Abstract
Multiple sclerosis (MS) is a polygenic disease characterized by inflammation and demyelination in the central nervous system (CNS), which can be modeled in experimental autoimmune encephalomyelitis (EAE). The Eae18b locus on rat chromosome 10 has previously been linked to regulation of beta-chemokine expression and severity of EAE. Moreover, the homologous chemokine cluster in humans showed evidence of association with susceptibility to MS. We here established a congenic rat strain with Eae18b locus containing a chemokine cluster (Ccl2, Ccl7, Ccl11, Ccl12 and Ccl1) from the EAE- resistant PVG rat strain on the susceptible DA background and utilized myelin oligodendrocyte glycoprotein (MOG)-induced EAE to characterize the mechanisms underlying the genetic regulation. Congenic rats developed a milder disease compared to the susceptible DA strain, and this was reflected in decreased demyelination and in reduced recruitment of inflammatory cells to the brain. The congenic strain also showed significantly increased Ccl11 mRNA expression in draining lymph nodes and spinal cord after EAE induction. In the lymph nodes, macrophages were the main producers of CCL11, whereas macrophages and lymphocytes expressed the main CCL11 receptor, namely CCR3. Accordingly, the congenic strain also showed significantly increased Ccr3 mRNA expression in lymph nodes. In the CNS, the main producers of CCL11 were neurons, whereas CCR3 was detected on neurons and CSF producing ependymal cells. This corresponded to increased levels of CCL11 protein in the cerebrospinal fluid of the congenic rats. Increased intrathecal production of CCL11 in congenic rats was accompanied by a tighter blood brain barrier, reflected by more occludin+ blood vessels. In addition, the congenic strain showed a reduced antigen specific response and a predominant anti-inflammatory Th2 phenotype. These results indicate novel mechanisms in the genetic regulation of neuroinflammation.
Introduction
Multiple sclerosis (MS) is an inflammatory demyelinating disease of the central nervous system (CNS), causing loss of sensory and motor functions in affected individuals. The disease is polygenic, and several genes contributing to susceptibility have been identified and confirmed [1], [2]. Characterization of the molecular mechanisms that mediate influence of MS risk genes can provide novel insights into disease pathogenesis. For example, genetically regulated expression of CD58, a cell adhesion molecule expressed primarily on macrophages, is protective due to enhanced activation of regulatory T cells [3]. However, the genes identified so far still explain only a part of the disease heritability [4]. In addition, the underlying mechanisms are described only for a limited set of genes. To further dissect the genetic and pathological mechanisms of neuroinflammation, several animal models are used. Myelin oligodendrocyte glycoprotein (MOG)-induced experimental autoimmune encephalomyelitis (EAE) is a well characterized animal model of MS, sharing several important features including T helper 1 (Th1), Th17 and B-cell involvement as well as histopathological characteristics [5], [6], [7], [8].
Previous linkage analyses in experimental crosses and congenic strains have identified a quantitative trait locus (QTL) on rat chromosome 10 [9], [10] and the homologous region on mouse chromosome 11 [11] as a candidate region for regulation of neuroinflammation. Initial linkage scans identified regions too large to allow for candidate gene investigation, but we have been able to restrict the QTL to a 0.88 Mb interval known as Eae18b, which includes a cluster of chemokine genes (Ccl2, Ccl7, Ccl11, Ccl12 and Ccl1) using high-resolution mapping approaches [12], [13]. Chemokines are small, secreted proteins involved in chemotaxis and activation of a variety of immune cells, acting through their corresponding receptors (reviewed in [14]). We have previously demonstrated that expression of these chemokine genes during EAE is regulated by the genotype in the region, i.e. cis-regulation [13].
In this study we sought to confirm the impact of the Eae18b locus on neuroinflammation and identify the chemokine(s) in the gene cluster that underlies the genetic regulation of EAE. Through selective backcrossing, we created a congenic strain (DA.PVG-Eae18b) that comprises alleles from the EAE-resistant Piebald Virol Glaxo (PVG) strain on the EAE-susceptible Dark Agouti (DA) background. These two strains differ in chemokine gene expression, but have the identical exonic sequences in the chemokine genes. We studied the impact of Eae18b locus on EAE development and observed a milder disease and a higher expression of CCL11 mRNA and protein levels in inguinal lymph nodes in the Eae18b congenic strain. The upregulation of CCL11 and its main receptor CCR3 was associated with a Th2-like immune response, which could skew the inflammatory response towards an anti-inflammatory state. In addition, we found increased levels of CCL11 in the cerebrospinal fluid (CSF) of the congenic strain, which possibly influences the blood brain barrier (BBB) integrity.
Results
Eae18b Congenic Animals Display Milder Disease
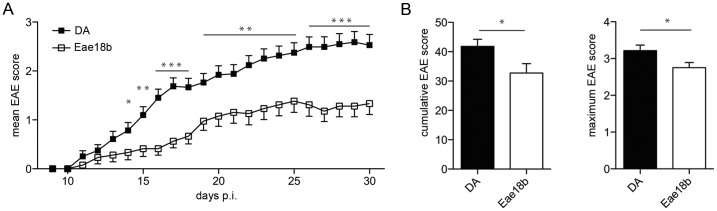
To confirm the biological effect of Eae18b in EAE, we immunized congenic DA.PVG-Eae18b and control DA rats with MOG. The congenic strain developed an overall milder disease with lower mean EAE score from day 14 post immunization (p.i.) until the end of the experiment compared to the DA strain (Figure 1A). The congenic strain showed a reduced incidence of disease with an overall incidence of 51% compared to 86% in the DA strain (p-value: 0.0004, Fischer exact test). In addition, the affected animals in the congenic strain also displayed a reduced severity of disease, reflected in significantly lower cumulative and maximum EAE scores (Figure 1B).
