Background: FOXC1 is associated with breast cancer aggressiveness and the basal-like breast cancer subtype but the mechanism through which FOXC1 increases aggressiveness has not been elucidated.
Results: FOXC1 induces expression of matrix metalloprotease 7 (MMP7).
Conclusion: The aggressive cancer phenotype imparted by FOXC1 is due, at least in part, to expression of MMP7.
Significance: MMP7 represents a putative target for the treatment of some basal-like breast cancers.
Keywords: Breast Cancer, Cancer, Cancer Biology, Invasion, Matrix Metalloproteinase (MMP), FOXC1, MMP7, Matrilysin, Basal-like Breast Cancer, Forkhead
Abstract
Therapeutic options for treatment of basal-like breast cancers are limited and identification of molecular targets for novel therapies to treat this aggressive cancer is urgently needed. Recently, FOXC1, a forkhead box transcription factor, was identified as a functionally important biomarker of breast cancer aggressiveness and the basal-like breast cancer subtype. However, the mechanism through which FOXC1 controls aggressiveness of basal-like breast cancer remains to be elucidated. Here, we identify matrix metalloprotease 7 (MMP7) as a key downstream effector of FOXC1-mediated invasiveness. Expression of FOXC1 and MMP7 is significantly correlated in breast cancer samples and cell lines at both the mRNA and protein levels. Transient expression of FOXC1 in nontransformed mammary epithelial cell lines resulted in significantly increased expression of MMP7 and an MMP7-dependent increase in invasiveness. In reciprocal experiments, silencing endogenous FOXC1 in basal-like breast cancer cell lines resulted in decreased expression of MMP7 without decreased expression of other matrix metalloproteinases. We also demonstrate that elevated co-expression of FOXC1 and MMP7 is an independent predictor of patient outcome in multivariate analyses of two breast cancer patient cohorts. Together, our findings identify MMP7 as a novel mechanism through which FOXC1 may regulate the basal-like breast cancer invasive phenotype and the propensity of these cancers to metastasize. Furthermore, our findings demonstrate for the first time a correlation between MMP7 expression and basal-like breast cancers, suggesting that MMP7 may be a useful therapeutic target for treatment of this disease.
Introduction
Targeted therapies have improved survival for many women diagnosed with breast cancer. Sex hormone receptor directed therapies have increased survival for women with estrogen receptor and/or progesterone receptor positive breast cancers, whereas treatments that disrupt human epidermal growth factor receptor 2 (HER2)2 signaling have increased life expectancy of women with HER2 amplified cancers (1–4). However, 15–20% of breast cancers do not express estrogen or progesterone receptors or HER2 (i.e. triple negative) and currently have no effective targeted treatment option (5, 6). The majority of triple negative breast cancers can be classified by gene expression profiling as belonging to the basal-like breast cancer (BLBC) subtype (7). These BLBC/triple negative tumors are among the most aggressive breast cancers and tend to be high grade, exhibit pushing boarders and are prone to metastasize (8–10). Identification of the essential regulators of BLBC is a critical step toward developing targeted therapies for this disease.
Members of the forkhead box (FOX) family of transcription factors regulate a wide array of biological processes including development, differentiation, and invasion (11). FOXC1 (Mf1, FKHL7, FREAC3) was originally identified as an important transcription factor that controls development of structures derived from the neural crest and FOXC1 mutations have long been recognized as a primary cause of Axenfeld-Rieger syndrome (12–14). In addition to its roles in normal function and development of the eye and meninges, FOXC1 has recently emerged as a possible master regulator of BLBC. Although one report has demonstrated decreased invasion and metastasis of breast cancer cells in response to FOXC1 expression (15), other groups have reported that FOXC1 increases invasion and metastasis in endometrial and breast cancer models (16–19). Stable overexpression of FOXC1 elicits changes in gene expression indicative of epithelial to mesenchymal transition (EMT) and increases cellular invasion, migration, and proliferation (16–19). In addition, FOXC1 expression correlates with the BLBC subtype and predicts poor breast cancer patient outcome (17, 20, 21). A better understanding of the mechanisms though which FOXC1 regulates aggressive cancer phenotypes should lead to the identification of new therapeutic targets for BLBC.
Although it has recently been suggested that FOXC1 may dictate the BLBC phenotype in part through NF-κB signaling (19), the mechanisms utilized by FOXC1 to increase cancer aggressiveness remain to be fully elucidated. Here, we show that transient expression of FOXC1, like the previously reported stable expression of FOXC1 (17, 18), increases invasion of nontransformed mammary epithelial cell lines. However, unlike stable FOXC1 expression, increased invasion imparted by transient FOXC1 expression is not accompanied by increased proliferation, migration, or EMT. These observations suggested that whereas FOXC1 can induce EMT, this transition was not the underlying cause of FOXC1-induced invasion. We postulated that FOXC1 increases cellular invasion through induction of a Matrigel degrading enzyme such as a member of the matrix metalloproteinase (MMP) family. Analyses of publicly available gene expression data sets as well as breast cancer cell line panels revealed a significant correlation between expression of FOXC1 and the matrix metalloproteinase, MMP7 (Matrilysin, PUMP-1). Enforced FOXC1 expression significantly increases MMP7 expression in nontransformed MCF10A and MCF12A cells. Furthermore, the increased invasion of MCF10A cells in response to FOXC1 expression required sustained expression of MMP7. Confirming the link between FOXC1 and MMP7, silencing endogenous FOXC1 in basal-like breast cancer cell lines results in decreased MMP7 expression. Last, high FOXC1 expression in breast cancers is associated with a significant decrease in patient survival when accompanied by high expression of MMP7 and elevated co-expression of FOXC1 and MMP7 is an independent predictor of patient outcome in multivariate analyses of two breast cancer patient cohorts. Together, these data suggest a novel mechanism though which FOXC1 increases cancer aggressiveness and implicate MMP7 as a putative therapeutic target in BLBC.
EXPERIMENTAL PROCEDURES
Cell Culture
Nontransformed mammary epithelial and breast cancer cell lines were acquired from American Type Culture Collection. MCF10A and MCF12A cells were grown in DMEM/F-12 medium supplemented with 5% horse serum, insulin (0.01 mg/ml), EGF (20 ng/ml), cholera toxin (100 ng/ml), and hydrocortisone (500 ng/ml). BT474, BT549, HCC70, HCC1143, HCC1187, HCC38, MDA-MB-231, and T47D cells were cultured in RPMI media supplemented with 10% FBS. MCF7, MDA-MB-435, MDA-MB-453, and MDA-MB-468 cells were cultured in DMEM supplemented with 10% FBS. SKBR3 cells were cultured in McCoy's 5A media supplemented with 10% FBS. All growth medias contained 1% l-glutamine, 1% penicillin, and 1% streptomycin.
Transient FOXC1 Overexpression
FOXC1 cDNA was cloned into pcDNA3.1 (Invitrogen). MCF10A and MCF12A cells were transiently transfected with empty pcDNA3.1 vector as control or pcDNA3.1/FOXC1 using Lipofectamine 2000 (Invitrogen) per the manufacturer's protocol. Adenoviruses encoding FOXC1 (AdFOXC1) or GFP (AdGFP) as a control were purchased from Welgen. Cells were transduced with 500 multiplicity of infection units of adenovirus unless otherwise noted.
FOXC1 Silencing
Endogenous FOXC1 and MMP7 were silenced with siRNA SMARTpools (Dharmacon). A nontargeting siRNA (sc-44230; Santa Cruz) served as control in all siRNA experiments. Lipofectamine 2000 (Invitrogen) was used as the siRNA delivery agent. In experiments in which MMP7 was silenced in conjunction with FOXC1 overexpression, control or MMP7-targeting siRNA was utilized at a concentration of 200 nm; otherwise transfections were conducted per the manufacturer's recommendations.
