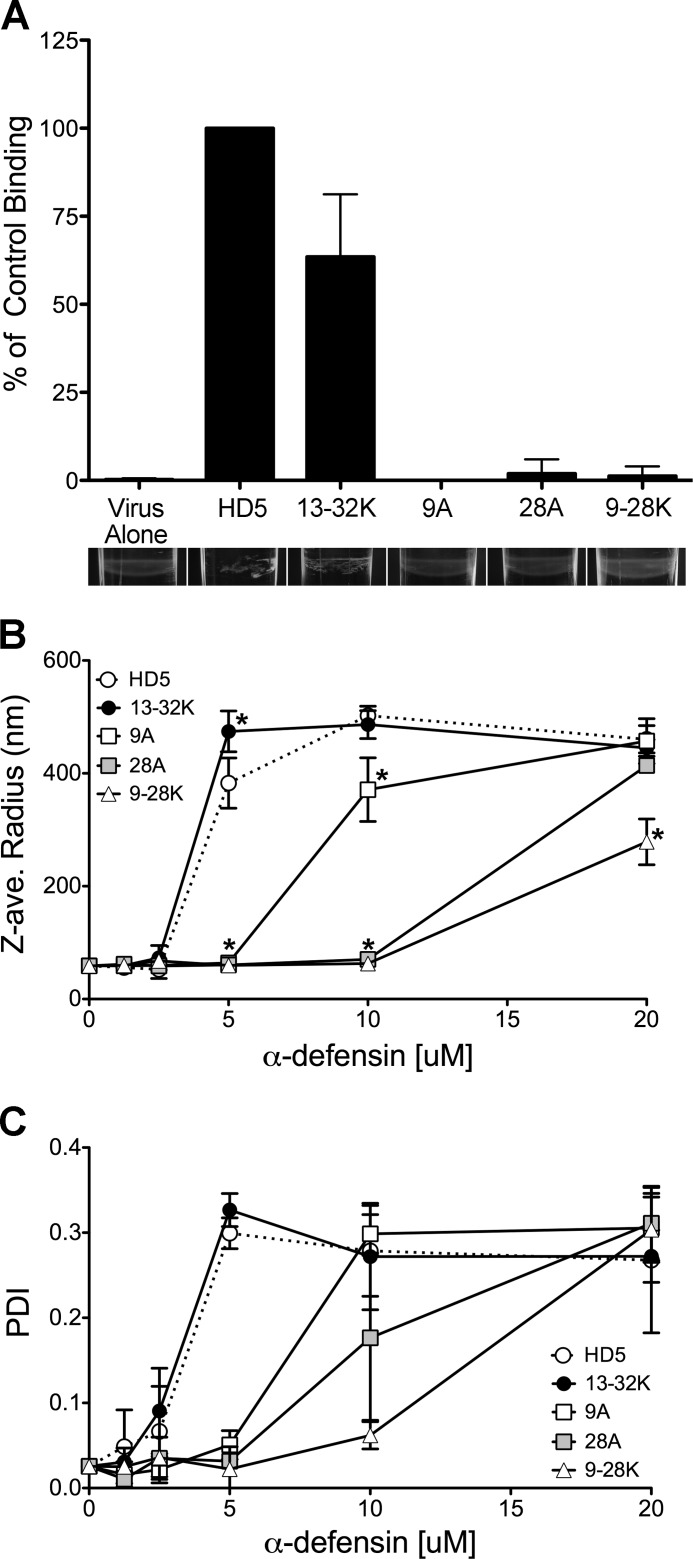
Binding of HD5 and HD5 analogs to AdV5.eGFP. A, binding of each HD5 analog to AdV5.eGFP after incubation at 10 μm on ice was quantified relative to that of wild type HD5 (set at 100%) and normalized to the amount of virus. Data are the means of at least three independent experiments ± S.D. HD5 analogs are labeled as in Fig. 1. Images below the graph for each α-defensin are an oblique view of a representative virus band in the Nycodenz gradient that was used to separate virus-bound and -unbound α-defensins. The Z-average (ave.) radius (B) and polydispersity index (PDI; C) generated from cumulant analysis of dynamic light scattering of AdV5.eGFP incubated with increasing concentrations of α-defensins labeled as in Fig. 1. Data are the means of at least three independent experiments ± S.D. *, p < 0.05.
FIGURE 5.