Figure 1. Congenic DA.PVG-Eae18b rats display clinically milder EAE.
(A) Mean daily EAE score from 51 DA and 39 DA.PVG-Eae18b (Eae18b) rats, immunized with MOG, p.i: post immunization. The graph includes daily mean scores from all immunized animals in two independent experiments. (B) Mean cumulative EAE score and mean EAE maximum score in affected rats (44 DA and 20 DA.PVG-Eae18b animals). Error bars represent SEM, and (*) indicates p-value <0.05, (**) indicates p-value <0.01, (***) indicates p-value <0.001.
Ccl11 is Highly Expressed in Eae18b Congenic Animals during EAE
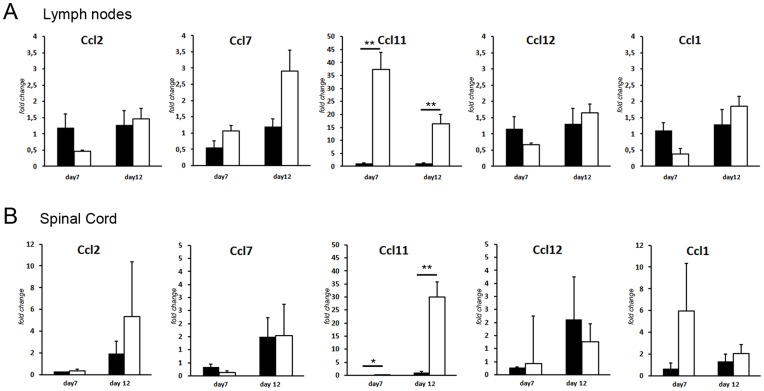
We have previously demonstrated a cis-regulation of chemokine transcription during EAE [13]. Therefore we sought to investigate the role of the chemokine expression in the Eae18b region in the regulation of inflammation. In order to study the kinetics of the expression of the chemokine genes during the induction of disease, mRNA levels of Ccl2, Ccl7, Ccl11, Ccl12 and Ccl1 were measured in lymph nodes and spinal cords from parental DA and PVG.av1 rats, at day 0, 5, 7 and 12 days after MOG immunization, using real-time quantitative PCR (qPCR). The expression analysis in the parental strains (DA and PVG.av1) revealed different regulation of Ccl11 and Ccl12 on day 7 and 12 p.i. in the lymph nodes and on day 12 p.i. in the spinal cord (Figure S1A and B). Having established the time points when the parental strains differ in expression, we further analyzed the mRNA expression of the chemokine genes in the DA.PVG-Eae18b congenic strain compared to littermate DA controls. In lymph nodes and spinal cord, Ccl11 was significantly upregulated in the congenic animals compared to the DA strain on both day 7 and 12 p.i. (Figure 2A and B). The other chemokine genes included in the cluster did not show any significant differential expression. These findings directly associate the regulation of Ccl11 expression to the genetic region defined by the congenic fragment.
Figure 2. Eae18b congenic animals significantly upregulate Ccl11 mRNA.
qPCR was used to measure mRNA expression of Ccl2, Ccl7, Ccl11, Ccl12 and Ccl1 in draining lymph nodes (A) and spinal cord tissue (B) on day 7 and 12 p.i. Ccl11 mRNA was significantly upregulated in the congenic strain in both lymph node and spinal cord tissue at both time-points. Five to eight animals per strain were used at each time-point and experiments were repeated three times. Error bars represent SEM, statistics were calculated using Mann-Whitney non-parametric test, (*) indicates p-value <0.05, (**) indicates p-value <0.01.
Ccl11 is Predominantly Expressed by Macrophages in the Lymph Nodes
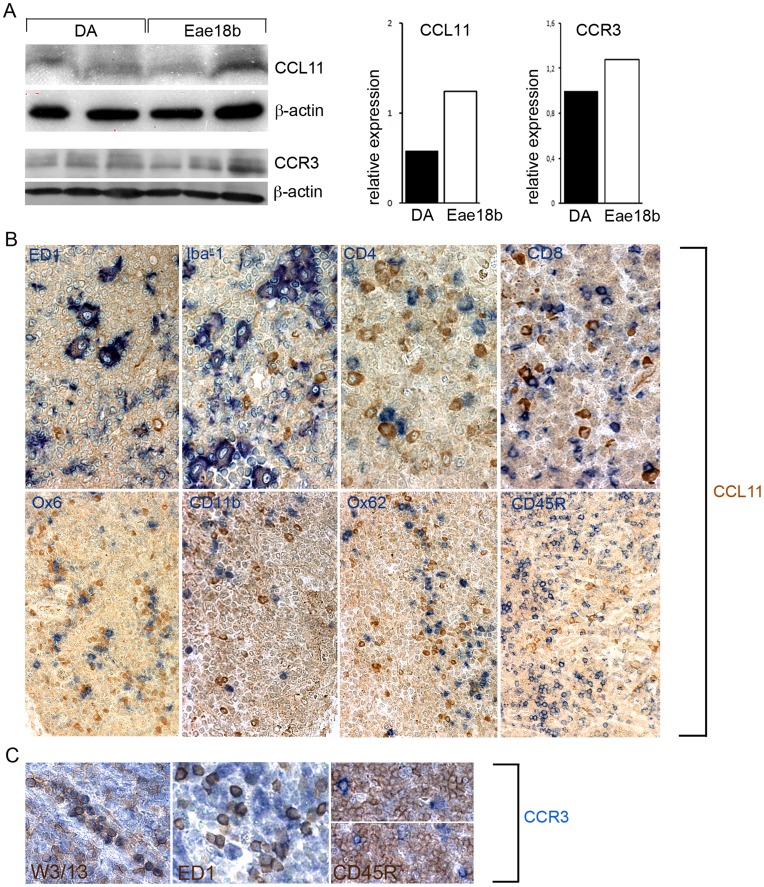
In order to confirm that the increased Ccl11 and Ccr3 mRNA expression also resulted in elevated CCL11 and CCR3 protein levels, respectively, we performed Western Blot and extensive immunohistochemical (IHC) analysis on cross-sections from draining inguinal lymph nodes harvested on day 7 p.i. Western blot analysis revealed that the lymph nodes of the congenic strain expressed increased levels of CCL11 and CCR3 protein (Figure 3A). Thereafter, we aimed to elucidate the cellular source of the increased CCL11 and CCR3 production in the congenic lymph nodes. IHC analysis revealed that the main producers of CCL11 in the lymph nodes were ED1+ and/or Iba-1+ macrophages, as well as plasma cells but none of the CCL11+ cells were Ox6+, CD11b+ or Ox62+ dendritic cells. We did not observe any CCL11 secreting CD3+ (CD4+ or CD8+) T lymphocytes or CD45RA+ B-cells (Figure 3B). CCR3 was expressed by W3/13+ lymphocytes, as well as by the ED1+ macrophage population. No CCR3+/CD45RA+ B-cells were detected in the peripheral lymph node tissue (Figure 3C).