FOXC1 was also silenced by shRNAs. Vectors encoding shFOXC1#2 (TRCN0000235692), shFOXC1#3 (TRCN0000235692), shFOXC1#4 (TRCN0000235693), and a control vector encoding an shRNA to luciferase, shluc (SHC007), were purchased from Sigma and used to generate lentiviral particles (Wistar Institute). Cells were transduced at 1 multiplicity of infection in the presence of Polybrene (5 μg/ml) for 24 h. Cells were then allowed to recover in normal growth medium for 48 h at which time RNA was harvested as previously described (22) or cells were selected for stable shRNA expression with puromycin.
Western Blotting and Immunodetection
Generation of protein lysates and Western blotting were performed as previously described (22). FOXC1 antibodies were purchased from Santa Cruz (C-18) or Cell Signaling (number 7415). MMP7 antibodies were purchased from Calbiochem (141-7B2) or R&D Systems (AF907). β-Actin antibody was purchased from Sigma (AC-15). HRP-conjugated secondary antibodies to mouse, rabbit, and goat were purchased from Santa Cruz (sc-2005, sc-2054, and sc-2020).
Quantitative Real-time PCR
Total RNA was extracted and used to generate cDNA as previously described (22). Quantitative real-time PCR was conducted using an Applied Biosystems (ABI) Step-One real-time PCR instrument by the comparative Ct method. GAPDH was used as an endogenous control for all experiments. The following ABI gene expression assays were used for quantitative RT-PCR: FOXC1 (Hs00559473_s1), MMP7 (Hs01042793_m1), MMP2 (Hs00234422-m1), MMP9 (Hs00957562_m1), SNAI1 (Hs00195591_m1), SNAI2 (Hs00950344_m1), TWIST1 (Hs00361186_m1), VIM (HS00185584_m1), FN1 (Hs00365052_m1), CDH2 (Hs00365052_m1), CDH1 (Hs00170423_m1), CCND1 (Hs00277039_m1), and GAPDH (Hs99999905_m1). Data were averaged from at least three independent experiments, each conducted in triplicate. Statistical differences in gene expression were assessed by Student's t test.
Migration and Invasion Assays
MCF10A and MCF12A cells were transfected and/or transduced 48 h prior to the start of migration or invasion protocols. Migration was assessed using culture well inserts from Costar. Invasion was assayed using Matrigel-coated Transwell chambers (BD Biosciences). For migration experiments, 5 × 104 cells in serum, insulin, and EGF-free media were plated on the upper chamber of the insert. For invasion experiments, 1.25 × 105 cells were seeded in the upper chamber. For invasion and migration experiments, the lower chamber was filled with complete growth medium as a chemoattractant. Migration experiments for MCF10A and MCF12A proceeded for 24 h. Invasion of MCF10A cells was evaluated at 24 h, whereas the less invasive MCF12A cell line was allowed to invade for 48 h. At the end of the migration or invasion experiment, cells that had migrated/invaded were stained with the Diff-Quik stain kit (BD Biosciences) and photographed under ×10 magnification. Cells were counted in quadruplicate fields of view in duplicate or triplicate membranes. Data were averaged from at least three independent experiments and statistical differences in migration and invasion were determined using Student's t test.
Casein and Gelatin Zymography
Cells were transfected as indicated and 48 h later were washed twice with PBS and once with serum-free media followed by 24 h incubation in serum-free media. Gelatin and casein zymography were performed as described previously (22, 23).
Gene Expression Microarray Analysis
Microarray data sets were retrieved from Oncomine (oncomine.org). For co-expression analysis between FOXC1 and members of the MMP and tissue inhibitor of metalloproteinase (TIMP) families, data sets were pared to those using Affymetrix HG-U133A or U133 Plus 2.0 arrays. The resulting 20 data sets utilized for further analysis are listed in supplemental Table S1. Pearson correlation coefficients between FOXC1 and members of the MMP and TIMP families were calculated and used to determine covariance of these factors in each data set. The overall significance of covariance between FOXC1 and each MMP and TIMP family member was determined by Fisher's combined probability test with values <0.05 considered significant.
Survival Analysis
Data from the Sørlie et al. (24) and van de Vijver et al. (25) data sets were retrieved from Oncomine, stratified by high (upper quartile) or low (remaining three quartiles) expression of FOXC1 and MMP7, and overall survival was analyzed over a five-year period. Kaplan-Meier survival curves were generated and the log-rank test was used for statistical comparison of survival curves between groups. Univariate and stepwise multivariate analyses were carried out using the Cox proportional hazards model. Clincopathological variables included in analysis of the Sørlie et al. (24) data set were M Stage, N stage, T stage, grade, and age, whereas N stage was the lone variable available for the van de Vijver et al. (25) data set.
RESULTS
Transient FOXC1 Expression Increases Invasion without Inducing EMT
Stable overexpression of FOXC1 has been shown to increase cancer cell invasion as well as elicit changes in gene expression that are indicative of EMT (17, 18), yet it is unclear whether EMT is required for the induction of invasion observed with stable FOXC1 expression as well as the association of FOXC1 with breast cancer aggressiveness. We hypothesized that analysis of early changes in gene expression associated with transient overexpression of FOXC1 would uncover immediate downstream transcriptional targets of FOXC1 that induce EMT and may reveal key effectors that increase invasion. To begin to test this hypothesis, we transiently overexpressed FOXC1 in the nontransformed mammary epithelial cell line, MCF10A, utilizing a cationic liposome-based reagent (Fig. 1A). Similar to the results reported for cells stably overexpressing FOXC1, transient FOXC1 overexpression causes a significant increase in cellular invasion (4.5-fold; p < 0.001) in modified Boyden chamber assays (Fig. 1B). Relative migration of MCF10A cells is also significantly increased by transient FOXC1 overexpression (1.4-fold; p < 0.01; Fig. 1C). However, the increase in migration was not nearly as robust as, and could not fully account for, the increase in invasion. In contrast to previous reports using stable FOXC1 overexpression, transient overexpression of FOXC1 did not increase the expression of genes associated with EMT (Fig. 1D). Cells were analyzed for changes in transcription factors known to mediate EMT, including SNAI1, SNAI2, and TWIST, with no significant changes observed. In addition, the expression of VIM or FN1, two genes frequently up-regulated during EMT whose expression is increased with stable FOXC1 expression (17, 18), were not changed. Finally, a “cadherin switch” (e.g. loss of epithelial E-cadherin with a concomitant increase in a mesenchymal cadherin), which is associated with EMT and has been observed with stable FOXC1 expression (17, 18), did not occur.
FIGURE 1.
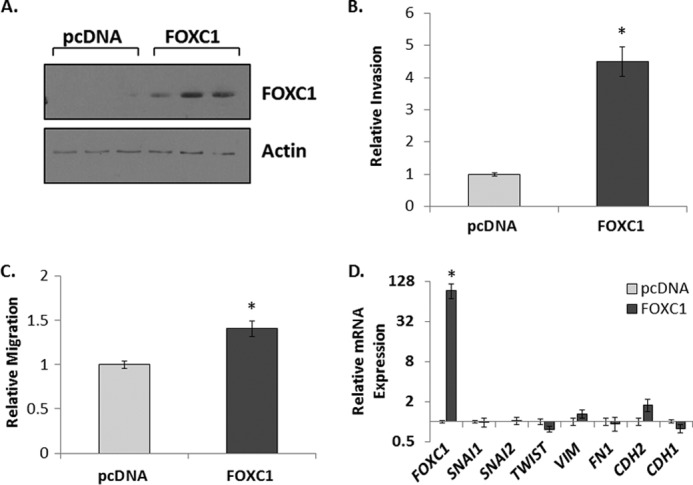
Transient expression of FOXC1 increases invasion of MCF10A Cells. A, representative immunoblots demonstrating transient overexpression of FOXC1 in MCF10A cells transfected with empty vector (pcDNA) or vector encoding FOXC1 (FOXC1). B, relative invasion of MCF10A cells transfected with pcDNA as control or pcDNA/FOXC1. C, relative migration of MCF10A cells transfected with pcDNA as control or pcDNA/FOXC1. D, relative transcript levels of EMT-related genes normalized to pcDNA control cells with GAPDH used as an endogenous control. Data represent at least three independent experiments and are presented as mean ± S.E. *, p < 0.05.