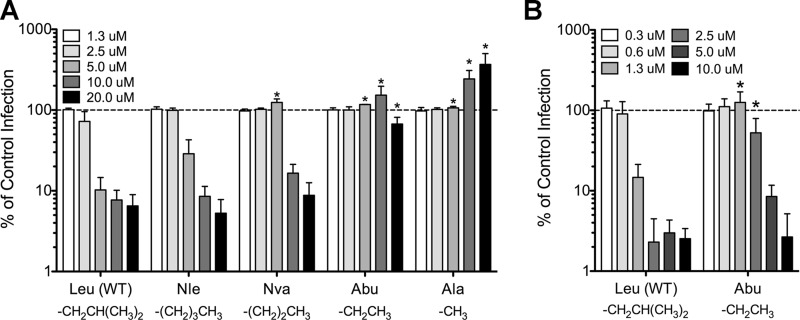
Effects of the hydrophobicity of HD5 residue 29 on antiviral activity. A, infection of A549 cells by AdV5.eGFP prebound to the indicated concentrations of HD5 analogs is expressed relative to control cells infected in the absence of α-defensin (100%, dashed line). The structure of each of the side chains is shown. Nle, norleucine; Nva, norvaline; Abu, α-aminobutyric acid. B, infection of HeLa cells by HPV16 prebound to the indicated concentrations of WT HD5 or the L29Abu analog. Data are the means of at least three independent experiments ± S.D. *, p < 0.05.
FIGURE 6.
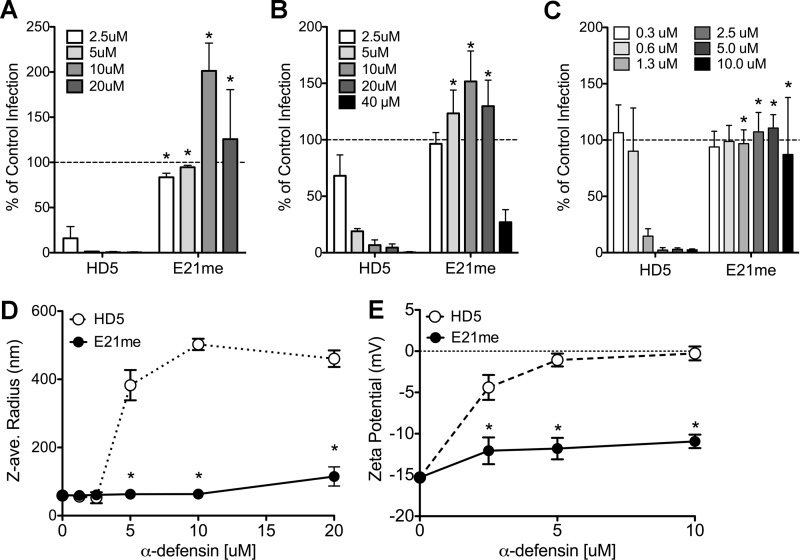
Effects of disrupting HD5 multimerization on antiviral activity and virus aggregation. A, infection of A549 cells by AdV5.eGFP pre-bound to the indicated concentrations of HD5 or an HD5 analog that is N-methylated on glutamate 21 (E21me); B, infection upon pre-binding of AdV.eGFP to A549 cells prior to addition of the indicated concentrations of HD5 or E21me; or C, infection of HeLa cells by HPV16 pre-bound to the indicated concentrations of E21me is expressed relative to control cells infected in the absence of α-defensin (100%, dashed line). Data are the means of at least three independent experiments ± S.D. D, the Z-average (ave.) radius generated from cumulant analysis of dynamic light scattering. E, the surface charge generated by M3-PALS analysis of AdV5.eGFP incubated with increasing concentrations of HD5 or E21me. Data are the means of at least three independent experiments ± S.D. The WT HD5 data in C and D are reproduced from Fig. 5B and 4B for comparison. *, p < 0.05.
FIGURE 3.
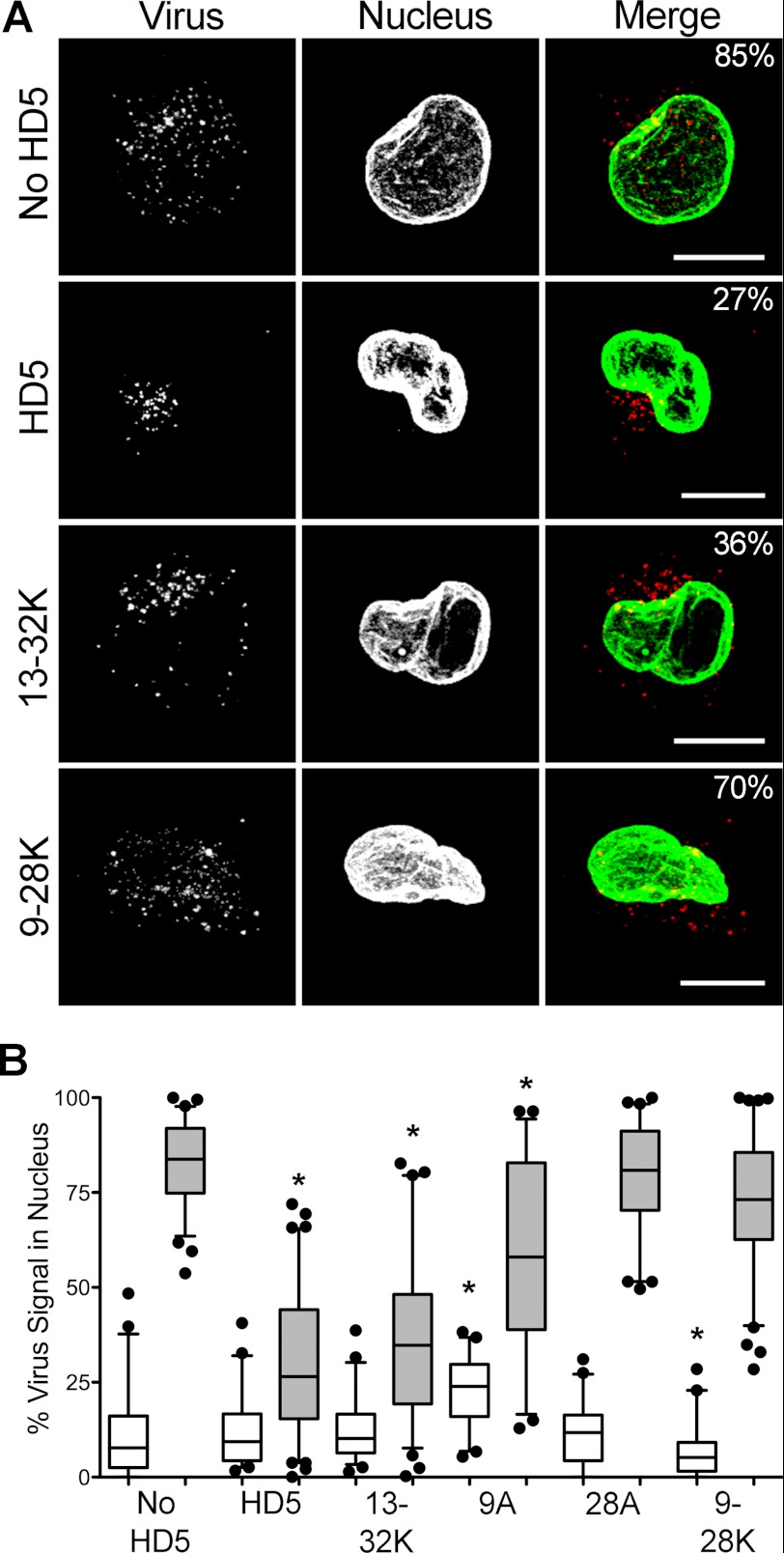
α-Defensins alter the subcellular localization of AdV5.eGFP during cell entry. A, images of cells 45 min post-infection with AdV5.eGFP, which was fluorescently labeled with Alexa Fluor 555 (red), in the presence or absence of 10 μm of the indicated α-defensins are representative of the mean viral nuclear localization for each data set. Signal above threshold for images in the z-stack that are co-planar with the nucleus are shown as maximum intensity z-projections. In the merge images, the virus is colored red, the nucleus is colored green, and the percent nuclear