Figure 3. CCL11 and CCR3 protein levels in the inguinal lymph nodes.
(A) Western blot analysis of CCL11 and its main receptor CCR3 in inguinal lymph nodes on day 7 p.i. Protein levels of both targets were increased in the congenic animals compared to the controls (data normalized to β- actin). CCL11, CCR3 and β- actin shown in original representative blots and graphically; three blots performed for each target within two separate EAE experiments. (B) Co-staining with antibodies directed against CCL11 and other immune cell types on paraffin cross sections of inguinal lymph nodes performed on day 7 p.i. in the congenic Eae18b strain. CCL11 co-localized with the macrophage/microglia markers ED1 and Iba-1 (first two images from the left, upper row). CCL11 was not expressed by CD4+ and CD8+ T cells (last two images, upper row). CCL11 was also not detected in MHC class II+ (Ox6+) and CD11b+ populations, neither in Ox62+ dendritic, nor in CD45RA+ B cells (second row, from left to right). (C) The main CCL11 receptor CCR3 was detected on W3/13+ T lymphocytes and ED1+ macrophages, but not on CD45RA+ B cells (five congenic and five control rats analyzed, two whole inguinal lymph nodes per each animal).
Eae18b Regulates the T Cell Response in EAE
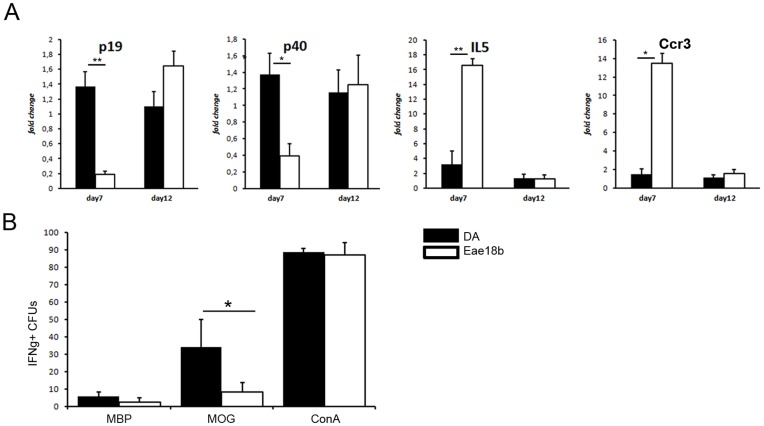
We further analyzed the mRNA expression levels of cytokines and receptors characteristic for either Th1/Th17 or Th2 response. IL-5, one of the key cytokines of the Th2 response, was upregulated in the lymph nodes of the congenic animals compared to DA controls (Figure 4A). In contrast, cytokines associated with a Th1/Th17 response, i.e. the IL-23 subunits p19 and p40, were upregulated in the DA strain compared to the congenic strain (Figure 4A). We also observed a tendency for higher expression of Gata-3 and IL-4 in the lymph nodes of congenic animals, but the difference was not significant (data not shown). Additionally, the peripheral lymph node tissue from the congenic animals harvested on day 7 also showed significant upregulation of Ccr3 mRNA (Figure 4A), the gene coding for the major CCL11 receptor [15]. Using flow cytometry, we analyzed the activation and recruitment of cells in the lymph nodes during the induction phase of the disease (day 7 p.i.), but could not detect any differences in major T cell subsets, B cells, macrophages or dendritic cells (Figures S2 and S3). Further, in vitro proliferative capability of lymphocytes (Figure S4), as well as specific anti-MOG antibody production (IgG1, IgG2b, IgG2c and total IgG, Figure S5) was also similar between the strains, indicating that the disease regulation by the congenic region is not primarily through regulation of these phenotypes. However, all those analysis have been performed during initial, or the early disease phase. In order to investigate if the specific response to MOG was altered in congenics in a later disease time-point, we performed an IFN-γ Elispot analysis on ex vivo splenocytes on day 15 and 21 p.i. The specific T cell response towards MOG antigen was 3-fold downregulated in the congenic rats compared to the controls (Figure 4B), whereas the response to the positive control Concavalin A and myelin basic protein (MBP), used as an unspecific antigen, was similar in both groups. These results indicate that Eae18b locus specifically modifies the T cell response by a Th2-like activation and suppression of the T cell expansion in response to MOG, thereby reducing the autoimmune response.
