Background: The Alzheimer Aβ peptide assembles into multiple small oligomers that are cytotoxic.
Results: Increased solvent exposure of hydrophobic residues within non-fibrillar Aβ oligomers of similar size increases cytotoxicity.
Conclusion: Aβ non-fibrillar oligomers display size-independent differences in toxicity that are strongly influenced by oligomer conformation.
Significance: Identifying the conformational determinants of Aβ-mediated toxicity is critical to understand and treat Alzheimer disease.
Keywords: Aggregation, Alzheimer Disease, Amyloid, Protein Misfolding, Protein Self-assembly, Toxicity
Abstract
Several protein conformational disorders (Parkinson and prion diseases) are linked to aberrant folding of proteins into prefibrillar oligomers and amyloid fibrils. Although prefibrillar oligomers are more toxic than their fibrillar counterparts, it is difficult to decouple the origin of their dissimilar toxicity because oligomers and fibrils differ both in terms of structure and size. Here we report the characterization of two oligomers of the 42-residue amyloid β (Aβ42) peptide associated with Alzheimer disease that possess similar size and dissimilar toxicity. We find that Aβ42 spontaneously forms prefibrillar oligomers at Aβ concentrations below 30 μm in the absence of agitation, whereas higher Aβ concentrations lead to rapid formation of fibrils. Interestingly, Aβ prefibrillar oligomers do not convert into fibrils under quiescent assembly conditions but instead convert into a second type of oligomer with size and morphology similar to those of Aβ prefibrillar oligomers. Strikingly, this alternative Aβ oligomer is non-toxic to mammalian cells relative to Aβ monomer. We find that two hydrophobic peptide segments within Aβ (residues 16–22 and 30–42) are more solvent-exposed in the more toxic Aβ oligomer. The less toxic oligomer is devoid of β-sheet structure, insoluble, and non-immunoreactive with oligomer- and fibril-specific antibodies. Moreover, the less toxic oligomer is incapable of disrupting lipid bilayers, in contrast to its more toxic oligomeric counterpart. Our results suggest that the ability of non-fibrillar Aβ oligomers to interact with and disrupt cellular membranes is linked to the degree of solvent exposure of their central and C-terminal hydrophobic peptide segments.
Introduction
The seminal role of protein misfolding in several aggregation disorders has motivated the identification of protein aggregates that are highly toxic relative to those that are either less toxic or non-toxic. One logical approach to accomplish this aim is to classify aggregated conformers based on their size, and evaluate the relationship between size and toxicity. Extensive work has convincingly demonstrated that aggregate size is a critical determinant of toxicity (1–6). For example, small oligomers of the Aβ peptide (as well as other misfolded polypeptides) are generally more toxic than large oligomers and amyloid fibrils (2, 7–11).
Nevertheless, it is becoming increasingly clear that protein aggregates of the same size can have unique structures and, therefore, unique toxicities (8, 12–15). An important advance in classifying misfolded proteins in terms of structure has been the development of conformation-specific antibodies that recognize unique misfolded isoforms (12, 16–24). For example, the A11 polyclonal antibody selectively recognizes prefibrillar oligomers of several amyloidogenic proteins relative to fibrils and monomers of the same proteins (17). Conversely, multiple fibril-specific antibodies have also been developed that selectively recognize amyloid fibrils of several aggregation-prone proteins (18, 19, 23). These antibodies have revealed that proteins can form multiple oligomeric and fibrillar conformers with similar sizes and unique conformations (8, 12, 17, 18, 25).
An attractive strategy for understanding how conformational differences between misfolded proteins mediate toxicity is to evaluate the structures of aggregated conformers of similar size and dissimilar toxicity. Chiti and co-workers (13) performed an elegant analysis of two oligomers of a bacterial protein (HypF-N) of the same size that differ in their cytotoxicity. HypF-N folds into two unique oligomers at different solution conditions that are indistinguishable in terms of size and morphology, yet only one oligomer is toxic to mammalian cells. The authors performed site-specific fluorescent labeling analysis of each oligomer and found that hydrophobic residues within the more toxic oligomer were less structured (and more solvent-exposed) than those within the less toxic oligomer. This important study of a non-pathogenic protein provides an important basis for similar studies of disease-linked misfolded polypeptides.
A previous report suggests that Aβ42 also forms two different oligomers of similar size and dissimilar toxicity (8). Klein and co-workers (8) found that Aβ42 forms toxic oligomers in cell culture media at 4 °C that are recognized by an oligomer-specific antibody. Notably, Aβ oligomers of similar size occasionally formed that were neither toxic nor recognized by oligomer-specific antibodies, revealing that Aβ oligomers can also possess size-independent differences in toxicity. We recently reported a reproducible procedure for forming Aβ oligomers that are weakly toxic and similar in size to their more toxic counterparts (14). Here we characterize the biochemical and structural properties of each Aβ oligomer relative to Aβ fibrils and monomers to understand the origins of their size-independent differences in toxicity.
EXPERIMENTAL PROCEDURES
Preparation of Aβ Conformers
Human Aβ42 (American Peptide) was dissolved in an aqueous, 50% acetonitrile solution (1 mg/ml), aliquoted, dried under vacuum and lyophilized, and then stored at −20 °C. Aβ oligomers were prepared by dissolving the peptide in 100% hexafluoroisopropanol (Fluka). The solvent was evaporated, and Aβ was dissolved in 50 mm NaOH (1 mg/ml Aβ), sonicated (30 s), and diluted in PBS (25 μm Aβ). The peptide was then centrifuged (22,000 × g for 30 min), and the pelleted fraction (5% of starting volume) was discarded. The supernatant was incubated at 25 °C for 0–6 days without agitation. Aβ fibrils were prepared via the same procedure except that monomers were mixed with preexisting fibrils (10–20 weight percent seed) without mixing for 24 h at 25 °C.
Thioflavin T (ThT)2 Assay
Aβ (25 μm) was diluted with ThT (44 μm; 1:19 volumetric ratio of Aβ/ThT solutions). The fluorescence was measured using a Tecan Safire2 plate reader (450/482-nm excitation/emission, 15-nm bandwidth). The seeding experiments were conducted with Aβ monomers (25 μm) and 5% preformed Aβ oligomers and fibrils.
Atomic Force Microscopy (AFM)
Aβ samples (25 μm) were spotted on cut mica mounted on glass slides. The samples were adsorbed (30 min), washed with water, and dried overnight. Images were taken using an Asylum Research MFP 3D AFM system with Olympus AC240TS silicon cantilevers in tapping mode (AC, scan rate of 0.5 Hz).
Cell Toxicity Assay
Rat adrenal medulla cells (PC12, ATCC) were cultured in Dulbecco's modified Eagle's medium (5% fetal bovine serum, 10% horse serum, and 1% penicillin/streptomycin). The cell suspension (90 μl) was incubated in 96-well microtiter plates (CellBIND, Corning) for 24 h. Afterward, Aβ or control samples (10 μl) were added to microtiter plates, and the cells were further incubated for 48 h at 37 °C.
The cell viability was then evaluated using two assays. In the first method, the media was removed, and fresh media (200 μl) and thiazolyl blue tetrazolium bromide (Sigma; 50 μl of 2.5 mg/ml) were added to each well for 3 h at 37 °C. These solutions were then discarded, 250 μl of DMSO was added, and the absorbance was measured at 562 nm. The toxicity values were normalized relative to the buffer (PBS).
The cell viability was also evaluated via the lactate dehydrogenase (LDH) assay (Sigma-Aldrich). The cell culture media was transferred to a clean 96-well, flat bottom plate. Equal volumes of LDH assay substrate, dye, and cofactor solutions were added to each well. The final volume of LDH assay solution added was equal to twice the volume of medium removed for testing. The microtiter plate was then covered and incubated at room temperature for 20 min, after which the reaction was terminated by the addition of 0.1 m HCl (final concentration). The absorbance was measured at 490 nm, and the LDH release values were normalized to the buffer (PBS).