Cationic liposome-based transfection agents can influence cell behavior and gene expression (26). To ensure that the apparent lack of EMT in response to FOXC1 was not dependent on the mode of transient transfection, we confirmed these results using adenovirus as a gene delivery agent. Invasion of MCF10A cells was increased following transduction with a FOXC1 encoding adenovirus (4-fold, p < 0.01; supplemental Fig. S1, A and B). In contrast, no significant change in migration (1.6-fold; p = 0.14) or changes in EMT-associated gene expression occurred in response to FOXC1 expression (supplemental Fig. S1, C and D). Last, these data were confirmed using a second nontransformed mammary epithelial cell line, MCF12A. Although MCF12A cells are less invasive in vitro than MCF10A cells, a significant increase in the invasive capacity of MCF12A cells occurs in response to FOXC1 overexpression compared with controls in a 48-h invasion assay (4.3-fold; p < 0.01; supplemental Fig. S2, A and B). Again, no significant change in migration (1.3-fold, p = 0.36) or EMT-associated gene expression was detected (supplemental Fig. S2, C and D). Furthermore, transient expression of FOXC1 in MCF10A or MCF12A cells did not induce the fibroblast-like morphology typically observed with EMT (data not shown). Stable expression of FOXC1 has been shown to increase cell proliferation (17, 19). To determine whether changes in cell number could account for the increase in invaded cells, we analyzed changes in cell number of MCF10A and MCF12A cells transduced with GFP control or FOXC1 expressing adenovirus over a 6-day period. Transient expression of FOXC1 in MF10A cells significantly (p < 0.05 at day 6) reduced the number of viable cells when compared with cells transduced by control virus (supplemental Fig. S3A) indicating that the increase in invasion imparted by FOXC1 expression was not due to an increase in cell number. In MCF12A cells, transient expression of FOXC1 had no effect on cell number (supplemental Fig. S3B), further supporting the conclusion that transient FOXC1 expression in MCF10A and MCF12A directly increases invasion of breast epithelial cells.
FOXC1 Expression Correlates with Expression of MMP7
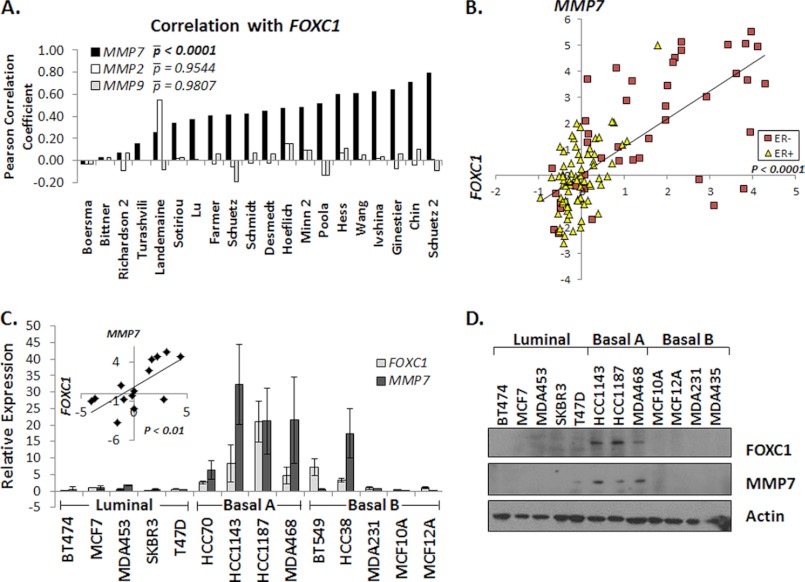
The observation that FOXC1 significantly increases invasion through Matrigel without increasing the cell number or robustly increasing migration suggested that transient FOXC1 expression increases expression and/or activity of an enzyme capable of degrading Matrigel such as members of the MMP family. To determine whether FOXC1 expression increases expression of one or more MMP or decreases expression of a member of the TIMP family, we evaluated correlations between expression of FOXC1 and 20 members of the MMP family along with four TIMPs by analyzing 20 publicly available human breast cancer gene expression microarray data sets (supplemental Table S1). To avoid platform-associated variance, all of the data sets included in the meta-analysis used Affymetrix U133A or U133 Plus 2.0 arrays. Twenty different datasets were used in this analysis, comprising >2,300 breast cancer samples. Although stable expression of FOXC1 in vitro has been shown to increase expression of the gelatinases MMP2 and -9 (20), no significant correlation between FOXC1 and either MMP2 or MMP9 was observed across the datasets (p = 0.9544 and p = 0.9807, respectively; Fig. 2A). In contrast, we found that MMP7 was the only MMP or TIMP family member whose expression was significantly correlated with the expression of FOXC1 (p < 0.0001; Fig. 2A and supplemental Table S2). Further analysis of individual data sets revealed not only a significant correlation between FOXC1 and MMP7 expression, but also that both FOXC1 and MMP7 were preferentially expressed in ER-negative breast cancers (Fig. 2B and supplemental Fig. S4A). Expression of FOXC1 and MMP7 was also significantly correlated (p < 0.01) in a panel of 14 breast cancer cell lines and nontransformed mammary epithelial cell lines (Fig. 2C). The correlated expression of FOXC1 and MMP7 in breast cancer cell lines was further confirmed in two public gene expression array datasets (27, 28) (supplemental Fig. S4, B and C). Furthermore, expression of FOXC1 and MMP7 proteins is highly correlated in 14 breast cancer and nontransformed mammary epithelial cell lines (Fig. 2D). Notably, the breast cancer cell lines that exhibited the highest expression of FOXC1 and MMP7 are those classified as basal A, whereas luminal and basal B/mesenchymal-like breast cancer cells lines showed low or moderate expression of FOXC1 and MMP7 (27, 28). Basal A cell lines are most like BLBC, whereas basal B lines are more similar to the rare claudin-low breast cancer subtype. Of note, there are several conflicting reports regarding the relative expression level of the FOXC1 protein in breast cancer cell lines (15, 17, 29). To exclude the possibility that the data presented herein was associated with poor antibody specificity, these data were confirmed using additional antibodies to FOXC1 and MMP7 (supplemental Fig. S4D). Together, these data demonstrate that FOXC1 and MMP7 expression is highly correlated in breast cancer tissues as well as cell lines.
FIGURE 2.
Expression of FOXC1 positively correlates with MMP7 expression in breast cancers and breast cancer cell lines. A, bar graph illustrating the Pearson correlation coefficients for FOXC1 and MMP7 (solid black bars), MMP2 (white bars), or MMP9 (gray bars) in 20 breast cancer gene expression data sets. Average p values (|p) were determined using Fisher's method of combined probabilities. B, log(2) transformed, median centered data from Chin et al. (see supplemental Table 1) was plotted by FOXC1 (x axis) and MMP7 (y axis) and colored coded according to estrogen receptor (ER) status. C, relative FOXC1 (light gray) and MMP7 (dark gray) expression in a panel of breast cancer and nontransformed mammary epithelial cell lines. Cell lines are grouped by subtype as defined by Neve et al. (27). Data represent at least three independent experiments and are presented as mean ± S.D. Average FOXC1 (x axis) and MMP7 (y axis) values were plotted (inset). D, representative Western blots demonstrating FOXC1 and MMP7 protein expression in a number of breast cancer and nontransformed mammary epithelial cell lines. p values less than 0.05 were considered statistically significant.