Figure 4. Eae18b modulates the immune response and reduces activation of MOG specific T cells.
(A) Pro-inflammatory cytokines p19, p40 were significantly upregulated in controls (DA), while congenic (Eae18b) animals upregulated anti-inflammatory IL-5 cytokine as well as CCR3, main receptor for CCL11 chemokine (mRNA analysis of the inguinal lymph node tissue collected day 7 and 12 p.i.). Five to eight animals per strain were used in all time points analyzed and experiments were repeated twice. Error bars represent SEM, statistics were calculated using Mann-Whitney non-parametric test, (*) indicates p-value <0.05, (**) indicates p-value <0.01. (B) Elispot analysis revealed less pathogenic IFN-γ producing MOG specific T cells in the Eae18b strain; rat splenocytes harvested on day 15 and 21 p.i., representative data shown.
Eae18b Reduces Cell Recruitment to the Inflammation Site
To assess the consequences of the lower disease score in the congenic animals undergoing EAE, we performed histopathological analysis of the brain and spinal cord on day 12 and 15 p.i. (Figure 5A; histopathological analysis for day 12 not shown). No congenic animal showed signs of inflammation/demyelination in the brain, although spinal cord tissue was affected. In contrast to the congenic group, 75% of the control animals developed brain lesions predominantly in medulla, pons and in optic nerves (Figure 5A). Congenic CNS showed overall significantly reduced inflammation (here referred to as inflammatory index I.I., according to [5]) and demyelination (presented as demyelination score, DM) compared to the controls (Figure 5B). Moreover, IHC analysis of the spinal cord cross-sections in both time-points showed that the control group had cumulatively more ED1+ and Iba-1+ macrophages/brain resident activated microglia, which correlated with their generally higher disease severity (Figure 6A–D). On day 15 p.i. one congenic animal with similar disease severity/neuroinflammation parameters as the controls, recruited surprisingly low amount of ED1+ peripheral macrophages to the demyelinated lesions, while the number of Iba-1+ activated brain resident microglia and macrophages was comparable to those observed in controls (data not shown).
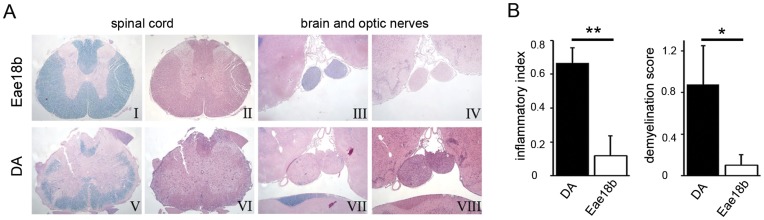
Figure 5. Congenic DA.PVG-Eae18b rats developed less severe neuroinflammation and demyelination.
(A) Histopathological analysis of the group representative paraffin embedded tissue cross-sections from congenic and control spinal cord (I, II and V, VI, respectively) or brain and optic nerves (III, IV and VII, VIII, respectively) on day 15 p.i. (five congenic and four control animals analyzed). Sections were stained with Klüver (I, III, V, VII) to reveal demyelination and Hämalaun & eosin (II, IV, VI, VIII) to detect inflammatory infiltrates. The congenic strain showed minor subpial demyelination and cell infiltration in the spinal cord (I, II), while the brain and optic nerves remained intact (III, IV). In contrast, the control strain displayed confluent subpial demyelinated lesion of the spinal cord white matter (V, VI) as well as an extensive myelin loss in both optic nerves (VII) accompanied by recruitment of inflammatory cells (VIII). (B) Analysis of the material shown in A. Inflammatory index (I.I., first graph from left) and demyelination score (DM, second graph) are both significantly higher in DA controls compared to Eae18b congenic rats. Error bars represent SEM, and (*) indicates p-value <0.05, (**) indicates p-value <0.01 calculated by Students t-test.