Aβ toxicity was also evaluated for primary cultures of embryonic rat cortical neurons, as described previously (11). Aβ42 peptide was added to the neuronal cell cultures at a concentration of 6 μm. The cells were incubated for 24 h, and then the cell viability was analyzed using the MTT assay (Sigma). After 4 h of incubation with MTT, the media were removed and replaced with DMSO. The fraction of viable cells were quantified using a SpectraMax M2 plate reader (Molecular Devices, Sunnyvale, CA) at a wavelength of 570 nm. The toxicity values were normalized relative to the buffer (PBS).
Gel Electrophoresis and Silver Staining
Aβ samples (25 μm) were diluted into sample buffer (Novex LDS, Invitrogen), sonicated, analyzed using 10% BisTris gels (Invitrogen), and silver-stained (SilverXpress kit, Invitrogen).
Antibody Dot Blot Analysis
Each Aβ conformer (25 μm) was spotted (2 μl) on nitrocellulose membranes (Hybond ECL, GE Healthcare). Afterward, the blots were blocked overnight (10% nonfat dry milk in PBST) and then probed with A11 (Invitrogen), OC (Millipore), or 6E10 (Millipore) antibodies. The blots were washed, incubated with the appropriate horseradish peroxidase-conjugated secondary antibody, and developed (ECL Western blotting substrate, Thermo Fisher).
Circular Dichroism Spectroscopy
Aβ conformers (25 μm in 0.1× PBS) were evaluated using a Jasco 815 spectrometer (1-mm path length cuvette) at 25 °C. Each sample spectrum is the average of at least 25 scans.
8-Anilinonaphthalene-1-sulfonate (ANS) Fluorescence Analysis
ANS (Sigma-Aldrich) was used at 7.5–12.5 μm to assay the conformation of Aβ (2.5 μm). The ANS fluorescence spectra (λex = 380 nm) were measured using a Tecan Safire2 plate reader.
Lipid Bilayer Conductance Analysis
All lipid bilayers were formed using l-α-phosphocholine (asolectin from soy, 20%; Sigma-Aldrich). Bilayers were formed using a modified version of the painting technique (26). A Delrin cup with a 250-μm aperture was inserted into a holding chamber (Warner Instruments), and both the cup and chamber were filled (10 mm HEPES, 100 mm KCl, pH 7). Asolectin was dissolved in n-decane (200 mg/ml asolectin) and applied to the exterior of the cup aperture using a fine tipped brush. Bilayer formation was detected via an increase in capacitance and the formation of a seal in excess of 1 gigaohm. Bilayers were equilibrated for at least 10 min prior to the sample addition. Aβ samples (250 nm) were added to a chamber reservoir on one side of the bilayer and equilibrated for 20 min. For some experiments, the A11 antibody (0.2–3 μm) was mixed with Aβ soluble oligomers (12.5 μm) for 4 h and then diluted 50 times into the chamber reservoir. Voltage sweeps were performed from −100 to +100 mV at a rate of 40 mV/s. The data were collected through a BC535 patch clamp amplifier (Warner Instruments), digitized using a Digidata 1440A digitizer, and analyzed using Clampex 10.1 software (Axon Instruments).
Proteolytic Fragmentation Analysis
Aβ (25 μm) was mixed with Proteinase K (0.5 μg/ml) in PBS (pH 7.4), and Aβ samples (2 μl) were deposited on nitrocellulose (Hybond ECL, GE Healthcare) periodically for 4 h. At the end of the fragmentation reaction, the blots were blocked overnight (10% nonfat dry milk in PBST) and then probed with sequence-specific antibodies against Aβ (6E10 against Aβ(3–10) from Sigma-Aldrich, BAM90.1 against Aβ(16–21) from Sigma-Aldrich, 4G8 against Aβ(18–22) from Covance, a polyclonal antibody against Aβ(30–36) from Sigma-Aldrich, 9F1 against Aβ(35–39) from Santa Cruz Biotechnology, Inc., and 12F4 against Aβ(37–42) from Abcam). The blots were then washed, incubated with the appropriate horseradish peroxidase-conjugated secondary antibody, and developed (ECL Western blotting Substrate, Thermo Fisher).
Size Exclusion Chromatography Analysis
Preformed Aβ oligomers (25 μm) were mixed with a single domain antibody (2.5 μm) specific for amyloid β presenting Aβ residues 33–42 in its third complementarity-determining region (24) and injected (100 μl) into an analytical size exclusion column (TSK Gel G3000SWxl column, 0.78 × 30 cm; Tosoh Bioscience). The elution profile of the Aβ-antibody complex was monitored at 280 nm.
RESULTS
Non-fibrillar Aβ Conformers Form at Low Aβ Concentrations
We first investigated the range of Aβ concentrations that promote formation of Aβ prefibrillar oligomers in PBS (pH 7.4). The assembly of Aβ42 (1–75 μm) can be readily evaluated via immunoblot analysis using antibodies specific for prefibrillar oligomers (A11) and fibrillar conformers (OC; Fig. 1) (12, 17, 18). We first confirmed that each conformation-specific antibody recognized prefibrillar oligomers and fibrils at submicromolar Aβ concentrations (supplemental Fig. S1). We also confirmed the proper loading of each Aβ conformer via the sequence-specific antibody 6E10, which recognizes the N terminus of Aβ (residues 3–10; Fig. 1).
FIGURE 1.
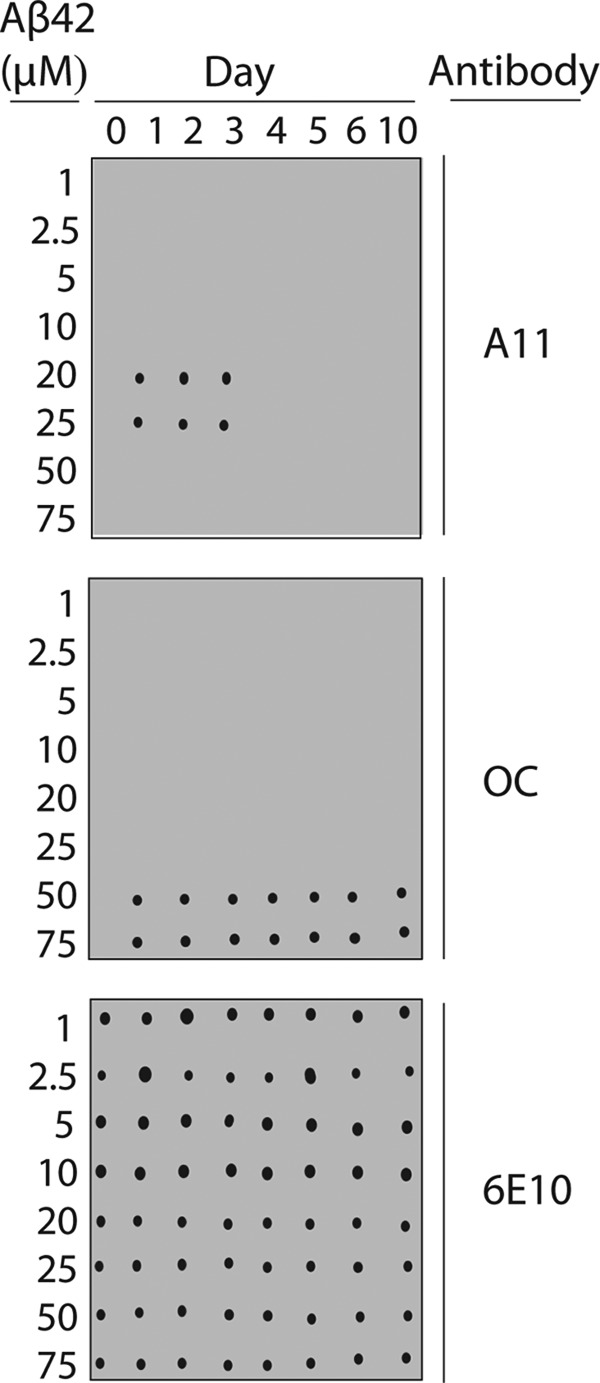
Conformation-specific antibody analysis of Aβ assembly. Aβ42 conformers (1–75 μm) were assembled for 10 days (without agitation), and periodically deposited on nitrocellulose membranes. Afterward, the membranes were probed with conformation-specific (A11, prefibrillar oligomers (top), and OC, fibrillar conformers (middle)) and sequence-specific (6E10, N terminus of Aβ (bottom)) antibodies.