Overexpression of FOXC1 Induces MMP7 Expression
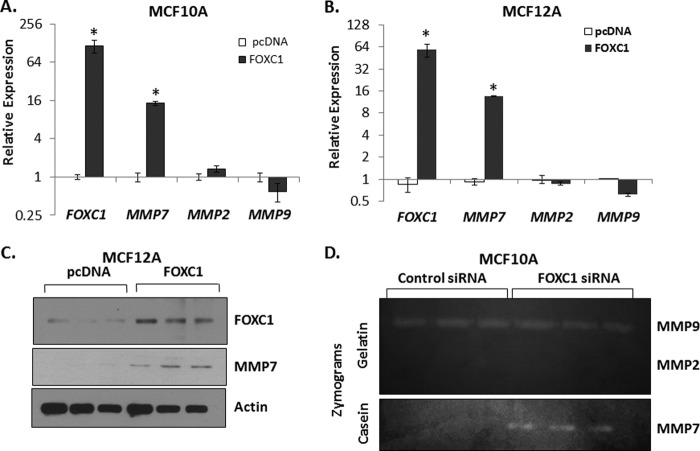
The significant correlation between FOXC1 and MMP7 expression in breast cancer samples and cell lines and the ability of FOXC1 overexpression to induce invasion suggested that FOXC1 regulates the expression of MMP7. To test this possibility, FOXC1 was transiently overexpressed and the impact on MMP7 expression was assessed. Transient overexpression of FOXC1 resulted in a significant increase in MMP7 mRNA in both MCF10A (14.5-fold; p < 0.05) and MCF12A (13.6-fold; p < 0.05) cells (Fig. 3, A and B), whereas MMP2 and MMP9 expression were unaffected by overexpression of FOXC1. MMP7 protein expression was also increased in response to FOXC1 overexpression in both MCF10A and MCF12A cells (Fig. 3C and data not shown). Zymography utilizing gelatin as a substrate for MMP2 and MMP9 or β-casein as an MMP7 substrate revealed that FOXC1 overexpression increases the amount of MMP7 protein in MCF10A- or MCF12A-conditioned media, but does not increase the amount of secreted MMP2 or MMP9 (Fig. 3D and data not shown). We confirmed that FOXC1 overexpression significantly increased the amount of MMP7 detectable in the conditioned media of MCF12A cells by ELISA (19.4-fold; p < 0.05; supplemental Fig. S5A). These data were further confirmed using adenoviral-mediated FOXC1 overexpression (supplemental Fig. S5, B and C). Last, the ability of FOXC1 to increase MMP7 expression was dose-dependent (supplemental Fig. S5D).
FIGURE 3.
Transient overexpression of FOXC1 in MCF10A or MCF12A cells results in increased expression of MMP7. Transcript levels of FOXC1, MMP7, MMP2, and MMP9 in MCF10A (A) or MCF12A (B) cells transiently transfected with empty vector (pcDNA) or vector encoding FOXC1. Data are relative to pcDNA; GAPDH was used as an endogenous control. Data summarize three independent experiments and are presented as mean ± S.D. C, representative Western blots demonstrating expression of FOXC1 and MMP7 in MCF12A cells transiently transfected with control (pcDNA) vector or vector encoding FOXC1. D, representative gelatin or casein zymograms demonstrating the relative levels of MMP9, MMP2, or MMP7 in conditioned media from MCF10A cells transfected with empty vector (pcDNA) or vector encoding FOXC1. *, p < 0.05.
Increased Invasion Imparted by FOXC1 Expression Requires Induction of MMP7
To determine whether the increase in invasion imparted by FOXC1 expression in MCF10A and MCF12A cells is dependent upon increased expression of MMP7, we overexpressed FOXC1 while blocking the induction of MMP7. This was accomplished by transducing MCF10A cells with control adenovirus encoding GFP or adenovirus encoding FOXC1 and simultaneously transfecting the cells with a nontargeting siRNA or siRNAs targeting MMP7 (Fig. 4A). Transfection with siRNA to MMP7 resulted in a 3.8-fold reduction (p < 0.01) in MMP7 expression compared with the nontargeting siRNA. More importantly, the siRNA targeting MMP7 significantly mitigated the increase in MMP7 expression that occurs in response to FOXC1 overexpression (3.5-fold decrease compared with AdFOXC1 + Control siRNA; p < 0.01). As shown above, transduction of MCF10A cells with FOXC1-expressing adenovirus resulted in a significant increase in invasion (Fig. 4B). However, MMP7 silencing significantly inhibited the increase in invasion imparted by FOXC1 overexpression and the extent of this inhibition was comparable with the ability of the MMP7 siRNA to block the induction of MMP7 expression (Fig. 4B). Combined, these results demonstrate that FOXC1 expression imparts a more invasive phenotype and that this increase in invasion is due, at least in part, to the increase in MMP7 expression that results upon FOXC1 overexpression.
FIGURE 4.
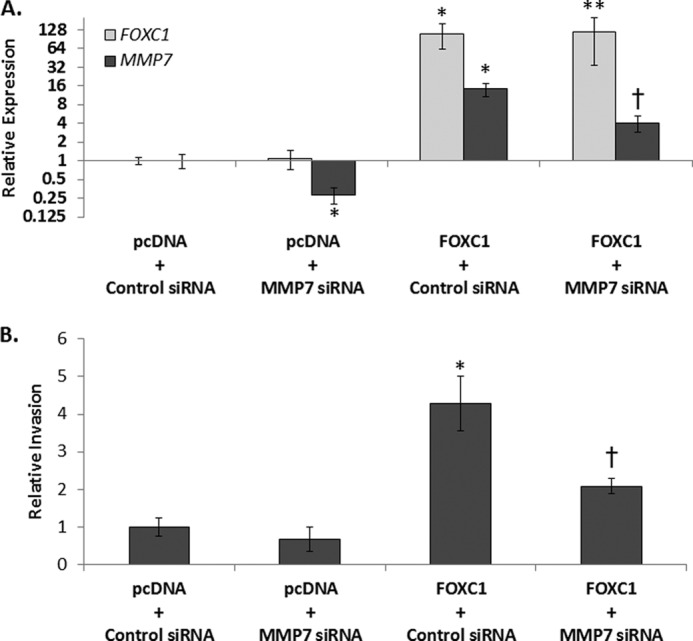
MMP7 is required for increased invasion of MCF10A cells overexpressing FOXC1. A, FOXC1 (light gray) or MMP7 (dark gray) transcript levels in MCF10A cells transfected with empty pcDNA vector or FOXC1 vector in combination with control siRNA or siRNA targeting MMP7. GAPDH was used as endogenous control. Data are relative to the pcDNA + Control siRNA-treated cells. B, relative invasion of MCF10A cells transfected with empty pcDNA vector or FOXC1 vector in combination with control siRNA or siRNA targeting MMP7. Data are relative to the pcDNA + Control siRNA-treated cells. Data summarize at least three independent experiments and are presented as mean ± S.D. *, p < 0.05 compared with pcDNA + Control siRNA-treated cells. **, p < 0.05 compared with pcDNA + MMP7 siRNA-treated cells but not significantly different from FOXC1 + Control siRNA-treated cells. †, p < 0.05 compared with either pcDNA + MMP7 siRNA or FOXC1 + control siRNA-treated cells.