localization of the virus for each cell is indicated. The scale bar is 10 μm. B, virus localization to the nucleus was quantified for 40–80 cells at 0 min (white) and 45 min (shaded) post-infection for cells infected in the presence of 10 μm of the indicated α-defensins (labeled as in Fig. 1). Whiskers are 5–95%, the horizontal line is the mean, and outliers are depicted as individual points. *, p < 0.05.
RESULTS
Antiviral Activity of HD5 Is Sequence-specific and Not Purely Charge-dependent
To identify residues critical for viral neutralization, we measured the activity of HD5 analogs with single or pair-wise substitutions of arginine residues to either lysine or alanine in antiviral assays against a vector based on the defensin-sensitive AdV-5 serotype (AdV5.eGFP). We chose HD5 concentrations that are within the physiologic range and non-cytotoxic (19, 20). Four of the six arginine residues in HD5 are paired on opposite sides of the molecule, whereas Arg-25 is unpaired and Arg-6 forms a salt bridge with Glu-14 (Fig. 1A) (21, 22). In one assay (protocol 1, Fig. 1B), the defensins were first incubated with purified AdV5.eGFP, and the defensin/virus mixture was then added to A549 cells. In the second assay (protocol 2, Fig. 1C), Ad5.eGPF was incubated with A549 cells at 4 °C, washed to remove unbound virus, exposed to increasing concentrations of defensins for 45 min at 4 °C, and then warmed to allow virus entry and infection. Expression of eGFP from A549 cells after infection with each defensin/virus combination was quantified relative to control cells infected in the absence of defensin. Of the single arginine substitutions for alanine, R13A had little effect; R9A, R25A, and R32A were similarly attenuating; and R28A was the most deleterious (Fig. 1, B and C). Substitution of both Arg-9 and Arg-28 for lysine (R9K/R28K) completely eliminated antiviral activity at all concentrations tested; however, the R9A/R28A analog retained some activity at 40 μm. In contrast, the R13A/R32A analog had no activity, whereas the R13K/R32K analog had wild type activity. We observed the same trend in relative antiviral activity of the mutants in both assays. In some cases, subinhibitory concentrations of these analogs enhanced infection up to 2.5-fold.