At Aβ concentrations of <20 μm, we find that both A11- and OC-positive conformers fail to form over 10 days (Fig. 1). In contrast, we find that higher concentrations of Aβ (20 and 25 μm) lead to formation of A11-positive conformers after 1 day, and these oligomers persist for an additional 2 days (Fig. 1). Importantly, the A11-positive conformers appear to be non-fibrillar because the OC antibody does not recognize them. On the fourth day, the A11-positive oligomers formed at 20 and 25 μm Aβ convert into an alternative conformer that is non-immunoreactive with either conformation-specific antibody. Longer times (5–10 days) do not promote formation of A11- or OC-positive conformers for Aβ samples at 20–25 μm (Fig. 1).
Because Aβ is well known to readily form amyloid fibrils, we suspected that Aβ concentrations above 25 μm would lead to formation of fibrils without agitation. Indeed, we found that Aβ at 50 and 75 μm formed OC-positive conformers after 1 day that were invariant over longer times (2–10 days; Fig. 1). These OC-positive conformers are A11-negative, consistent with fibrillar intermediates and mature amyloid fibrils (14, 18).
We also evaluated the homogeneity of A11-positive oligomers formed at 25 μm Aβ (supplemental Fig. S1). We doped Aβ fibrils into preparations of Aβ A11-positive oligomers and evaluated the minimum fibril detection limit of the OC antibody. We found that fibrils could be detected at ≥0.6 μm (supplemental Fig. S1), which represents 5% of the Aβ peptide at 25 μm. We observed the same sensitivity of the A11 antibody when we doped fibril samples with A11-positive oligomers. Therefore, we estimate that the fraction of Aβ fibrils in our non-fibrillar oligomer preparations was below 5%.
Aβ Oligomers Possess Size-independent Differences in Toxicity
The unique immunoreactivity of Aβ conformers formed at intermediate Aβ concentrations (20 and 25 μm) led us to evaluate the size and toxicity of both A11-positive and A11-negative Aβ conformers. Using AFM, we found that A11-positive oligomers (25 μm Aβ) formed on days 1–3 were globular and possessed similar size (6.2 ± 0.5 nm in height; Fig. 2 and supplemental Fig. S2). Importantly, the A11-negative conformers formed on days 4–6 were also globular and of similar size (6.1 ± 0.6 nm in height) relative to A11-positive oligomers (Fig. 2 and supplemental Fig. S2). We also used size exclusion chromatography to evaluate the size of both Aβ oligomers. Because each Aβ oligomer sticks to the column matrix and fails to elute in non-denaturing buffers, we evaluated the size of Aβ oligomers when complexed to a small antibody domain specific for Aβ (supplemental Fig. S3). Importantly, we found that the elution times of Aβ oligomers bound to the same antibody were indistinguishable. We herein refer to A11-positive Aβ oligomers as A+ oligomers and oligomers of the same size that are non-reactive with either conformation-specific antibody as A− oligomers. Finally, AFM analysis revealed that higher concentrations of Aβ (50 μm) led to the formation of fibrillar structures after 1 day (supplemental Fig. S4) as expected based on their reactivity with the OC antibody (Fig. 1).
FIGURE 2.
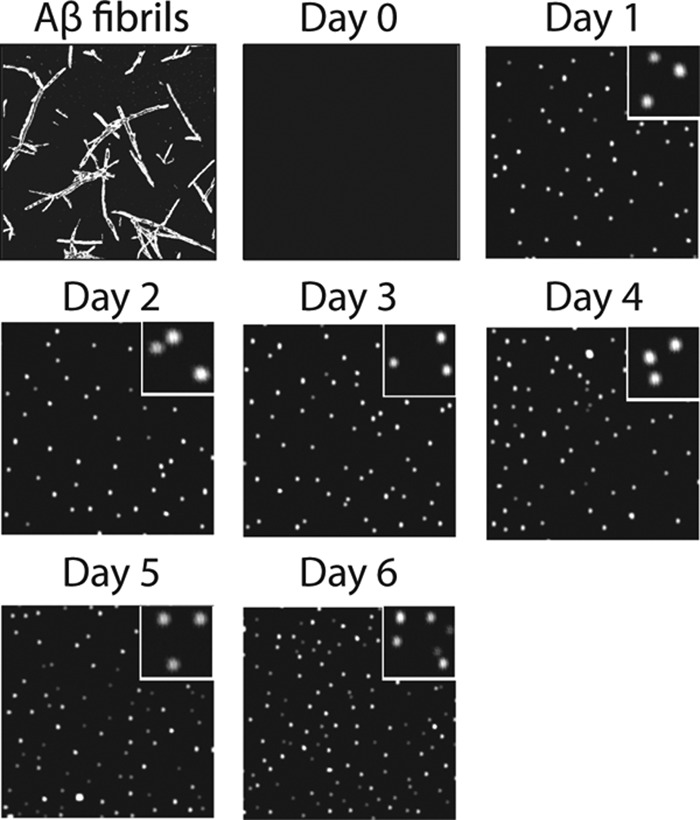
AFM analysis of Aβ oligomerization. Aβ42 (25 μm) was assembled without agitation, deposited on mica substrates, and imaged using AFM. Each image is 3 × 3 μm, and the inset images are 0.5 × 0.5 μm.
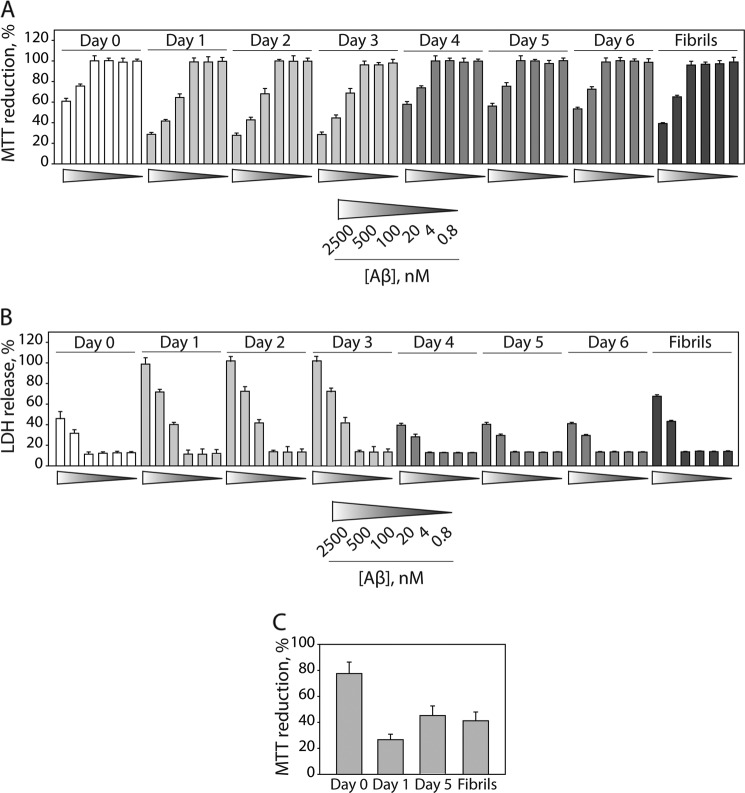
We next evaluated the toxicity of each Aβ oligomer using multiple mammalian cell culture assays (14, 27, 28). We expected that A+ oligomers (formed at 20–25 μm; days 1–3) would be more toxic than A− oligomers (formed at 20–25 μm; days 4–10) and OC-positive conformers (formed at 50–75 μm; days 1–6). We found that A+ oligomers were highly toxic to differentiated PC12 cells (Fig. 3A), whereas fibrils formed at elevated Aβ concentrations (50 μm) were mildly toxic (supplemental Fig. S5). In contrast, A− oligomers formed at 25 μm Aβ (days 4–6) were non-toxic relative to Aβ monomers (Fig. 3A). We also confirmed that these toxicity results were similar when evaluating metabolic activity (MTT reduction; Fig. 3A) or membrane integrity (LDH activity; Fig. 3B). Finally, we evaluated the toxicity of each Aβ oligomer to primary cultures of embryonic rat cortical neurons and also found that A+ oligomers were more toxic than A− oligomers and OC-positive fibrillar conformers (Fig. 3C).