Silencing Endogenous FOXC1 Results in Decreased MMP7 Expression
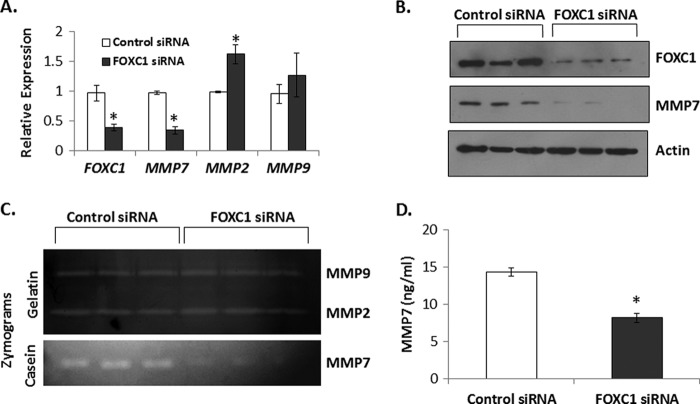
Basal A breast cancer cell lines (27) exhibit the highest expression of both FOXC1 and MMP7 (Fig. 2). We next determined if FOXC1 is necessary for MMP7 expression in these cells. Transient silencing of FOXC1 by siRNA in HCC1187 cells, the basal A line with the highest level of endogenous FOXC1 mRNA and the third highest level of MMP7 mRNA, reduced FOXC1 mRNA expression by 2.5-fold (p < 0.05). In response to this change in FOXC1 expression, MMP7 mRNA was also decreased to a similar extent (2.86-fold; p < 0.05) (Fig. 5A). At the protein level, FOXC1 silencing resulted in a 70% decrease in FOXC1 and 90% decrease in MMP7 expression (Fig. 5B). In contrast, silencing of FOXC1 in HCC1187 cells did not result in decreased expression of either MMP2 or MMP9 although a modest but significant increase in MMP2 mRNA expression was observed (1.6-fold; p < 0.05; Fig. 5A). Furthermore, silencing of FOXC1 in HCC1187 cells resulted in decreased levels of secreted MMP7 as detected by casein zymography, whereas no changes in MMP2 or MMP9 could be detected by gelatin zymography (Fig. 5C). We confirmed that HCC1187 cells treated with siRNA against FOXC1 secreted less MMP7 into conditioned media by ELISA (1.75-fold decrease; p < 0.05; Fig. 5D). Finally, we confirmed these data using shRNAs directed to FOXC1 to exclude off-target effects of the siRNA. Transient transduction of HCC1187 cells with three different shRNAs to FOXC1 resulted in significantly decreased FOXC1 expression 72 h post-transduction with commensurate significant decreases in MMP7 expression (supplemental Fig. S6A).
FIGURE 5.
Silencing of FOXC1 reduces MMP7 expression in the basal-like breast cancer cell line HCC1187. A, transcript levels of FOXC1, MMP7, MMP2, and MMP9 in HCC1187 cells transfected with nontargeting control siRNA or siRNA targeting FOXC1. Data are relative to control siRNA-transfected cells; GAPDH was used as endogenous control. Data summarize three independent experiments and are presented as mean ± S.D. B, representative Western blots demonstrating expression of FOXC1 and MMP7 in HCC1187 cells transiently transfected with control siRNA or siRNA targeting FOXC1. C, representative gelatin (upper panel) or casein (lower panel) zymograms demonstrating the relative levels of MMP9, MMP2, or MMP7 in conditioned media from HCC1187 cells transfected with control siRNA or siRNA targeting FOXC1. D, conditioned media from HCC1187 cells transfected with control siRNA or siRNA targeting FOXC1 was used in an ELISA for MMP7. Bar graph represents mean ± S.D. for three independent experiments. *, p < 0.05.
To confirm that the requirement for sustained expression of FOXC1 to maintain MMP7 expression in basal-like breast cancer cells was not cell line dependent, we transiently silenced FOXC1 in two additional basal A breast cancer cell lines: HCC1143 and MDA-MB-468. FOXC1-targeted shRNAs decreased FOXC1 expression in these cells 72 h after transduction with corresponding decreases in MMP7 expression (supplemental Fig. S6, B and C). Furthermore, stable silencing of FOXC1 in MDA-MB-468 cells resulted in significant reductions in both FOXC1 and MMP7 expression (supplemental Fig. S6D). These data confirm that FOXC1 consistently plays an important role in maintaining elevated MMP7 expression in basal-like breast cancer cells.
Silencing of FOXC1 has previously been shown to decrease viability or proliferation in a number of cell types (29, 30). In agreement with these previous findings, FOXC1 silencing in basal A breast cancer cell lines resulted in a substantial decrease in cell number compared with control treated cells (data not shown). This prohibited the utilization of these lines in invasion assays, but suggests that FOXC1 plays multiple roles in regulating the behaviors of basal breast cancer cells.
High FOXC1 Expression in Combination with High MMP7 Expression Predicts Poor Breast Cancer Patient Outcome
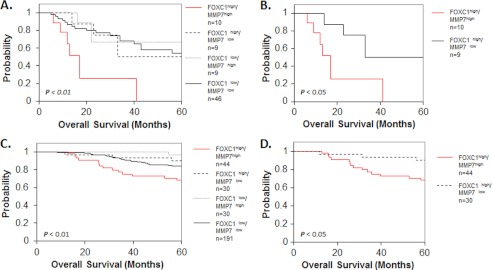
Expression of FOXC1 correlates with the BLBC subtype and poor prognosis in breast cancer patients (17, 20, 21). Given the significant functional correlation between FOXC1, MMP7, and invasion in our in vitro findings, we postulated that the association between FOXC1 expression and poor prognosis in breast cancer patients might be explained by the association between FOXC1 and MMP7 expression. To test this hypothesis we obtained gene expression data from a well studied breast cancer patient cohort, Sørlie et al. (24) and stratified patient samples into groups based upon high (upper quartile) or low (lower three quartiles) expression of FOXC1 and MMP7. This revealed that high FOXC1 expression alone was associated with poor outcome in this dataset (p < 0.05; supplemental Fig. S7A), whereas high MMP7 expression alone trended toward, but was not significantly associated with, poor outcome (p = 0.094; supplemental Fig. S7B). We next divided patients into four groups based on the pairwise expression of either high or low FOXC1 and high or low MMP7. Kaplan-Meier estimates revealed a significant difference in overall survival between the four resulting groups (p < 0.01; Fig. 6A). When the FOXC1high/MMP7high group was removed from the analysis, the Kaplan-Meier curves for the remaining three groups were not statistically different (p = 0.935). Importantly, direct comparison of survival curves for the FOXC1high/MMP7high and FOXC1high/MMP7low groups revealed that the FOXC1high/MMP7high group had a significantly worse outcome (p < 0.05; Fig. 6B) than the FOXC1high/MMP7low group. These findings were confirmed using a gene expression dataset from a second, larger patient cohort reported by van de Vijver et al. (25). In this group of patients we found that, whereas there was a trend between high FOXC1 expression and outcome, elevated expression of neither FOXC1 nor MMP7 alone was significantly associated with poor outcome when stratified by high (upper quartile) versus low (lower three quartiles) expression (p = 0.063 for FOXC1 and p = 0.29 for MMP7, respectively; supplemental Fig. S7, C and D). However, after stratifying this cohort into four groups based on the expression of both FOXC1 and MMP7, a significant difference in overall survival between the four resulting groups was revealed (p < 0.01, Fig. 6C). If the FOXC1high/MMP7high group was removed from the analysis, there was no statistical difference between survival times of the remaining three groups (p = 0.145). Furthermore, direct comparison of the survival curves from the FOXC1high/MMP7high and FOXC1high/MMP7low groups confirmed that the FOXC1high/MMP7high group had a significantly worse outcome (p < 0.05, Fig. 6D) than did patients from the FOXC1high/MMP7low group. Interestingly, high expression of MMP7 in the absence of elevated FOXC1 was not significantly associated with poor outcome in either patient cohort suggesting that MMP7 may be an important mediator of breast cancer aggressiveness only within the context of basal-like breast cancers, or when it is expressed in combination with FOXC1.