To determine whether these results were true for other non-enveloped viruses, we measured the antiviral activities of the HD5 analogs against HPV16 PsVs. For these experiments (Fig. 1D), HPV16 PsVs were premixed with the defensins prior to being added to HeLa cells, as in protocol 1 above. Consistent with previous studies, the IC50 of wild type HD5 for HPV16 was 0.6–1.25 μm, ∼4-fold less than for AdV (3). Like for AdV, R28A was the most deleterious single substitution, R25A and R32A were moderately attenuating, and R9K/R28K was the most strongly attenuated. However, unlike AdV, R9A had little effect, R13A and R13K/R32K were moderately deleterious, and R13A/R32A retained some activity at high concentrations. Taken together, these data indicate that Arg-28 and, to a lesser extent, Arg-9 are critical for antiviral activity and that interactions with AdV and HPV are not purely charge-dependent, as the charge-neutral substitution of lysine for these arginine residues abrogated HD5-mediated neutralization of infection.
HD5 Enhances AdV Binding to Cells
Our current model of HD5-mediated neutralization of AdV is that HD5 binding to the capsid blocks escape of the virus from the endosome (6, 7). Rather than reducing cell binding, the virus-defensin interaction increases association of the virus with the cell surface compared with virus alone, in both a receptor-dependent and -independent manner, and has no effect on the kinetics of virus internalization (5, 6). Similarly, HD5 has been shown to increase HIV binding to the cell surface (23). To determine whether the HD5 analogs affect receptor interactions, we compared the amount of fluorescently labeled AdV5.eGFP bound with cells in the presence or absence of wild type HD5 or HD5 analogs representative of a range of antiviral activity (Fig. 2). As in previous studies, HD5 enhanced cell binding 2.4–2.8-fold compared with virus alone (5). This effect was dose-dependent, and a subinhibitory concentration of HD5 (2.5 μm) was still enhancing. Despite having wild type antiviral activity, R13K/R32K was less potent in this assay, although more cell binding was still observed at all concentrations ≥2.5 μm. R9A also increased binding at ≥2.5 μm. In contrast, incubation of virus with subinhibitory concentrations of R28A and R9K/R28K resulted in a small but reproducible reduction in cell binding; however, both of these analogs enhanced binding at concentrations for which we observed more infection (Fig. 1, B and C). Thus, like for wild type HD5, the HD5 analogs increase virus binding to the cell rather than block receptor interactions. Enhanced binding is observed at subinhibitory concentrations and likely contributes to increased infection.
HD5 Binding Alters Intracellular Trafficking of AdV
In previous studies, we established that HD5 prevents viral escape from the endosome by blocking viral uncoating and release of the membrane lytic internal capsid protein VI (6). Thus, despite enhanced binding to the cell, infection is blocked once a threshold concentration of HD5 capable of inhibiting uncoating is reached. To determine whether the changes in neutralization activity that we observed for the HD5 analogs impact this mechanism, we examined the intracellular trafficking of fluorescently labeled AdV5.eGFP in A549 cells in the presence and absence of 10 μm HD5 or selected HD5 analogs (Fig. 3). Cells were fixed immediately after warming to initiate virus internalization (0 min) and 45 min post-internalization, stained for the nuclear marker lamin B1, and analyzed by confocal microscopy for colocalization of the virus with the nucleus. In the absence of HD5, ∼83% of virus was colocalized with the nucleus at 45 min post-infection, whereas virus incubated with HD5 was concentrated in a perinuclear compartment with a concomitant reduction in nuclear colocalization (∼29%), consistent with previous studies (6, 7). The effects of the analogs on intracellular virus trafficking were closely correlated with their antiviral activities (Fig. 1C). At 45 min post-infection, the nuclear colocalization of virus incubated with R13K/R32K was equivalent to that of virus incubated with HD5. In contrast, virus trafficking in the presence of R28A or R9K/R28K was equivalent to infection in the absence of defensin. R9A had an intermediate effect on the ability of the virus to reach the nucleus. The close correlation between the impact of the HD5 mutations on this assay and on infection supports our model that HD5 exerts its antiviral effect by preventing escape of the virus from the endosome. Moreover, within each cell population, there was a continuum of cells containing virus partially colocalized with the nucleus, rather than a biphasic distribution of cells into those with virus either strongly colocalized with the nucleus or not. Thus, defensin binding seems to affect the phenotype of individual virions rather than individual cells, supporting our conclusion that the target of HD5 is the viral capsid and not the cell.