FIGURE 3.
Toxicity analysis of Aβ oligomers and fibrils. Aβ42 (25 μm) was assembled without agitation and added to rat adrenal medulla cells (A and B) and rat primary cortical neuronal cells (C), and the relative toxicity was evaluated (n = 3). Error bars, S.D.
Aβ Oligomers Display Dissimilar Activity for Disrupting Lipid Membranes
Aβ A+ oligomers have been shown to permeabilize reconstituted lipid membranes (29), which has been posited to be integral to their toxic activity in vivo. This observation led us to hypothesize that A− oligomers would be inactive at disrupting lipid membranes. To investigate this hypothesis, we evaluated the conductance of lipid bilayers in the absence and presence of both Aβ oligomers. We found that A+ oligomers permeabilized lipid bilayers and increased their conductance (Fig. 4A), as observed previously (29). In contrast, A− oligomers (as well as Aβ fibrils and monomers) failed to increase membrane conductance at the same Aβ concentrations (250 nm). We also evaluated whether the oligomer-specific (A11) antibody could antagonize the ability of A+ oligomers to permeabilize lipid bilayers (Fig. 4B). We found that the anti-oligomer antibody reduced membrane conductance in a dose-dependent manner. Our results reveal that the same Aβ conformer recognized by the A11 antibody is responsible for interacting with lipid bilayers and altering their structure.
FIGURE 4.
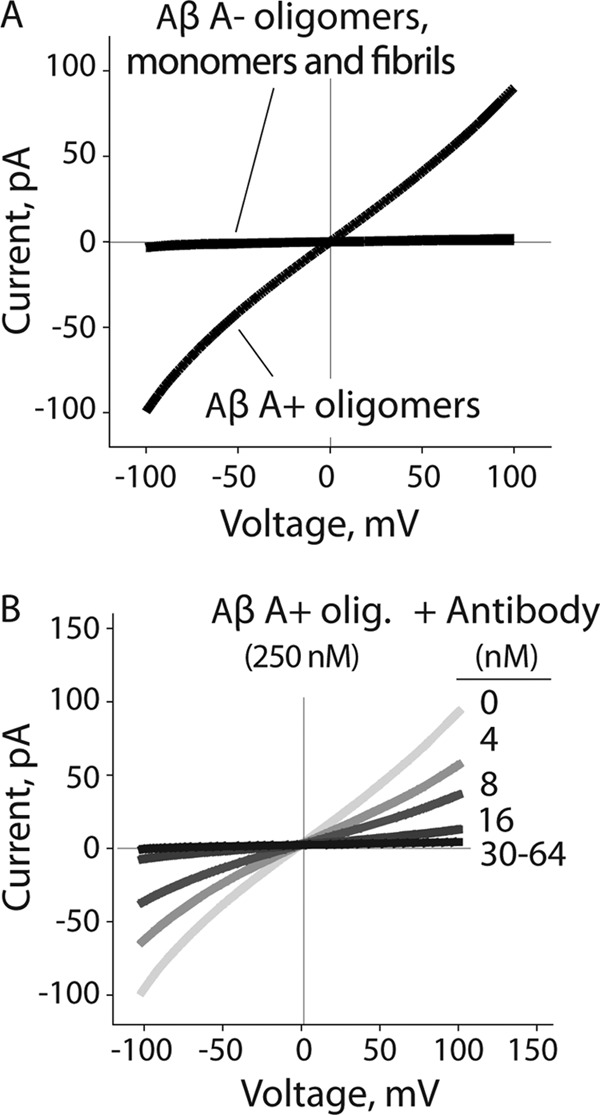
Impact of Aβ oligomers on lipid bilayer conductance. A, Aβ42 conformers (250 nm) were added to a reservoir on one side of the lipid bilayer (l-α-phosphocholine), and the membrane conductance was measured. B, Aβ A+ oligomers (12.5 μm) were mixed with the A11 antibody (0.2–3 μm) and diluted into a reservoir on one side of the lipid bilayer (50× dilution), and the membrane conductance was measured.
Aβ A+ Oligomers Are More Hydrophobic than A− Oligomers
We next sought to define the biochemical and structural differences between A+ and A− Aβ oligomers. As a possible mechanism for the low toxicity of A− oligomers, we hypothesized that A− oligomers are folded in a manner in which their hydrophobic residues are less solvent-exposed than in A+ oligomers. To evaluate this hypothesis, we developed a proteolytic assay that uses sequence-specific antibodies to interrogate the relative degree of solvent exposure of multiple sequence epitopes within Aβ oligomers and fibrils. This assay is based on the premise that linear sequence epitopes within Aβ conformers are cleaved by the protease at a rate proportional to their degree of solvent exposure. Solvent-exposed sequences are more accessible to proteolysis and will exhibit more rapid loss of antibody binding with time. We used six sequence-specific Aβ antibodies directed against different epitopes distributed throughout Aβ (Aβ residues 3–10, 16–21, 18–22, 30–35, 35–39, and 37–42). We posited that the hydrophobic middle (Aβ(17–21)) and C-terminal (Aβ(30–42)) peptides within Aβ oligomers and fibrils would be cleaved more slowly than the hydrophilic N terminus (Aβ(1–16)).
We chose to use Proteinase K because it cuts at 19 positions that are distributed throughout the N-terminal, middle, and C-terminal regions of Aβ. We added Proteinase K (0.5 μg/ml) to each Aβ conformer (25 μm) and deposited Aβ samples on nitrocellulose membranes every 10–30 min during the fragmentation reaction (150 min). Because the deposited Aβ samples dry quickly (within seconds) once spotted on nitrocellulose, the proteolytic reaction is quenched rapidly and can be interrogated after the fragmentation reaction is complete. We found that the rate of proteolytic cleavage for the hydrophilic N terminus (Aβ(3–10)) was the same for Aβ monomers, oligomers, and fibrils (Fig. 5). In contrast, the fragmentation of the central Aβ peptide segment (Aβ(18–22)) was slower for Aβ oligomers and fibrils than for monomers, suggesting that this central region is more solvent-protected in aggregated Aβ conformers than in Aβ monomers. Importantly, this same region was more protected from proteolytic fragmentation within A− oligomers than within A+ oligomers and most protected within fibrils (Fig. 5). We obtained identical results using a second antibody against an overlapping Aβ peptide segment (Aβ residues 16–21; supplemental Fig. S6). We also confirmed that both antibodies bound to each Aβ conformer with the same apparent affinity (supplemental Fig. S7).