FIGURE 6.
High FOXC1 in combination with high MMP7 is associated with decreased breast cancer patient survival. Kaplan-Meier curves of overall survival using data from the Sørlie et al. (24) (A and B) or van de Vijver et al. (25) (C and D) datasets. Samples were stratified into four groups by high (upper quartile) or low (lower three quartiles) FOXC1 and MMP7 expression (A and C). Survival curves of FOXC1high/MMP7high and FOXC1high/MMP7low groups were directly compared in B and D. p values less than 0.05 were considered statistically significant.
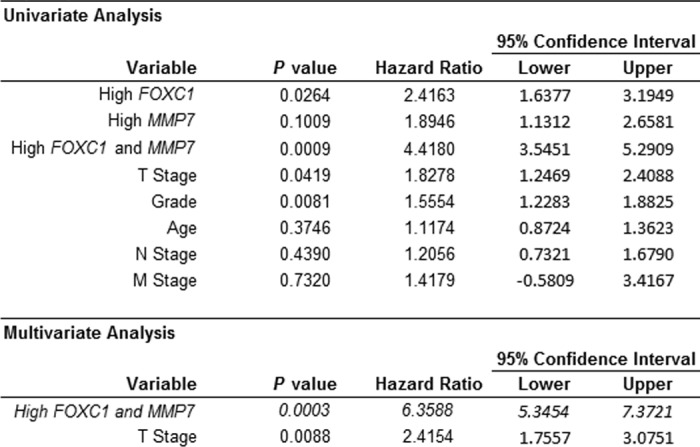
We next performed COX regression analysis on the Sørlie et al. (24) data set to assess the independent prognostic ability of combined FOXC1 and MMP7 expression (Table 1). As expected, univariate analysis showed that high (upper quartile) FOXC1 expression in combination with high MMP7 expression was a significant predictor (p = 0.009) of poor outcome with the highest hazard ratio (HR) of the factors examined (HR = 4.418). High FOXC1 expression alone (p = 0.026; HR = 2.416), T Stage (p = 0.042; HR = 1.828), and Grade (p = 0.008; HR = 1.555) were also significantly associated with outcome in univariate analysis (Table 1). In stepwise multivariate analysis, high combined expression of both FOXC1 and MMP7 was found to be a significant (p = 0.0003) independent predictor of poor outcome (HR = 6.3588) (Table 1). In a second patient cohort (van de Vijver et al. (25)) we confirmed that high expression of both FOXC1 and MMP7 was a significant predictor of poor outcome in both univariate and multivariate analyses (p = 0.002; HR = 2.6674), whereas neither high FOXC1 expression (p = 0.0667; HR = 1.7393) nor high MMP7 expression (p = 0.2924; HR = 1.388) alone were predictive of outcome in this dataset (supplemental Table S3). Together, these findings suggest a novel mechanism through which FOXC1 increases cancer cell invasion and that the relationship between FOXC1 and MMP7 has functional consequences on breast cancer patient outcome.
TABLE 1.
High combined expression of FOXC1 and MMP7 is an independent predictor of poor patient outcome in the Sorlie et al. (24) dataset
DISCUSSION
Improved treatment options for women with BLBC/triple negative breast cancer will require the identification of targetable molecules that are functionally responsible for the proliferation, survival, and metastasis of these cancers. Here, working downstream of FOXC1, a transcription factor known to correlate with the BLBC subtype, we identified MMP7 as a possible therapeutic target in BLBC treatment. Although previous studies have shown that FOXC1 increases invasion and metastasis of cancer cells, its mechanism of action is not fully understood. The increased invasive capacity imparted by FOXC1 in previous studies could be attributed to induction of EMT by FOXC1 expression (17, 18). However, the studies reported herein demonstrate that FOXC1 can increase the invasive capacity of mammary cell lines in the absence of EMT. Furthermore, whereas FOXC1 has previously been associated with increased expression of MMP2 and MMP9, we utilized meta-analysis of 20 gene expression datasets to demonstrate for the first time that a different MMP family member, MMP7, significantly correlates with FOXC1 both in vitro and in human breast cancer samples, whereas neither MMP2 nor MMP9 are associated with FOXC1. MMP7 is an established instigator of aggressive behavior in a number of cancer types including colorectal tumors (31–36). A role for MMP7 in breast cancer aggressiveness has been investigated (37–39) but to our knowledge this is the first report of a critical role for MMP7 in BLBC. Thus, MMP7 may be a promising druggable target for the treatment of at least a subset of BLBC.
Our studies identified a number of cell lines with high endogenous expression of FOXC1 and MMP7. Interestingly, the cell lines with the greatest FOXC1 and MMP7 expression were basal A breast cancer lines (27). These cell lines have gene expression patterns that most closely correlate with BLBC (27, 28). Hence, basal A breast cancer cell lines such as HCC1187 may serve as superior models than basal B lines such as MDA-MB-231 for understanding BLBC and the roles of FOXC1 and MMP7 in this disease. For this reason, we utilized basal A breast cancer cell lines in our current study. In addition, we found that FOXC1 was sufficient to induce expression of MMP7 in nontransformed mammary epithelial cells. Of note, enforced expression of FOXC1 did not robustly induce MMP7 expression in models of other breast cancer subtypes, including luminal (MCF-7 and T47D) or basal B (MDA-MB-231) (data not shown). Several previous studies have overexpressed FOXC1 in luminal or basal B breast cancer cell lines such as MCF7 or MDA-MB-231 to gain insight into the role of FOXC1 in breast cancers. However, these cell lines may not be ideal models in which to study the role of FOXC1 in BLBC. It is postulated that luminal, basal-like, and claudin-low breast cancers originate from different mammary cell populations (40, 41) and therefore may retain some of the epigenetic imprinting of their cells of origin. As a result, key transcriptional targets of FOXC1, including those that dictate BLBC aggressiveness such as MMP7, may be inaccessible to FOXC1 in the chromatin of luminal or basal B breast cancer cells. In support of the possibility that the MMP7 promoter may be inaccessible to FOXC1 in luminal and basal B cell lines, this promoter is silenced by hypermethylation in a number of pancreatic cancer cell lines (42). With these possible epigenetic shortcomings in mind we choose to utilize nontransformed mammary epithelial cell lines, rather than luminal or claudin-low breast cancer cell lines, in our FOXC1 overexpression experiments. Unlike breast cancer cell lines, nontransformed mammary epithelial cell lines such as MCF10A cells retain a level of plasticity allowing them to form acinar structures with apico-basal polarity in three-dimensional culture (43). The inherent luminal-basal plasticity of these cells may explain their ability to increase MMP7 expression in response to FOXC1. Future studies are necessary to determine whether the MMP7 promoter is methylated in a subtype-dependent manner in breast cancer cells and if it is directly or indirectly regulated by FOXC1. Intriguingly, the MMP7 proximal promoter contains a single consensus binding element for FOXC1 as well as numerous binding sites for other forkhead box transcription factors suggesting a direct role for FOXC1, as well as other forkheads, in MMP7 regulation. Supporting a role for forkheads in the regulation of MMP7 expression, a polymorphism that results in a novel binding site for FOXA2 in the MMP7 promoter was recently shown to result in FOXA2-dependent up-regulation of MMP7 expression (44).