AdV Neutralization Correlates with HD5 Binding
To measure the defensin interaction with virus directly, HD5 and its analogs were incubated with AdV5.eGFP to allow binding, and virus-defensin complexes were separated from unbound defensin in a Nycodenz density gradient. Bound defensin was visualized after acid-urea PAGE using a fluorescent total protein stain and quantified against a standard curve (Fig. 4A). In control experiments, all of the defensin analogs were visible with the fluorescent stain (data not shown). Binding of R13K/R32K to AdV5.eGFP was reduced ∼40% compared with HD5, whereas that of R9A, R28A, and R9K/R28K was at or below the limit of detection of this assay. Furthermore, binding correlated with visible aggregation of the virus band in the Nycodenz gradient (Fig. 4A). Thus, α-defensin binding to the virus correlates with neutralization activity; however, the sensitivity of this assay is not sufficient to detect the small amounts of R9A, R28A, and R9K/R28K that contribute to enhanced cell binding and enhanced infection (Figs. 1 and 2).
Given the limited sensitivity of the previous assay, we quantified virus aggregation by dynamic light scattering analysis as an alternative measure of the interaction between AdV5.eGFP and the α-defensins. (Fig. 4, B and C). In the absence of α-defensin, the hydrodynamic (Z-) radius of AdV5.eGFP was 58 nm, which closely approximates the size of the viral particle. The particles were of uniform size and well dispersed in solution with a polydispersity index of ∼0.02. Upon incubation with HD5, we observed a shift in the average Z-radius of the population to a maximum of 475 nm and an increase in the polydispersity index to >0.27 at antiviral defensin concentrations. The entire population shifted, with >95% of the particles in the major peak. The aggregation properties of R13K/R32K were equivalent to that of HD5, consistent with its antiviral activity. R9A-mediated aggregation was absent at 5 μm, intermediate at 10 μm, and equivalent to HD5 at 20 μm. R28A and R9K/R28K were only capable of aggregating AdV5.eGFP at 20 μm. The aggregates consist of both virus and defensin, as there was no dose-dependent aggregation in solutions of HD5 alone. These results support our model that HD5 activity is directly proportional to its ability to bind to the virus capsid. In addition, although aggregation may contribute to neutralization, it is not sufficient, as R9K/R28K aggregates viral particles at 20 μm but fails to neutralize infection at this concentration.
Hydrophobicity of Residue 29 Is a Strong Determinant of Antiviral Activity
We also tested the contribution of HD5 hydrophobicity to antiviral activity against AdV. In the myeloid α-defensin HNP1, an alanine scan identified Trp-26 as a critical residue for antibacterial activity (8). A similar study identified Leu-29 in HD5 as critical (12). We tested HD5 analogs containing natural and artificial amino acid residues with side chains of increasing hydrocarbon chain length at position 29 (Fig. 5). We observed a positive correlation between hydrophobicity and anti-adenoviral activity. The norleucine (Nle) analog had wild type activity. Substitution of norvaline (Nva) was ∼2-fold attenuating, and the α-aminobutyric acid (Abu) analog retained some activity but was >8-fold less potent (Fig. 5A). Remarkably, L29A had no antiviral activity and enhanced AdV5.eGFP infection ∼3.5-fold at the highest concentrations. For HPV16, L29Abu was ∼4-fold less potent (Fig. 5B). Thus, hydrophobicity at this position is crucial for antiviral activity.
Neutralization Is Dependent on HD5 Multimerization
HD5 self-associates in solution and upon ligand binding into higher-order oligomers (14). The pair-wise substitutions of alanine for arginine and changes in hydrophobicity at position 29 in HD5 have been shown to affect HD5 self-association, in addition to their potential to perturb the initial binding of HD5 to the viral capsid (12, 14). Because our assays are unable to distinguish the contribution of these two binding events to virus neutralization, we tested the antiviral activity of an HD5 analog with selective disruption of the HD5 dimer interface. N-methylation of Glu-21 (E21me) disrupts HD5-HD5 hydrogen bonding, analogous to N-methylation of isoleucine 20 in HNP1 (13). E21me crystallizes as a monomer but has a charge and conformation unaltered from HD5 (12). We observed >8-fold reduction in antiviral activity for E21me against AdV5.eGFP (Fig. 6, A and B) and >16-fold reduction for HPV16 (Fig. 6C).
We were unable to detect direct E21me binding to the capsid (data not shown), and dynamic light scattering analysis revealed minimal aggregation of virus (to double the size of unbound virus) only upon incubation with 20 μm E21me (Fig. 6D). To determine whether disruption of dimerization precludes all binding of HD5 to the capsid or only accumulation of HD5 on the capsid through HD5-HD5 multimerization, we measured the effect of increasing concentrations of HD5 and the E21me analog on the zeta potential or surface charge of AdV5.eGFP (Fig. 6E). With wild type HD5, we observed a marked increase in the surface charge of AdV5.eGFP from an initial value of −15 mV. This effect was half-maximal below 2.5 μm, saturated near neutrality at 5 μm HD5 and was coincident with aggregation (Fig. 6D). In contrast, the E21me mutant produced a moderate neutralization of the capsid to −12 mV, which saturated at 2.5 μm and was not coincident with aggregation. Taken together, these results suggest that monomeric HD5 is capable of binding to the adenoviral capsid but that self-association is critical for virus aggregation and antiviral activity.