FIGURE 5.
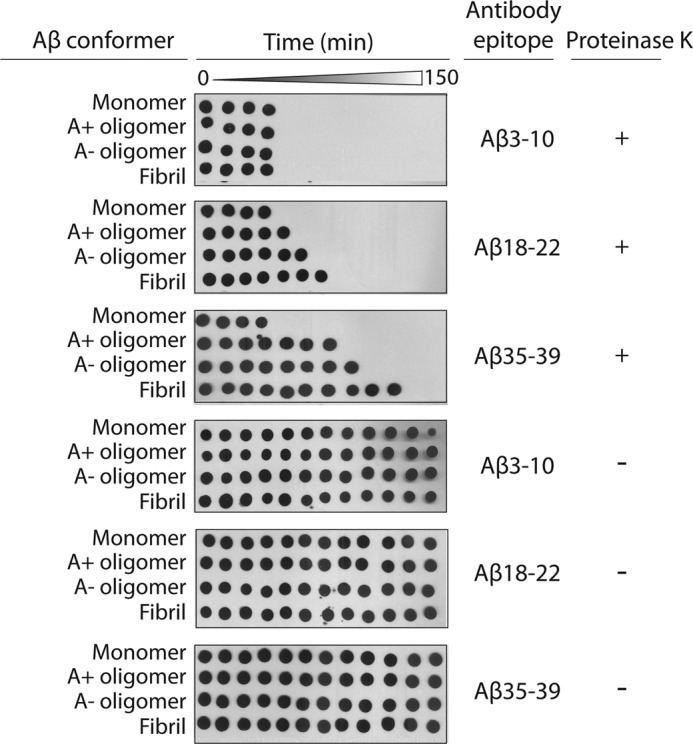
Rate of proteolytic fragmentation of Aβ peptide segments within Aβ oligomers. Aβ42 (25 μm) was incubated with Proteinase K (0.5 μg/ml), deposited periodically on nitrocellulose (every 10–30 min), and probed with antibodies specific for N-terminal (Aβ(3–10); 6E10), central (Aβ(18–22); 4G8), and C-terminal (Aβ(35–39); 9F1) Aβ epitopes. The time intervals were 0, 10, 20, 30, 40, 50, 60, 70, 80, 90, 120, and 150 min.
Next we investigated the rate of proteolytic fragmentation of a C-terminal hydrophobic peptide segment of Aβ (residues 35–39) within each Aβ conformer (Fig. 5). Importantly, this C-terminal hydrophobic motif was more protected from the protease than the central (Aβ(18–22)) and N-terminal (Aβ(3–10)) peptide segments within Aβ oligomers and fibrils. Moreover, the same C-terminal Aβ peptide (Aβ(35–39)) was more protected within A− oligomers than within A+ oligomers and most protected within Aβ fibrils. We obtained identical results for two additional antibodies directed against overlapping C-terminal Aβ peptides (Aβ(30–35) and Aβ(37–42); supplemental Fig. S6). Collectively, our results reveal that hydrophobic residues within the central and C-terminal regions of Aβ42 are more solvent-protected within A− Aβ oligomers than within A+ oligomers.
These proteolytic fragmentation results that suggest specific differences in the extent of solvent exposure of hydrophobic Aβ peptides within A+ and A− oligomers led us to evaluate the overall hydrophobicity of each Aβ conformer using the hydrophobic dye ANS (Fig. 6A and supplemental Fig. S8). As expected, we found that Aβ monomers (day 0) were most hydrophobic, as judged by their blue-shifted spectra (λmax = 455 ± 1 nm). Moreover, we found that the hydrophobicity of the aggregated Aβ conformers was highest for A+ oligomers (λmax = 483 ± 1 nm; days 1–3), intermediate for A− oligomers (λmax = 502 ± 1 nm; days 4–6), and lowest for fibrils (λmax = 527 ± 1 nm). We also confirmed that the same patterns of hydrophobicity for each Aβ conformer were obtained at multiple ratios of ANS to Aβ (supplemental Fig. S8). Moreover, we confirmed that each Aβ conformer possesses a high level of homogeneity, as evidenced by a single peak in the ANS emission spectra (supplemental Fig. S8) relative to the multiple ANS peaks obtained when mixing Aβ oligomers and fibrils (supplemental Fig. S9). In summary, the increased hydrophobicity of A+ oligomers relative to A− oligomers and fibrils revealed by ANS is consistent with our proteolytic fragmentation results (Fig. 5).
FIGURE 6.
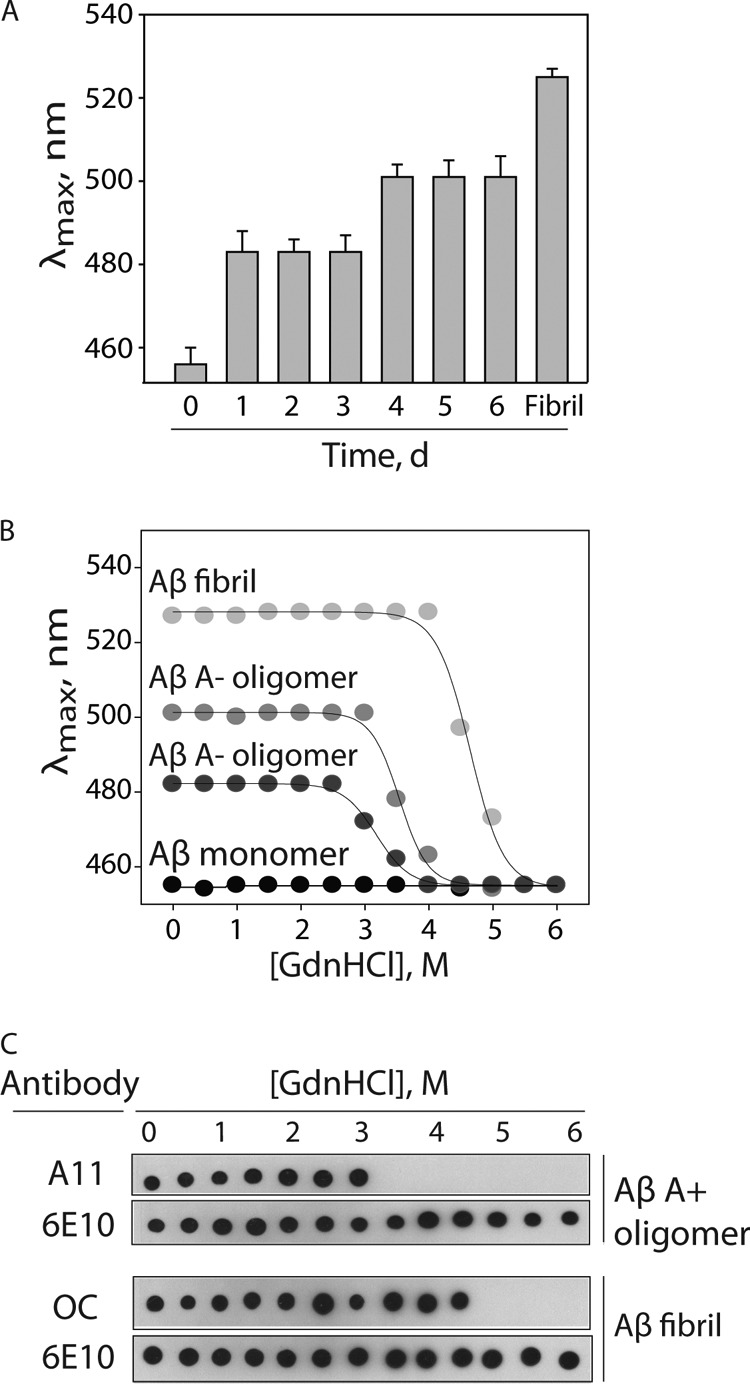
Analysis of hydrophobicity and conformational stability of Aβ oligomers. A, Aβ42 (25 μm) was assembled without agitation, and the hydrophobicity of Aβ conformers formed each day was evaluated using ANS fluorescence. The wavelength corresponding to the maximum ANS fluorescence is reported on the y axis. B and C, Aβ conformers (25 μm) were incubated with variable amounts of guanidine hydrochloride, and then their relative degree of unfolding was evaluated using ANS fluorescence (B) and antibodies (A11, prefibrillar oligomers; OC, fibrillar conformers; 6E10, N terminus of Aβ) (C). Error bars, S.D.