Last, we demonstrated that the association between FOXC1 and MMP7 expression has important implications in breast cancer patient outcome. FOXC1 has previously been shown to correlate with a poor outcome in a number of patient cohorts (17, 20, 21), and our analyses of survival data provide novel insight into the mechanism underlying this correlation. We analyzed the association between combined FOXC1 and MMP7 expression with patient outcome and found that the correlation between high FOXC1 and poor outcome may depend, in part, upon high MMP7 expression. High FOXC1 expression was associated with significantly worse patient outcome when accompanied by elevated MMP7 in our analyses. Furthermore, combined expression of FOXC1 and MMP7 identified a unique population of breast cancers that convey poor patient survival in multivariate analyses. Together, these data demonstrate a novel role for MMP7 in the increased aggression imparted by FOXC1 expression. More importantly, these data suggest that inhibiting MMP7 may be a therapeutic strategy in treatment of BLBC.
Supplementary Material
Acknowledgments
We thank the Case Comprehensive Cancer Center (P30CA043703) for providing access to the Oncomine data base, David Schultz, Wistar Inc., for generous assistance and advice on lentivirus development and production, and Michael Walter, University of Alberta, Department of Ophthalmology, for supplying the FOXC1 cDNA.
This work was supported, in whole or in part, by National Institutes of Health Training Grant T32CA059366 (to S. T. S.) and Department of Defense Grants W81XWH-09-1-0696 (to S. T. S.) and W81XWH-08-1-0347 (to R. A. K.).

This article contains supplemental Figs. S1–S7 and Tables S1–S3.
- HER2
- human epidermal growth factor receptor 2
- BLBC
- basal-like breast cancer
- FOX
- forkhead box
- EMT
- epithelial to mesenchymal transition
- MMP
- matrix metalloproteinase
- TIMP
- tissue inhibitor of metalloproteinase
- HR
- hazard ratio.
REFERENCES
- 1. Cuzick J., Powles T., Veronesi U., Forbes J., Edwards R., Ashley S., Boyle P. (2003) Overview of the main outcomes in breast cancer prevention trials. Lancet 361, 296–300 [DOI] [PubMed] [Google Scholar]
- 2. Early Breast Cancer Trialists' Collaborative Group (1998) Tamoxifen for early breast cancer. An overview of the randomised trials. Lancet 351, 1451–1467 [PubMed] [Google Scholar]
- 3. Pegram M. D., Lipton A., Hayes D. F., Weber B. L., Baselga J. M., Tripathy D., Baly D., Baughman S. A., Twaddell T., Glaspy J. A., Slamon D. J. (1998) Phase II study of receptor-enhanced chemosensitivity using recombinant humanized anti-p185 HER2/neu monoclonal antibody plus cisplatin in patients with HER2/neu-overexpressing metastatic breast cancer refractory to chemotherapy treatment. J. Clin. Oncol. 16, 2659–2671 [DOI] [PubMed] [Google Scholar]
- 4. Slamon D. J., Leyland-Jones B., Shak S., Fuchs H., Paton V., Bajamonde A., Fleming T., Eiermann W., Wolter J., Pegram M., Baselga J., Norton L. (2001) Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 344, 783–792 [DOI] [PubMed] [Google Scholar]
- 5. Gucalp A., Traina T. A. (2011) Triple negative breast cancer. Adjuvant therapeutic options. Chemother. Res. Pract. 696208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pal S. K., Childs B. H., Pegram M. (2011) Triple negative breast cancer. Unmet medical needs. Breast Cancer Res. Treat. 125, 627–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bertucci F., Finetti P., Cervera N., Esterni B., Hermitte F., Viens P., Birnbaum D. (2008) How basal are triple negative breast cancers? Int. J. Cancer 123, 236–240 [DOI] [PubMed] [Google Scholar]
- 8. Dent R., Trudeau M., Pritchard K. I., Hanna W. M., Kahn H. K., Sawka C. A., Lickley L. A., Rawlinson E., Sun P., Narod S. A. (2007) Triple negative breast cancer. Clinical features and patterns of recurrence. Clin. Cancer Res. 13, 4429–4434 [DOI] [PubMed] [Google Scholar]
- 9. Kassam F., Enright K., Dent R., Dranitsaris G., Myers J., Flynn C., Fralick M., Kumar R., Clemons M. (2009) Survival outcomes for patients with metastatic triple negative breast cancer. Implications for clinical practice and trial design. Clin. Breast Cancer 9, 29–33 [DOI] [PubMed] [Google Scholar]
- 10. Hicks D. G., Short S. M., Prescott N. L., Tarr S. M., Coleman K. A., Yoder B. J., Crowe J. P., Choueiri T. K., Dawson A. E., Budd G. T., Tubbs R. R., Casey G., Weil R. J. (2006) Breast cancers with brain metastases are more likely to be estrogen receptor negative, express the basal cytokeratin CK5/6, and overexpress HER2 or EGFR. Am. J. Surg. Pathol. 30, 1097–1104 [DOI] [PubMed] [Google Scholar]
- 11. Myatt S. S., Lam E. W. (2007) The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer 7, 847–859 [DOI] [PubMed] [Google Scholar]
- 12. Kidson S. H., Kume T., Deng K., Winfrey V., Hogan B. L. (1999) The forkhead/winged-helix gene, Mf1, is necessary for the normal development of the cornea and formation of the anterior chamber in the mouse eye. Dev. Biol. 211, 306–322 [DOI] [PubMed] [Google Scholar]
- 13. Kume T., Deng K. Y., Winfrey V., Gould D. B., Walter M. A., Hogan B. L. (1998) The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell 93, 985–996 [DOI] [PubMed] [Google Scholar]
- 14. Mears A. J., Jordan T., Mirzayans F., Dubois S., Kume T., Parlee M., Ritch R., Koop B., Kuo W. L., Collins C., Marshall J., Gould D. B., Pearce W., Carlsson P., Enerbäck S., Morissette J., Bhattacharya S., Hogan B., Raymond V., Walter M. A. (1998) Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am. J. Hum. Genet. 63, 1316–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Du J., Li L., Ou Z., Kong C., Zhang Y., Dong Z., Zhu S., Jiang H., Shao Z., Huang B., Lu J. (2012) FOXC1, a target of polycomb, inhibits metastasis of breast cancer cells. Breast Cancer Res. Treat. 131, 65–73 [DOI] [PubMed] [Google Scholar]
- 16. Chung T. K., Lau T. S., Cheung T. H., Yim S. F., Lo K. W., Siu N. S., Chan L. K., Yu M. Y., Kwong J., Doran G., Barroilhet L. M., Ng A. S., Wong R. R., Wang V. W., Mok S. C., Smith D. I., Berkowitz R. S., Wong Y. F. (2012) Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int. J. Cancer 130, 1036–1045 [DOI] [PubMed] [Google Scholar]
- 17. Ray P. S., Wang J., Qu Y., Sim M. S., Shamonki J., Bagaria S. P., Ye X., Liu B., Elashoff D., Hoon D. S., Walter M. A., Martens J. W., Richardson A. L., Giuliano A. E., Cui X. (2010) FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res. 70, 3870–3876 [DOI] [PubMed] [Google Scholar]
- 18. Bloushtain-Qimron N., Yao J., Snyder E. L., Shipitsin M., Campbell L. L., Mani S. A., Hu M., Chen H., Ustyansky V., Antosiewicz J. E., Argani P., Halushka M. K., Thomson J. A., Pharoah P., Porgador A., Sukumar S., Parsons R., Richardson A. L., Stampfer M. R., Gelman R. S., Nikolskaya T., Nikolsky Y., Polyak K. (2008) Cell type-specific DNA methylation patterns in the human breast. Proc. Natl. Acad. Sci. U.S.A. 105, 14076–14081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang J., Ray P. S., Sim M. S., Zhou X. Z., Lu K. P., Lee A. V., Lin X., Bagaria S. P., Giuliano A. E., Cui X. (2012) FOXC1 regulates the functions of human basal-like breast cancer cells by activating NF-κB signaling. Oncogene, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ray P. S., Bagaria S. P., Wang J., Shamonki J. M., Ye X., Sim M. S., Steen S., Qu Y., Cui X., Giuliano A. E. (2011) Basal-like breast cancer defined by FOXC1 expression offers superior prognostic value. A retrospective immunohistochemical study. Ann. Surg. Oncol. 18, 3839–3847 [DOI] [PubMed] [Google Scholar]