DISCUSSION
These studies provide the first identification of α-defensin residues that are critical for non-enveloped virus neutralization. Our initial assumption, based in part on previous studies demonstrating that super-physiologic salt concentrations disrupt both the interaction of defensins with AdV and antiviral activity (6), was that binding to viral capsids would be predominantly dependent upon charge-charge interactions mediated by the positively charged arginine residues in HD5. In support of this hypothesis, each of the individual substitutions of alanine for arginine was moderately to severely attenuating, with the exception of R9A for HPV16 and R13A for both viruses. However, the charge-neutral substitution of lysine for arginine had opposite effects depending on the pair of arginine residues for which lysine was substituted. Thus, although charge-charge interactions likely contribute to virus binding and antiviral activity, other non-covalent interactions are also important. Because all of the HD5 analogs for which structures have been obtained retain the wild type conformation with only minor perturbations due to side chain substitutions, it is unlikely that reduced functionality is due to misfolding (9, 12, 22). Arginine and lysine differ in their capacity to form hydrogen bonds, as the decentralized charge of arginine makes it a better hydrogen bond partner (24). Furthermore, the two residues differ in their hydrophobicity (25), which changes the energetics of solvent exclusion or surface burial during folding, ligand binding, and multimerization. Thus, the strong selection for arginine over lysine among human α-defensins may reflect the importance of these non-covalent interactions in HD5 function (10). Taken together, these studies highlight the selectivity of the defensin-AdV interaction, which is likely mediated by charge-charge interactions and hydrogen bonding.
Our analysis identified the same residues in HD5 as critical for antiviral activity that were identified previously as important for antibacterial and lectin activity (9, 12, 14). Given that the modes of inhibition of non-enveloped viruses and bacteria are thought to be distinct, it is surprising that similar residues are important for both processes. One possibility is that the interaction of HD5 with viral capsids is based on carbohydrate recognition rather than protein binding. A subset of AdV-5 fibers is O-glycosylated; however, as AdV serotypes that lack glycosylation also are neutralized, it is unlikely that this modification is an important determinant (5, 6, 26). Furthermore, the HPV16 capsid is not glycosylated. A more likely explanation is that the effect of the mutations is predominantly on HD5 multimerization, which is required for both antiviral and antibacterial activity, albeit through different mechanisms. Wild type HD5 self-associates in solution with a Kd of 2.2 μm (14). The pair-wise substitutions of alanine for Arg-13/Arg-32 and Arg-9/Arg-28 have been shown to impair HD5 self-association by ∼8-fold and ∼13-fold, respectively (14). In addition, reduction of hydrophobicity at Trp-26 in HNP1 dramatically destabilizes the dimer interface (8, 13), and altering the hydrophobicity of Leu-29 in HD5 changes the geometry of dimerization (12). Thus, mutation of these residues could affect both monomer binding of HD5 to the capsid and HD5 self-association, making it difficult to dissect the individual contributions of these two binding activities to antiviral function. To address this issue, we analyzed the E21me mutant that is defective in self-association but otherwise unaltered compared with wild type HD5 (12). Our finding that the E21me mutant has greatly attenuated antiviral activity provides evidence that HD5 multimerization is required for virus neutralization, and an intimate link between these two activities is supported by the close correlation between the IC50 of HD5 (2.5–5 μm) against AdV and its Kd of HD5 self-association (2.2 μm). Consistent with this model, the activity of the R9A/R28A analog is restored at 40 μm, which is above the Kd of self-association, although this does not hold true for the R13A/R32A analog. The differing capacities of wild type HD5 and the E21me mutant to neutralize the surface charge of AdV suggest that monomeric HD5 can bind to the capsid, but that accumulation through self-association is required for antiviral activity. Moreover, AdV aggregation occurs upon HD5 accumulation due to almost complete neutralization of the net negative capsid, which does not occur in the absence of HD5-HD5 binding. Intriguingly, HPV16 inhibition by HD5 occurs at submicromolar concentrations, although disruption of multimerization still abrogates HD5 antiviral activity. Thus, HPV binding may alter the conformation of HD5 in a manner that increases its affinity for itself. Our results, in combination with previous studies, indicate that the importance of multimerization may be a general feature of HD5 activity.
We have shown that infection by non-neutralized serotypes of AdV is often enhanced by HD5 and that non-neutralizing HD5 analogs enhance AdV5.eGFP infection (5). We have also shown that antiviral HD5 analogs counter-intuitively enhance virus binding to the cell. This could be due in part to charge neutralization that facilitates apposition of the virus and the cell surface, similar to the enhanced transduction and cell-binding phenotype of poly-lysine-coated AdV-5 (27); however, other mechanisms cannot be excluded. Thus, binding sites that enhance infection may differ from neutralizing sites. Identification of these sites remains unresolved.