Aβ A+ Oligomers Are Less Stably Folded than A− Oligomers
Based on our ANS and proteolytic results, we posited that A+ oligomers are less stably folded than A− oligomers and fibrils. To evaluate this hypothesis, we analyzed the conformational stability of each Aβ conformer in guanidine hydrochloride (GdnHCl) using ANS fluorescence analysis (Fig. 6B). We found that A+ oligomers unfolded at lower denaturant concentrations (D½ = 3.2 ± 0.03 m GdnHCl) than A− oligomers (D½ = 3.5 ± 0.03 m GdnHCl) and fibrils (D½ = 4.6 ± 0.04 m GdnHCl; Fig. 6B). The increased stability of A− oligomers relative to A+ oligomers is consistent with our finding that A− oligomers were insoluble in a strong surfactant (0.5% lithium dodecyl sulfate), whereas A+ oligomers were soluble in lithium dodecyl sulfate (supplemental Fig. S10).
It is also possible that A+ oligomers possess a small fraction of highly toxic oligomers with unique biochemical properties. This hypothesis would predict that the bulk biochemical properties of A+ oligomers (as judged by nonspecific dyes, such as ANS) would be different from those of the specific toxic oligomers themselves (as judged by specific probes, such as the A11 antibody). Therefore, we asked whether the conformational stability of A+ oligomers evaluated by ANS and the A11 antibody are similar (Fig. 6C). Indeed, we found that the A11 epitope was eliminated at 3.5 m GdnHCl, consistent with the ANS results (D½ = 3.2 ± 0.03 m GdnHCl). We also performed a similar analysis for Aβ fibrils using the OC antibody. We found that the OC epitope was eliminated at 5 m GdnHCl (Fig. 6C), which is also consistent with the ANS results (D½ = 4.6 ± 0.04 M GdnHCl). These results reveal that the folding stabilities of A11 (oligomer)- and OC (fibrillar)-positive conformers are weakly dependent on the method used to measure them.
Aβ Oligomers Lack β-Sheet Structure
We next investigated whether differences in secondary structure between A+ and A− oligomers explain their differential toxicity. We first asked whether A− oligomers are positive for the amyloid dye ThT, as would be expected for β-sheet-rich conformers (Fig. 7A). In contrast to Aβ fibrils, A− oligomers display low ThT fluorescence that is indistinguishable from the ThT fluorescence of Aβ monomers and A+ oligomers. We also evaluated the secondary structure of each Aβ oligomer using circular dichroism spectroscopy (Fig. 7B). Importantly, both Aβ oligomers possess random-coil structure that is indistinguishable from Aβ monomers and significantly different than the β-sheet structure of Aβ fibrils. Finally, we asked whether the Aβ oligomers are competent for seeding Aβ monomers into ThT-positive conformers in a manner similar to Aβ fibrils (Fig. 7C). Importantly, neither Aβ oligomer seeds Aβ monomers. We conclude that the secondary structure of A− oligomers is similar to that of A+ oligomers and significantly different from that of Aβ fibrils.
FIGURE 7.
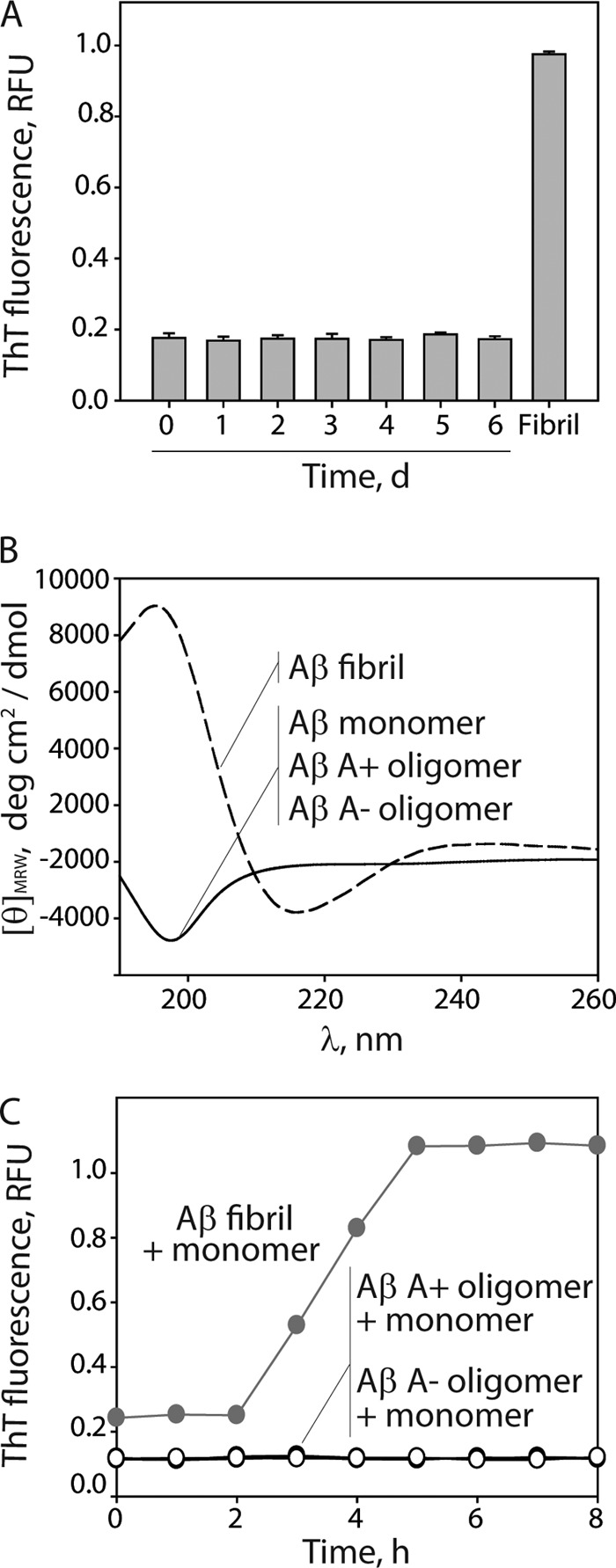
Characterization of the secondary structure and seeding activity of Aβ oligomers. Aβ42 (25 μm) was assembled without agitation (0–6 days), and its extent of fibrillization and secondary structure were evaluated using ThT fluorescence (A) and circular dichroism (B). C, Aβ fibrils and oligomers were mixed with Aβ monomers (25 μm, 5% seed), and their ThT fluorescence was monitored. RFU, relative fluorescence units. Error bars, S.D.