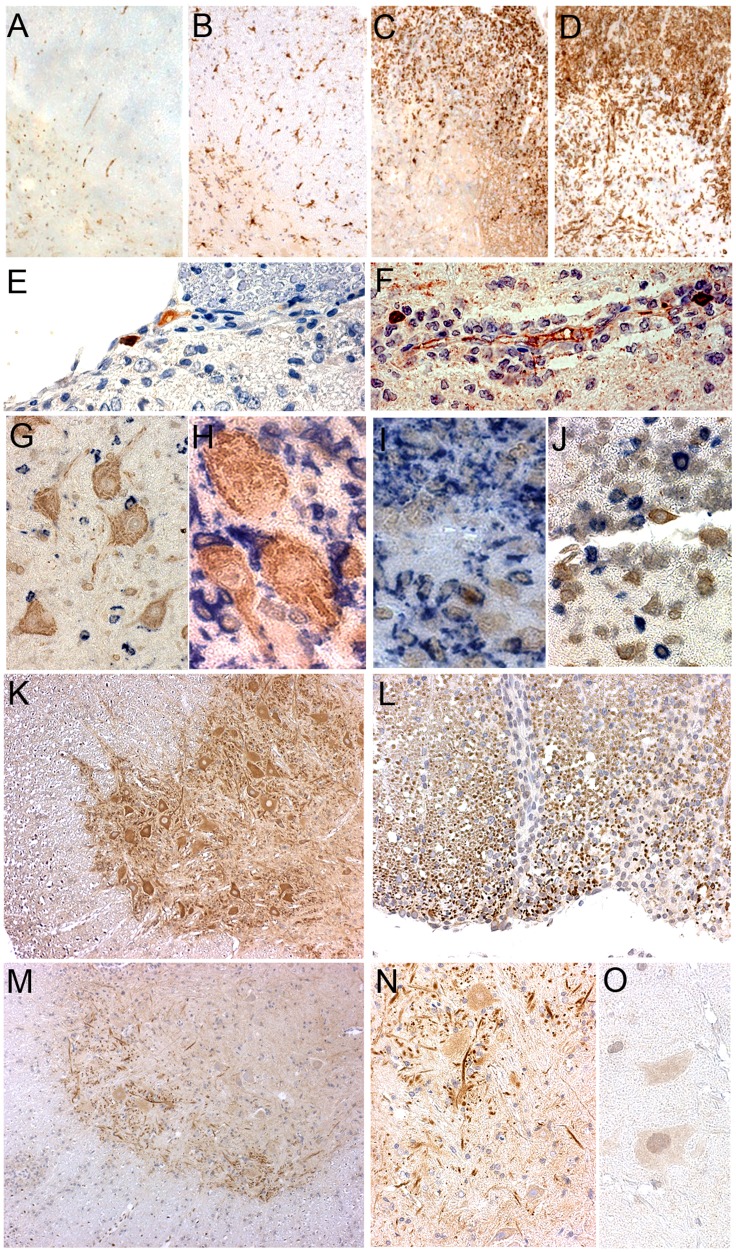
Figure 6. CCL11 and CCR3 expression in the central nervous system of Eae18b congenic rats.
(A–D) Congenic and control group representative spinal cord cross sections stained for ED1 (A, C) and Iba-1 (B, D), both hallmarks for activation of microglia/macrophages (brown). Control rats (C, D) showed upregulation of both markers in contrast to congenics (A, B). (E, F) Portions of the control spinal cord cross section showing IgG+ single positive (golden brown, E) plasma cell and CCL11+/IgG+ double positive (maroon; E, F) plasma cells. (G, H) Congenic spinal cord; ventral horn gray matter portion showing CCL11 in neuronal pericarya, dendrites and axons (brown) together with co-stained ED1+ (G) or Iba-1+ (H) cells (both in blue). (I) Congenic spinal cord portion presenting solitary CCL11+ (golden brown)/ED1+ (blue) macrophage on the inflammation site. (J) CCL11+ plasma cell (golden brown) not co-localizing with CD3+ T cells (blue), here in the control spinal cord cross section. (K) Ventral horn of the congenic spinal cord gray matter not undergoing inflammation showing CCR3 upregulation in neurons, dendrites and axons (brown). (L) Dorsal spinal cord white matter expressing CCR3 (dark brown dots), partially downregulated within demyelinated lesion on the right side of the image. (M, N) Ventral horn of the congenic spinal cord gray matter undergoing inflammation revealing CCR3 downregulation in neurons (brown, in the lower (M, 40×) and a chosen detail in the higher magnification (N, 160×). (O) Negative control; a detail representing resident cells in the ventral horn of the spinal cord gray matter.
In both strains we observed CCL11+ neurons located in the ventral horns of the spinal cord gray matter. CCL11 protein was detected in neuronal pericarya, dendrites and axons (Figure 6G and H). Furthermore, rare CCL11+ plasma cells were also detected in the inflamed spinal cord tissue (Figure 6E and F). No CCL11+ plasma cells were detected in the congenics on day 12 p.i. while control spinal cord already started harboring these cells. On day 15 p.i. we could also find CCL11+ plasma cells in the congenic animals. However, they were very rare and significantly less numerous compared to controls (1±0,45 in the congenic vs. 14,75±3,17 in the controls; p = 0,0009; MV±SEM, number of cells per spinal cord cross section). We also observed several CCL11-producing ED1+ macrophages recruited to inflammatory lesions, predominantly in the congenic group (Figure 6I), while no CCL11 co-localization was observed with CD3+ T cells (Figure 6J).
It has been described that eosinophils are the main responders to CCL11 at inflammatory sites in asthma [16], but also in neuromyelitis optica [17]. In order to investigate if the increased CCL11 expression in the congenic animals also correlated with eosinophils recruitment in our animal model, we stained the spinal cord cross sections with an antibody directed against eosinophil peroxidase (EPX). For that purpose we analyzed spinal cord cross-sections from both animal groups harvested on day 7–8, 12 and 15 p.i. The earlier time points were included in the analysis since eosinophils enter the CNS in a relatively short time-window, prior to massive invasion of the other inflammatory cells. However, we could not detect any eosinophils at any time-point analyzed (data not shown). Absence of eosinophils was also confirmed in sections stained for Hematoxylin & eosin. More interestingly, we observed significantly higher numbers of neutrophile granulocytes invading demyelinated lesions in the control animals (0,25±0,25 in the congenic vs. 27±11,02; p = 0,0168; MV±SEM, number of cells in whole spinal cord). Thus, in our congenic strain CCL11 seems to be involved in a distinct signaling pathway not related to the well-described eosinophil recruitment.