Our studies support the hypothesis that the relevant target for HD5 that mediates neutralization is the viral capsid. However, for some viruses, α-defensins block infection through inhibition of a cellular target rather than through a direct interaction with the virus. One example is protein kinase C inhibition by HNP1, which impairs HIV infection (28). The correlation between binding to the virus and neutralization activity from our previous studies provided strong evidence that the virus rather than the cell is the target for HD5 inhibition (6). The resistance of certain AdV serotypes and chimeric viruses supports this conclusion but may also be due to differences in the cellular entry pathways of these viruses rather than their direct interactions with HD5 (5). Although we have not formally excluded the possibility that the HD5 mutations that affect virus binding in our current study also impact binding to a cellular target, the strong correlation between the capacity of the HD5 analogs to bind to virus, block translocation to the nucleus, and attenuate infection provides compelling corroborating evidence that the virus is the relevant target of HD5.
HD5 aggregates AdV-5, providing an indirect measure of HD5 binding to the virus capsid; however, aggregation alone does not block infection. In contrast, for the polyomavirus BK, aggregation has been proposed as the major mechanism of neutralization (4). For AdV, aggregation is dependent upon defensin multimerization and co-incident with capsid neutralization. However, we also observe aggregation at α-defensin concentrations that do not block infection (e.g. 10 μm R9A and 20 μm R9A/R28K). Furthermore, both preincubation of virus with HD5 prior to addition to cells and preincubation of virus with cells prior to addition of HD5 result in reduced infection. In the latter case, the virus is monodispersed on the cell surface and bound to its cellular receptor prior to the addition of HD5, and the virus in these samples is not visibly aggregated (data not shown and Ref. 7). Thus, aggregation often correlates with neutralization but is neither necessary nor sufficient to block virus infection.
These studies provide the first description of the molecular determinants for α-defensin binding to and neutralization of non-enveloped viruses. The importance of multimerization to antiviral activity may be critical for other α-defensin interactions with proteins. Although it is tempting to generalize these findings, additional mechanistic studies of HD5 with other defensin-sensitive viruses and of AdV and HPV16 with additional α-defensins are required to define common principles that dictate the broadly antiviral activity of these innate immune effectors.
Acknowledgments
We thank John D. Scott, Kim A. Woodrow, and Suzie H. Pun of the University of Washington for generously sharing equipment.
This work was supported, in whole or in part, by National Institutes of Health Grants K22 AI081870 (to J. G. S.), T32 AI083203 (to S. S. W.), and R01 AI072732 (to W. L.).
- HNP
- human neutrophil peptide
- AdV
- human adenovirus
- eGFP
- enhanced green fluorescent protein
- HD
- human defensin
- HPV
- human papillomavirus
- PALS
- phase analysis light scattering
- Z-radius
- hydrodynamic radius
- SFM
- serum-free DMEM
- Abu
- α-aminobutyric acid
- E21me
- N-methylation of Glu-21.
REFERENCES
- 1. Lehrer R. I., Lu W. (2012) α-Defensins in human innate immunity. Immunol. Rev. 245, 84–112 [DOI] [PubMed] [Google Scholar]
- 2. Klotman M. E., Chang T. L. (2006) Defensins in innate antiviral immunity. Nat. Rev. Immunol. 6, 447–456 [DOI] [PubMed] [Google Scholar]
- 3. Buck C. B., Day P. M., Thompson C. D., Lubkowski J., Lu W., Lowy D. R., Schiller J. T. (2006) Human α-defensins block papillomavirus infection. Proc. Natl. Acad. Sci. U.S.A. 103, 1516–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dugan A. S., Maginnis M. S., Jordan J. A., Gasparovic M. L., Manley K., Page R., Williams G., Porter E., O'Hara B. A., Atwood W. J. (2008) Human α-defensins inhibit BK virus infection by aggregating virions and blocking binding to host cells. J. Biol. Chem. 283, 31125–31132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith J. G., Silvestry M., Lindert S., Lu W., Nemerow G. R., Stewart P. L. (2010) Insight into the mechanisms of adenovirus capsid disassembly from studies of defensin neutralization. PLoS Pathog. 6, e1000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith J. G., Nemerow G. R. (2008) Mechanism of adenovirus neutralization by human α-defensins. Cell Host Microbe 3, 11–19 [DOI] [PubMed] [Google Scholar]