DISCUSSION
We have evaluated the conformational differences between two Aβ oligomers of similar size in order to carefully separate the contributions of aggregate size and conformation to cellular toxicity. An important finding of our studies is that increased solvent exposure of two hydrophobic Aβ peptide motifs within non-fibrillar Aβ oligomers is linked to increased cellular toxicity and disruption of lipid membranes. Our studies share important similarities with and differences from a previous study of two oligomers of a non-pathological bacterial protein (HypF-N) that possess similar size and dissimilar toxicity (13). Although HypF-N and Aβ possess little sequence similarity (6%), our finding that less well folded Aβ oligomers that are more toxic is consistent with the structural data for HypF-N oligomers. A notable difference between our work and this previous study is that the HypF-N oligomers are β-sheet-rich, whereas our Aβ oligomers lack β-sheets. The similar size-independent differences in toxicity for both β-sheet HypF-N oligomers and non-β-sheet Aβ oligomers may suggest that the toxicity of small amyloidogenic oligomers is governed primarily by the degree of solvent exposure of hydrophobic residues and is weakly influenced by their secondary structures. It will be important in the future to compare the toxicity of the non-β-sheet A+ oligomers formed in this work with β-sheet-rich Aβ oligomers reported previously (2, 11, 16).
Our identification of two Aβ42 oligomers of similar size and dissimilar toxicity is reminiscent of two Aβ oligomers (Aβ-derived diffusible ligands) identified previously (8). An important advance in our work is the development of reproducible methods for forming low toxicity Aβ oligomers, which were reported previously to form sporadically (8). It is notable in this previous study that Aβ42 was assembled at high concentration (100 μm) and low temperature (4 °C) in cell culture media. We find that lower Aβ concentrations (25 μm) are essential to control the formation of A11- and OC-negative Aβ oligomers, and that the low temperature and cell culture media are unnecessary because we conducted our studies at 25 °C in PBS. We posit that the lack of reproducibility in the previous study was due to the elevated Aβ concentration and/or the use of cell culture media that would significantly accelerate aggregation and potentially promote heterogeneous nucleation with components in the media.
The intriguing concentration dependence we observed for Aβ oligomerization deserves further consideration. We find that A− oligomers form at Aβ concentrations of 20–25 μm without agitation, whereas fibrils form at higher Aβ concentrations (50–75 μm). Moreover, at Aβ concentrations of <20 μm, we find that Aβ fails to form oligomers or fibrils over 10 days in the absence of mixing. Our findings are consistent with two primary aggregation pathways for quiescent assembly: one that is dominant at Aβ concentrations near the threshold concentration necessary for aggregation and a second one that is dominant at elevated Aβ concentrations. At 20–25 μm Aβ, we find that Aβ A+ oligomers mature into “off-pathway” oligomers that possess similarities (e.g. insoluble in strong surfactant) and differences (e.g. non-β-sheet structure) relative to Aβ fibrils. We have previously shown that A+ oligomers at 25 μm mature into fibrils instead of A− oligomers when agitated (14, 28), revealing that quiescent assembly is necessary to favor formation of A− oligomers. We posit that relatively low Aβ concentrations and/or the lack of agitation favor A+ oligomers to undergo a modest conformational change that results in decreased solvent exposure of hydrophobic residues without fully maturing into β-sheet fibrils. In contrast, we posit that higher Aβ concentrations and/or agitation favor A+ oligomers to mature into β-sheet fibrils without being trapped in the A− oligomer conformation.
It will be interesting to evaluate if Aβ A− oligomers that we formed in vitro also form in vivo. Although the range of Aβ concentrations used in our study (20–75 μm) are much higher than the concentrations found in extracellular fluid (which are as low as 1 nm) (30–33), it is well established that A+ oligomers form in vivo (17, 34, 35). One possible explanation for the ability of Aβ to oligomerize in vivo is that Aβ can be concentrated significantly (>2.5 μm) within endosomes and lysosomes (36). High concentrations of Aβ within such intracellular compartments coupled with molecular crowding effects (37) may explain the ability of Aβ to oligomerize in vivo despite extremely low levels of Aβ in extracellular fluid. The antibody A11 has been invaluable for detecting A+ oligomers that form in vivo (17, 34, 35), and it will be necessary to also raise antibodies against A− oligomers to evaluate the biological relevance of such oligomers. Nevertheless, our characterization of a less toxic Aβ oligomeric isoform (A−) that is similar in size to the biologically relevant A+ Aβ oligomer provides important insights into size-independent mechanisms of Aβ-mediated cytotoxicity. We expect that our findings will motivate future site-specific structural studies to elucidate in more detail the conformational differences encoding the dissimilar toxicities of non-fibrillar A+ and A− Aβ oligomers.
Supplementary Material
Acknowledgments
We thank Anne Marie Hickey and Jonathan S. Dordick for providing Proteinase K and x-ray films.
This work was supported, in whole or in part, by National Institutes of Health Grant AG-27317 (to S. O. S.). This work was also supported by Alzheimer's Association Grant NIRG-08-90967 (to P. M. T.), American Health Assistance Foundation Grant ADR-A2011355 (to P. M. T.), National Science Foundation Grant CBET NIRT 0608978 (to R. S. K.), and the P. K. Lashmet Chair (to R. S. K.).

This article contains supplemental Figs. S1–S10.
- ThT
- thioflavin T
- ANS
- 8-anilinonaphthalene-1-sulfonate
- GdnHCl
- guanidine hydrochloride
- LDH
- lactate dehydrogenase
- AFM
- atomic force microscopy
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Kirkitadze M. D., Bitan G., Teplow D. B. (2002) Paradigm shifts in Alzheimer's disease and other neurodegenerative disorders. The emerging role of oligomeric assemblies. J. Neurosci. Res. 69, 567–577 [DOI] [PubMed] [Google Scholar]
- 2. Ono K., Condron M. M., Teplow D. B. (2009) Structure-neurotoxicity relationships of amyloid β-protein oligomers. Proc. Natl. Acad. Sci. U.S.A. 106, 14745–14750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hoshi M., Sato M., Matsumoto S., Noguchi A., Yasutake K., Yoshida N., Sato K. (2003) Spherical aggregates of β-amyloid (amylospheroid) show high neurotoxicity and activate Tau protein kinase I/glycogen synthase kinase-3β. Proc. Natl. Acad. Sci. U.S.A. 100, 6370–6375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cizas P., Budvytyte R., Morkuniene R., Moldovan R., Broccio M., Lösche M., Niaura G., Valincius G., Borutaite V. (2010) Size-dependent neurotoxicity of β-amyloid oligomers. Arch. Biochem. Biophys. 496, 84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sakono M., Zako T. (2010) Amyloid oligomers. Formation and toxicity of Aβ oligomers. FEBS J. 277, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 6. Keshet B., Yang I. H., Good T. A. (2010) Can size alone explain some of the differences in toxicity between β-amyloid oligomers and fibrils? Biotechnol. Bioeng. 106, 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., Selkoe D. J. (2002) Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416, 535–539 [DOI] [PubMed] [Google Scholar]