The presence of CCL11’s main receptor CCR3 in the CNS was observed in the ventral horns of the spinal cord gray matter as well as in the subpial white matter portion. CCR3 staining was restricted to neurons, dendrites and axons as well as to ependymal glia cells and was particularly intense in the ventral horn neurons of the congenic animals not undergoing neuroinflammation (Figure 6K). However, affected spinal cord tissue from both strains tended to downregulate CCR3+ expression in response to inflammation (Figure 6L–N). Additionally, we could not detect co-localization of CCR3 with any other brain resident or infiltrating cells (data not shown). Overall, in our MS-model congenic animals developed milder disease, associated with a reduced recruitment of ED1+and W3/13+ cells. The lower clinical score was reflected in a smaller size of less numerous demyelinated lesions in the CNS of the protected congenic strain, which also harbored significantly less granulocytes and plasma cells compared to EAE-susceptible DA controls.
Eae18b Influences BBB Homeostasis
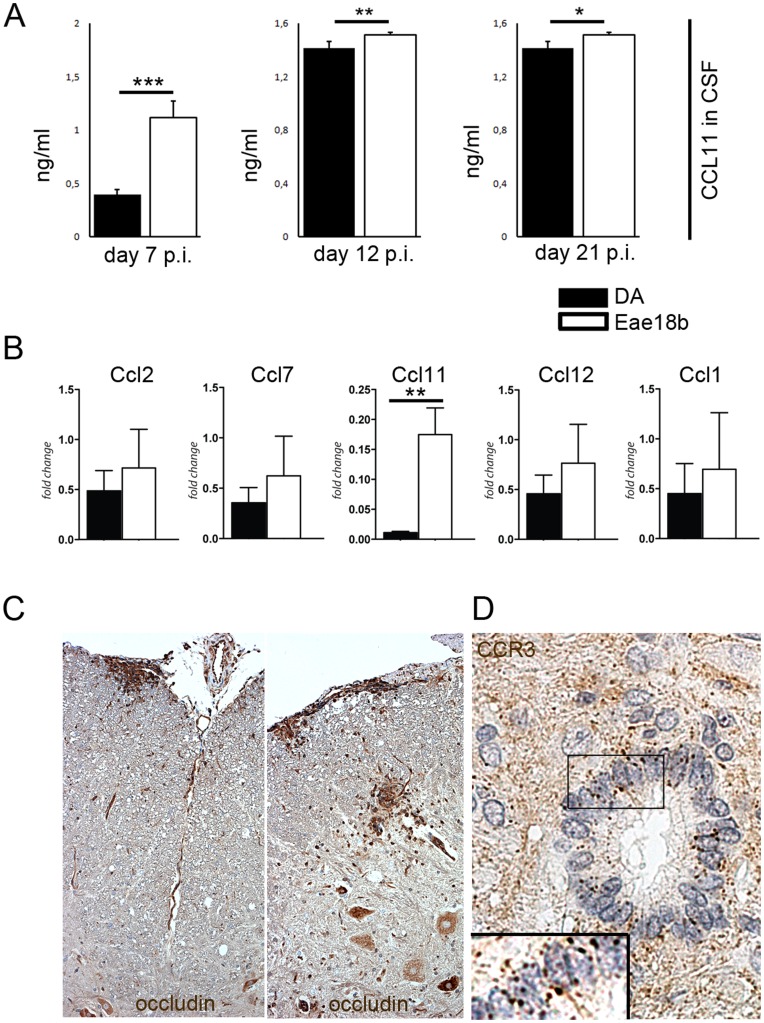
In order to identify possible mechanism how elevated CCL11 levels in the CNS protect against EAE, we analyzed CCL11 concentration in serum and CSF on days 7, 12 and 21 p.i. by enzyme-linked immunosorbent assay (ELISA). We detected significantly higher levels of CCL11 in congenic CSF in all analysed time points (Figure 7A) while CCL11 levels in blood serum were comparable in both groups through the whole disease course (Figure S6). Higher amounts of CCL11 in the congenic CSF correlated with increased mRNA expression in spinal cord on day 7, 12 (Figure 2B) and 23 p.i. (Figure 7B). This suggests a central protective effect of the congenic fragment through an enhancement of intrathecal chemokine production. Additionally, we detected the main CCL11 receptor, CCR3, expressed on CSF- producing ependymal cells of the spinal cord’s canalis centralis (Figure 7D).
Figure 7. Eae18b regulates intrathecal production of CCL11 and influences the BBB integrity.
(A) During the whole disease course CCL11 protein levels were increased in the cerebrospinal fluid (CSF), in the congenics (Eae18b) compared to controls (DA). Experiments repeated twice, three to five animals per group analyzed. (B) mRNA levels of Ccl11 measured in the late disease time point (day 23 p.i.) showed upregulation in the congenic spinal cord. Error bars represent SEM, and (*) indicates p-value <0.05, (**) indicates p-value <0.01, (***) indicates p-value <0.001 calculated by Student’s t-test in (A) and by Mann-Whitney non-parametrical test in (B). (C) Representative images of occludin, a marker for the BBB tight junction components; more abundant in the congenic spinal cord (cross sections of seven congenic and eight control animals analyzed on day 12 p.i.). (D) Representative image of CCR3, the main receptor for CCL11, expressed on ependymal cells of the spinal cord canalis centralis, including a detail in high magnification (160×). Five congenic and four control animals analyzed on day 15 p.i.