- 7. Nguyen E. K., Nemerow G. R., Smith J. G. (2010) Direct evidence from single-cell analysis that human α-defensins block adenovirus uncoating to neutralize infection. J. Virol. 84, 4041–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wei G., Pazgier M., de Leeuw E., Rajabi M., Li J., Zou G., Jung G., Yuan W., Lu W. Y., Lehrer R. I., Lu W. (2010) Trp-26 imparts functional versatility to human α-defensin HNP1. J. Biol. Chem. 285, 16275–16285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Leeuw E., Rajabi M., Zou G., Pazgier M., Lu W. (2009) Selective arginines are important for the antibacterial activity and host cell interaction of human α-defensin 5. FEBS Lett. 583, 2507–2512 [DOI] [PubMed] [Google Scholar]
- 10. Zou G., de Leeuw E., Li C., Pazgier M., Li C., Zeng P., Lu W. Y., Lubkowski J., Lu W. (2007) Toward understanding the cationicity of defensins. Arg and Lys versus their noncoded analogs. J. Biol. Chem. 282, 19653–19665 [DOI] [PubMed] [Google Scholar]
- 11. Llenado R. A., Weeks C. S., Cocco M. J., Ouellette A. J. (2009) Electropositive charge in α-defensin bactericidal activity: functional effects of Lys-for-Arg substitutions vary with the peptide primary structure. Infect. Immun. 77, 5035–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rajabi M., Ericksen B., Wu X., de Leeuw E., Zhao L., Pazgier M., Lu W. (2012) Functional determinants of human enteric α-defensin HD5: Crucial role for hydrophobicity at dimer interface. J. Biol. Chem. 287, 21615–21627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pazgier M., Wei G., Ericksen B., Jung G., Wu Z., de Leeuw E., Yuan W., Szmacinski H., Lu W. Y., Lubkowski J., Lehrer R. I., Lu W. (2012) Sometimes it takes two to tango: Contributions of dimerization to functions of human α-defensin HNP1 peptide. J. Biol. Chem. 287, 8944–8953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lehrer R. I., Jung G., Ruchala P., Andre S., Gabius H. J., Lu W. (2009) Multivalent binding of carbohydrates by the human alpha-defensin, HD5. J. Immunol. 183, 480–490 [DOI] [PubMed] [Google Scholar]
- 15. Buck C. B., Pastrana D. V., Lowy D. R., Schiller J. T. (2004) Efficient intracellular assembly of papillomaviral vectors. J. Virol. 78, 751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Buck C. B., Thompson C. D., Pang Y. Y., Lowy D. R., Schiller J. T. (2005) Maturation of papillomavirus capsids. J. Virol. 79, 2839–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu Z., Ericksen B., Tucker K., Lubkowski J., Lu W. (2004) Synthesis and characterization of human α-defensins 4–6. J. Pept. Res. 64, 118–125 [DOI] [PubMed] [Google Scholar]
- 18. Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghosh D., Porter E., Shen B., Lee S. K., Wilk D., Drazba J., Yadav S. P., Crabb J. W., Ganz T., Bevins C. L. (2002) Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol 3, 583–590 [DOI] [PubMed] [Google Scholar]
- 20. Ayabe T., Satchell D. P., Wilson C. L., Parks W. C., Selsted M. E., Ouellette A. J. (2000) Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 1, 113–118 [DOI] [PubMed] [Google Scholar]
- 21. Rajabi M., de Leeuw E., Pazgier M., Li J., Lubkowski J., Lu W. (2008) The conserved salt bridge in human α-defensin 5 is required for its precursor processing and proteolytic stability. J. Biol. Chem. 283, 21509–21518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Szyk A., Wu Z., Tucker K., Yang D., Lu W., Lubkowski J. (2006) Crystal structures of human α-defensins HNP4, HD5, and HD6. Protein Sci 15, 2749–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rapista A., Ding J., Benito B., Lo Y. T., Neiditch M. B., Lu W., Chang T. L. (2011) Human defensins 5 and 6 enhance HIV-1 infectivity through promoting HIV attachment. Retrovirology 8, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Borders C. L., Jr., Broadwater J. A., Bekeny P. A., Salmon J. E., Lee A. S., Eldridge A. M., Pett V. B. (1994) A structural role for arginine in proteins: Multiple hydrogen bonds to backbone carbonyl oxygens. Protein Sci. 3, 541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karplus P. A. (1997) Hydrophobicity regained. Protein Sci. 6, 1302–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cauet G., Strub J. M., Leize E., Wagner E., Van Dorsselaer A., Lusky M. (2005) Identification of the glycosylation site of the adenovirus type 5 fiber protein. Biochemistry 44, 5453–5460 [DOI] [PubMed] [Google Scholar]
- 27. Wang C. H., Chan L. W., Johnson R. N., Chu D. S., Shi J., Schellinger J. G., Lieber A., Pun S. H. (2011) The transduction of Coxsackie and adenovirus receptor-negative cells and protection against neutralizing antibodies by HPMA-co-oligolysine copolymer-coated adenovirus. Biomaterials 32, 9536–9545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chang T. L., Vargas J., Jr., DelPortillo A., Klotman M. E. (2005) Dual role of α-defensin-1 in anti-HIV-1 innate immunity. J. Clin. Invest. 115, 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]