- 8. Chromy B. A., Nowak R. J., Lambert M. P., Viola K. L., Chang L., Velasco P. T., Jones B. W., Fernandez S. J., Lacor P. N., Horowitz P., Finch C. E., Krafft G. A., Klein W. L. (2003) Self-assembly of Aβ(1–42) into globular neurotoxins. Biochemistry 42, 12749–12760 [DOI] [PubMed] [Google Scholar]
- 9. Walsh D. M., Selkoe D. J. (2007) Aβ oligomers. A decade of discovery. J. Neurochem. 101, 1172–1184 [DOI] [PubMed] [Google Scholar]
- 10. Lee S., Fernandez E. J., Good T. A. (2007) Role of aggregation conditions in structure, stability, and toxicity of intermediates in the Aβ fibril formation pathway. Protein Sci. 16, 723–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ahmed M., Davis J., Aucoin D., Sato T., Ahuja S., Aimoto S., Elliott J. I., Van Nostrand W. E., Smith S. O. (2010) Structural conversion of neurotoxic amyloid β(1–42) oligomers to fibrils. Nat. Struct. Mol. Biol. 17, 561–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Glabe C. G. (2008) Structural classification of toxic amyloid oligomers. J. Biol. Chem. 283, 29639–29643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Campioni S., Mannini B., Zampagni M., Pensalfini A., Parrini C., Evangelisti E., Relini A., Stefani M., Dobson C. M., Cecchi C., Chiti F. (2010) A causative link between the structure of aberrant protein oligomers and their toxicity. Nat. Chem. Biol. 6, 140–147 [DOI] [PubMed] [Google Scholar]
- 14. Ladiwala A. R., Lin J. C., Bale S. S., Marcelino-Cruz A. M., Bhattacharya M., Dordick J. S., Tessier P. M. (2010) Resveratrol selectively remodels soluble oligomers and fibrils of amyloid Abeta into off-pathway conformers. J. Biol. Chem. 285, 24228–24237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petkova A. T., Leapman R. D., Guo Z., Yau W. M., Mattson M. P., Tycko R. (2005) Self-propagating, molecular level polymorphism in Alzheimer's β-amyloid fibrils. Science 307, 262–265 [DOI] [PubMed] [Google Scholar]
- 16. Kayed R., Glabe C. G. (2006) Conformation-dependent anti-amyloid oligomer antibodies. Methods Enzymol. 413, 326–344 [DOI] [PubMed] [Google Scholar]
- 17. Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G. (2003) Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 [DOI] [PubMed] [Google Scholar]
- 18. Kayed R., Head E., Sarsoza F., Saing T., Cotman C. W., Necula M., Margol L., Wu J., Breydo L., Thompson J. L., Rasool S., Gurlo T., Butler P., Glabe C. G. (2007) Fibril-specific, conformation-dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol. Neurodegener. 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O'Nuallain B., Wetzel R. (2002) Conformational Abs recognizing a generic amyloid fibril epitope. Proc. Natl. Acad. Sci. U.S.A. 99, 1485–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lambert M. P., Velasco P. T., Chang L., Viola K. L., Fernandez S., Lacor P. N., Khuon D., Gong Y., Bigio E. H., Shaw P., De Felice F. G., Krafft G. A., Klein W. L. (2007) Monoclonal antibodies that target pathological assemblies of Aβ. J. Neurochem. 100, 23–35 [DOI] [PubMed] [Google Scholar]
- 21. Lambert M. P., Viola K. L., Chromy B. A., Chang L., Morgan T. E., Yu J., Venton D. L., Krafft G. A., Finch C. E., Klein W. L. (2001) Vaccination with soluble Aβ oligomers generates toxicity-neutralizing antibodies. J. Neurochem. 79, 595–605 [DOI] [PubMed] [Google Scholar]
- 22. Zameer A., Kasturirangan S., Emadi S., Nimmagadda S. V., Sierks M. R. (2008) Anti-oligomeric Aβ single-chain variable domain antibody blocks Aβ-induced toxicity against human neuroblastoma cells. J. Mol. Biol. 384, 917–928 [DOI] [PubMed] [Google Scholar]
- 23. Habicht G., Haupt C., Friedrich R. P., Hortschansky P., Sachse C., Meinhardt J., Wieligmann K., Gellermann G. P., Brodhun M., Götz J., Halbhuber K. J., Röcken C., Horn U., Fändrich M. (2007) Directed selection of a conformational antibody domain that prevents mature amyloid fibril formation by stabilizing Aβ protofibrils. Proc. Natl. Acad. Sci. U.S.A. 104, 19232–19237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perchiacca J. M., Ladiwala A. R., Bhattacharya M., Tessier P. M. (2012) Structure-based design of conformation- and sequence-specific antibodies against amyloid β. Proc. Natl. Acad. Sci. U.S.A. 109, 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kayed R., Canto I., Breydo L., Rasool S., Lukacsovich T., Wu J., Albay R., 3rd, Pensalfini A., Yeung S., Head E., Marsh J. L., Glabe C. (2010) Conformation-dependent monoclonal antibodies distinguish different replicating strains or conformers of prefibrillar Aβ oligomers. Mol. Neurodegener 5, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mueller P., Rudin D. O., Tien H. T., Wescott W. C. (1962) Reconstitution of cell membrane structure in vitro and its transformation into an excitable system. Nature 194, 979–980 [DOI] [PubMed] [Google Scholar]
- 27. Ladiwala A. R., Mora-Pale M., Lin J. C., Bale S. S., Fishman Z. S., Dordick J. S., Tessier P. M. (2011) Polyphenolic glycosides and aglycones utilize opposing pathways to selectively remodel and inactivate toxic oligomers of amyloid β. Chembiochem 12, 1749–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ladiwala A. R., Dordick J. S., Tessier P. M. (2011) Aromatic small molecules remodel toxic soluble oligomers of amyloid beta through three independent pathways. J. Biol. Chem. 286, 3209–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kayed R., Sokolov Y., Edmonds B., McIntire T. M., Milton S. C., Hall J. E., Glabe C. G. (2004) Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J. Biol. Chem. 279, 46363–46366 [DOI] [PubMed] [Google Scholar]
- 30. Seubert P., Vigo-Pelfrey C., Esch F., Lee M., Dovey H., Davis D., Sinha S., Schlossmacher M., Whaley J., Swindlehurst C. (1992) Isolation and quantification of soluble Alzheimer's β-peptide from biological fluids. Nature 359, 325–327 [DOI] [PubMed] [Google Scholar]
- 31. Galasko D., Chang L., Motter R., Clark C. M., Kaye J., Knopman D., Thomas R., Kholodenko D., Schenk D., Lieberburg I., Miller B., Green R., Basherad R., Kertiles L., Boss M. A., Seubert P. (1998) High cerebrospinal fluid Tau and low amyloid β42 levels in the clinical diagnosis of Alzheimer disease and relation to apolipoprotein E genotype. Arch. Neurol. 55, 937–945 [DOI] [PubMed] [Google Scholar]
- 32. Andreasen N., Hesse C., Davidsson P., Minthon L., Wallin A., Winblad B., Vanderstichele H., Vanmechelen E., Blennow K. (1999) Cerebrospinal fluid β-amyloid(1–42) in Alzheimer disease. Differences between early- and late-onset Alzheimer disease and stability during the course of disease. Arch. Neurol. 56, 673–680 [DOI] [PubMed] [Google Scholar]
- 33. Strozyk D., Blennow K., White L. R., Launer L. J. (2003) CSF Aβ 42 levels correlate with amyloid neuropathology in a population-based autopsy study. Neurology 60, 652–656 [DOI] [PubMed] [Google Scholar]
- 34. Lesné S., Koh M. T., Kotilinek L., Kayed R., Glabe C. G., Yang A., Gallagher M., Ashe K. H. (2006) A specific amyloid-β protein assembly in the brain impairs memory. Nature 440, 352–357 [DOI] [PubMed] [Google Scholar]
- 35. Tomic J. L., Pensalfini A., Head E., Glabe C. G. (2009) Soluble fibrillar oligomer levels are elevated in Alzheimer's disease brain and correlate with cognitive dysfunction. Neurobiol. Dis. 35, 352–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu X., Crick S. L., Bu G., Frieden C., Pappu R. V., Lee J. M. (2009) Amyloid seeds formed by cellular uptake, concentration, and aggregation of the amyloid-β peptide. Proc. Natl. Acad. Sci. U.S.A. 106, 20324–20329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Munishkina L. A., Cooper E. M., Uversky V. N., Fink A. L. (2004) The effect of macromolecular crowding on protein aggregation and amyloid fibril formation. J. Mol. Recognit. 17, 456–464 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.