We suspected that differential levels of CCL11 measured in the CSF might have an impact on the BBB integrity. As a read-out for determination of the BBB property, we used dysferlin, a sensitive marker for “leaky” brain blood vessels [18], and occludin, a marker frequently used for detecting one of the main tight junction components [19]. IHC analysis of the CNS performed on day 12 p.i., i.e. prior to the infiltration of inflammatory cells from the periphery to the inflammatory lesions, revealed more occludin+ blood vessels in the congenic animals (12±5,26 in the congenic vs. 3±1,25 in the controls; p = 0,04; MW± SEM, number of cells per spinal cord cross section; Figure 7C), while the blood vessels in controls tended to express higher levels of dysferlin (data not shown). Occludin upregulation was also observed in perivascular and subpial regions related to inflammatory lesions. Our observations suggest that the Eae18b locus regulates intrathecal production of CCL11, and that the increased chemokine levels in the congenic animals are associated with maintenance of the integrity of the blood-brain barrier.
Discussion
Previous linkage analyses identified the Eae18b QTL on rat chromosome 10, which regulates severity of MOG-EAE [9], [12]. The region includes a cluster of chemokine genes that are also associated with susceptibility to MS [12], [13], [20], [21], [22]. Although this region has not been confirmed in recent whole genome studies in MS [2], we believe that detailed studies of biological effects of naturally occurring genetic variants can identify important regulatory mechanisms in neuroinflammation. In this study we developed and tested a congenic strain carrying the resistant PVG alleles in the Eae18b region on the susceptible DA background, to gain further insights in the effects of genetically determined immune regulation by the chemokine genes in the cluster (Ccl2, Ccl7, Ccl11, Ccl12 and Ccl1). The congenic strain exhibited milder disease with less severe inflammation and demyelination in the CNS after MOG immunization. Experiments on the congenic strain thereby confirmed previous statistical linkage analysis and provided a biological system to test immunological mechanisms regulated by the susceptible and resistant Eae18b alleles.
We have previously shown that the DA and PVG.av1 strains have identical sequence in the exons of the chemokine genes, indicating that genetically determined expression differences underlie the protection against EAE [13]. Therefore we focused first on the gene expression of chemokines present in the Eae18b congenic region. Ccl11 expression, but no other chemokine genes in the cluster, was significantly upregulated in spinal cords and peripheral lymph nodes of the EAE-protected DA.PVG-Eae18b congenic strain compared to the susceptible DA control. To unravel in more detail how the defined genetic region mediates the protective effect, we performed extensive IHC analysis to determine which cells produce CCL11 in the CNS, and in the peripheral lymph nodes. In EAE, the autoimmune activation is initiated in the draining inguinal lymph nodes after immunization with MOG. Lymph node tissue analysis prior to clinical disease onset (day 7 p.i.) showed upregulation of CCL11 chemokine protein in the congenic strain. Co-staining for CCL11 with markers for various immune cell types showed that CCL11 is mainly produced by ED1+ and Iba1+ macrophages in the local draining lymph nodes whereas T cells (CD4+ and CD8+), and CD45RA+ B cells did not produce CCL11. Additionally, we could not detect CCL11 production in any of lymph node resident CD11b+ or Ox62+ dendritic cells. The absence of MHC class II expression on the surface of the CCL11-producing macrophages suggests that in our MS model, these cells are involved in modulation of the local microenvironment, rather than specific antigen presentation.
CCL11 and its main receptor CCR3 are effectors of the Th2 response [15] and CCR3 has been shown to be expressed on Th2 cells, but also on eosinophils, dendritic cells, basophils and neurons [15], [23], [24], [25], [26], [27]. We observed upregulated Ccr3 mRNA and protein in the congenic lymph nodes, expressed on ED1+ macrophages and W3/13+ T lymphocytes. Previous studies have shown that CCL11 can inhibit dendritic cell maturation in vitro and reduce the endocytotic capacity of these cells, through induction of SOCS1 and 3 [28]. Even though the effects of CCL11 on monocyte-derived cell populations in EAE needs to be investigated further, similar mechanisms could be involved in the protection against disease that is seen in the congenic strain. In addition, our data suggest that CCL11 and CCR3 contribute to a preferential Th2 recruitment and/or activation in the congenic lymph tissue, exemplified by the upregulation of IL-5 and downregulation of IL-23 in the congenic strain. In contrast, the susceptible DA strain showed a typical Th1/Th17 inflammatory response, including high expression of both IL-23 subunits, previously implicated in EAE development [29], [30]. In vitro proliferation and anti-MOG IgG levels in serum were similar in the both strains, whereas we observed a 3-fold reduction of IFN-γ producing MOG specific T cells in the congenic strain. Taken together, the data indicate that both strains have the ability to respond to inflammatory stimuli but the response is qualitatively different, reflected by the differential expression of pro- and anti-inflammatory cytokines in the lymph nodes as well as by the reduced number of IFN-γ producing MOG specific T cells in the congenic animals.