- 21. Taube J. H., Herschkowitz J. I., Komurov K., Zhou A. Y., Gupta S., Yang J., Hartwell K., Onder T. T., Gupta P. B., Evans K. W., Hollier B. G., Ram P. T., Lander E. S., Rosen J. M., Weinberg R. A., Mani S. A. (2010) Core epithelial-to-mesenchymal transition interactome gene expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. U.S.A. 107, 15449–15454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sizemore S., Cicek M., Sizemore N., Ng K. P., Casey G. (2007) Podocalyxin increases the aggressive phenotype of breast and prostate cancer cells in vitro through its interaction with ezrin. Cancer Res. 67, 6183–6191 [DOI] [PubMed] [Google Scholar]
- 23. Fernández-Resa P., Mira E., Quesada A. R. (1995) Enhanced detection of casein zymography of matrix metalloproteinases. Anal. Biochem. 224, 434–435 [DOI] [PubMed] [Google Scholar]
- 24. Sørlie T., Perou C. M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M. B., van de Rijn M., Jeffrey S. S., Thorsen T., Quist H., Matese J. C., Brown P. O., Botstein D., Lønning P. E., Børresen-Dale A. L. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U.S.A. 98, 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van de Vijver M. J., He Y. D., van't Veer L. J., Dai H., Hart A. A., Voskuil D. W., Schreiber G. J., Peterse J. L., Roberts C., Marton M. J., Parrish M., Atsma D., Witteveen A., Glas A., Delahaye L., van der Velde T., Bartelink H., Rodenhuis S., Rutgers E. T., Friend S. H., Bernards R. (2002) A gene expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009 [DOI] [PubMed] [Google Scholar]
- 26. Jacobsen L., Calvin S., Lobenhofer E. (2009) Transcriptional effects of transfection. The potential for misinterpretation of gene expression data generated from transiently transfected cells. BioTechniques 47, 617–624 [DOI] [PubMed] [Google Scholar]
- 27. Neve R. M., Chin K., Fridlyand J., Yeh J., Baehner F. L., Fevr T., Clark L., Bayani N., Coppe J. P., Tong F., Speed T., Spellman P. T., DeVries S., Lapuk A., Wang N. J., Kuo W. L., Stilwell J. L., Pinkel D., Albertson D. G., Waldman F. M., McCormick F., Dickson R. B., Johnson M. D., Lippman M., Ethier S., Gazdar A., Gray J. W. (2006) A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Charafe-Jauffret E., Ginestier C., Monville F., Finetti P., Adélaïde J., Cervera N., Fekairi S., Xerri L., Jacquemier J., Birnbaum D., Bertucci F. (2006) Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene 25, 2273–2284 [DOI] [PubMed] [Google Scholar]
- 29. Tkocz D., Crawford N. T., Buckley N. E., Berry F. B., Kennedy R. D., Gorski J. J., Harkin D. P., Mullan P. B. (2011) BRCA1 and GATA3 corepress FOXC1 to inhibit the pathogenesis of basal-like breast cancers. Oncogene, in press [DOI] [PubMed] [Google Scholar]
- 30. Berry F. B., Skarie J. M., Mirzayans F., Fortin Y., Hudson T. J., Raymond V., Link B. A., Walter M. A. (2008) FOXC1 is required for cell viability and resistance to oxidative stress in the eye through the transcriptional regulation of FOXO1A. Hum. Mol. Genet. 17, 490–505 [DOI] [PubMed] [Google Scholar]
- 31. Matrisian L. M., Wright J., Newell K., Witty J. P. (1994) Matrix-degrading metalloproteinases in tumor progression. Princess Takamatsu Symp. 24, 152–161 [PubMed] [Google Scholar]
- 32. Powell W. C., Knox J. D., Navre M., Grogan T. M., Kittelson J., Nagle R. B., Bowden G. T. (1993) Expression of the metalloproteinase matrilysin in DU-145 cells increases their invasive potential in severe combined immunodeficient mice. Cancer Res. 53, 417–422 [PubMed] [Google Scholar]
- 33. Witty J. P., McDonnell S., Newell K. J., Cannon P., Navre M., Tressler R. J., Matrisian L. M. (1994) Modulation of matrilysin levels in colon carcinoma cell lines affects tumorigenicity in vivo. Cancer Res. 54, 4805–4812 [PubMed] [Google Scholar]
- 34. Yamamoto H., Itoh F., Hinoda Y., Imai K. (1995) Suppression of matrilysin inhibits colon cancer cell invasion in vitro. Int. J. Cancer 61, 218–222 [DOI] [PubMed] [Google Scholar]
- 35. Fukushima H., Yamamoto H., Itoh F., Nakamura H., Min Y., Horiuchi S., Iku S., Sasaki S., Imai K. (2001) Association of matrilysin mRNA expression with K-ras mutations and progression in pancreatic ductal adenocarcinomas. Carcinogenesis 22, 1049–1052 [DOI] [PubMed] [Google Scholar]
- 36. Nakamura H., Horita S., Senmaru N., Miyasaka Y., Gohda T., Inoue Y., Fujita M., Meguro T., Morita T., Nagashima K. (2002) Association of matrilysin expression with progression and poor prognosis in human pancreatic adenocarcinoma. Oncol. Rep. 9, 751–755 [PubMed] [Google Scholar]
- 37. Wang W. S., Chen P. M., Wang H. S., Liang W. Y., Su Y. (2006) Matrix metalloproteinase-7 increases resistance to Fas-mediated apoptosis and is a poor prognostic factor of patients with colorectal carcinoma. Carcinogenesis 27, 1113–1120 [DOI] [PubMed] [Google Scholar]
- 38. Jiang W. G., Davies G., Martin T. A., Parr C., Watkins G., Mason M. D., Mokbel K., Mansel R. E. (2005) Targeting matrilysin and its impact on tumor growth I. The potential implications in breast cancer therapy. Clin. Cancer Res. 11, 6012–6019 [DOI] [PubMed] [Google Scholar]
- 39. Lynch C. C., Vargo-Gogola T., Martin M. D., Fingleton B., Crawford H. C., Matrisian L. M. (2007) Matrix metalloproteinase 7 mediates mammary epithelial cell tumorigenesis through the ErbB4 receptor. Cancer Res. 67, 6760–6767 [DOI] [PubMed] [Google Scholar]
- 40. Prat A., Parker J. S., Karginova O., Fan C., Livasy C., Herschkowitz J. I., He X., Perou C. M. (2010) Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 12, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prat A., Perou C. M. (2011) Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 5, 5–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sato N., Maehara N., Su G. H., Goggins M. (2003) Effects of 5-aza-2′-deoxycytidine on matrix metalloproteinase expression and pancreatic cancer cell invasiveness. J. Natl. Cancer Inst. 95, 327–330 [DOI] [PubMed] [Google Scholar]
- 43. Debnath J., Brugge J. S. (2005) Modeling glandular epithelial cancers in three-dimensional cultures. Nat. Rev. Cancer 5, 675–688 [DOI] [PubMed] [Google Scholar]
- 44. Richards T., Park C., Chen Y., Gibson K. F., Di Y. P., Pardo A., Watkins S. C., Choi A. M., Selman M., Pilewski J. M., Kaminski N., Zhang Y. (2012) Allele-specific transactivation of matrix metalloproteinase 7 by FOXA2 and correlation with plasma levels in idiopathic pulmonary fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 302, L746–L754 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.