Analysis of the CNS, the target organ for EAE revealed that the milder EAE phenotype in the congenic strain was accompanied by significantly lower myelin loss, reduced cumulative microglia activation, and a lower macrophage and lymphocyte recruitment to the brain and spinal cord of the congenic animals. CCL11 is traditionally associated with eosinophil recruitment and pro-inflammatory response, as seen in asthma [16] or associated with highly destructive CNS lesions observed in neuromyelitis optica [17]. In contrast to these observations, CCL11 has also been implicated as a regulator of monocyte activation and differentiation, apart from chemotaxis [28]. In our model, we did not observe any eosinophil invasion and related CNS tissue destruction, but instead we observed a significant increase in neutrophil granulocytes in inflammatory lesions of the control compared to the congenic animals. The upregulation of CCL11 in the CNS might instead have influenced the neuronal function and homeostasis, as these cells produced both CCL11 and CCR3. It has previously been shown that CCR3 might have a role in neuroprotection, Ccr3−/− mice have significantly reduced survival after facial nerve axotomy, possibly through a Th2-related mechanism [31].
The levels of CCL11 measured in the CSF revealed significantly increased intrathecal production of this chemokine in the congenic strain during the whole disease course (day 7, 12 and 21 p.i.), while the levels in blood serum were comparable between the strains in all analyzed time-points. Higher amounts of CCL11 in the congenic CSF correlated with increased mRNA levels in spinal cord on day 7, 12 and 23 p.i. These data suggest that continued high CCL11 secretion through the whole disease course contributed to a milder disease profile observed in the congenic strain. In this context we also analyzed the influence on BBB integrity and could observe an increased expression of occludin in the lesions and lesion related blood vessels in the congenic strain, which indicates maintenance of BBB integrity and thus reduced invasion of inflammatory cells into the CNS.
In conclusion, we present a biological confirmation of the genetic regulation of EAE by the Eae18b locus on rat chromosome 10, and implicate differential regulation of Ccl11 in CNS and periphery as a mechanism underlying the disease protection. We propose that the increased CCL11 expression and secretion by macrophages in the peripheral lymph nodes represent an early step in an active signaling pathway involving the CCR3 receptor, which leads to a preferential activation of the Th2 immune response. In addition, the congenic region also contributes to high levels of CCL11 in the CSF, maintained BBB integrity and reduced number of inflammatory cells in the CNS, indicating additional regulatory mechanisms in the target organ. Further studies are needed do define the details of the regulatory pathways involved in this diseases protection, but this study indicates that modulation of Ccl11 expression and downstream signaling through CCR3 could provide efficient strategies for immuno-modulation in MS and other inflammatory diseases.
Materials and Methods
Ethics Statement
All experiments in this study were approved by the North Stockholm Animal Ethics Committee (Stockholms Norra djurförsöksetiska nämnd) (permit numbers: N332/06, N338/09, N15/10, N65/10) and performed in accordance with the guidelines from the Swedish National Board for Laboratory Animals and the European Community Council Directive (86/609/EEC). In accordance with the ethical permits, the animals were examined at least daily for clinical signs of EAE and signs of distress, dehydration or underfeeding. To ensure proper food intake in partially paralyzed animals, all cages were supplemented with wet chow throughout the experiments. In addition, animals were euthanized if they lost more than 25% body weight during the experiment, or remained tetraplegic (score 4, see below) or showed signs of severe balance disturbance for 2 consecutive days.
Rats were routinely tested according to a health-monitoring program at the National Veterinary Institute (Statens Veterinärmedicinska Anstalt, SVA) in Uppsala, Sweden. All animals were bred and housed at the Karolinska University Hospital (Stockholm, Sweden), in polystyrene cages containing aspen wood shavings and with free access to water and standard rodent chow, with a 12-hour light/dark cycle.
Animals
Inbred DA rats were originally obtained from the Zentralinstitut für Versuchstierzucht (Hannover, Germany) and PVG.av1 rats from Harlan UK Limited (Blackthorn, UK). Animals for congenic breeding were selected from the 10th generation of an advanced intercross line (AIL) [32] originating from the EAE-susceptible DA and EAE-resistant PVG.av1 rat strains. Selected males containing a PVG.av1 fragment in the Eae18b region (from OT24.18 to D10Rat93 marker, at 68.36 Mb and 81.9 Mb, respectively) were backcrossed with DA females for 8 generations, and offspring were genotyped using microsatellite markers in each generation. Genotyping of animals was performed on DNA extracted from ear tissues according to a standard protocol. PCR amplification was performed with VIC, NED, FAM or PET end-labeled forward primers (Applied Biosystems, Carlsbad, CA). The PCR products were size fractioned using an ABI 3730 capillary sequencer and analyzed in GeneMapper v3.7 (Applied Biosystems). In the 8th generation, heterozygous males and females were crossed to obtain the experimental population of homozygous congenic rats and littermate controls.
EAE Induction and Phenotypic Evaluation
Recombinant MOG (aa 1–125 from the N-terminus) was expressed in Escherichia coli and purified to homogeneity by chelate chromatography as previously described [33]. The purified protein, dissolved in 6 M urea, was dialyzed against PBS to obtain a semi-precipitated preparation that was stored at −20°C. Rats between 10 and 14 weeks of age were anesthetized with isofluorane (Forene, Abbott Laboratories, Abbot Park, IL) and immunized with a single subcutaneous injection in the dorsal base of the tail with 200 µl of inoculum containing rMOG (aa 1–125), 15 µg/rat (females) or 30 µg/rat (males), in saline emulsified with IFA (Sigma Aldrich, St. Louis, MO), ratio 1∶1. The rats were weighed and monitored daily for clinical signs of EAE. The clinical score was graded as follows: 0, no clinical signs of EAE; 1, tail weakness or tail paralysis; 2, hind leg paraparesis or hemiparesis; 3, hind leg paralysis or hemiparalysis; 4, tetraplegia or moribund; 5, death. The following clinical parameters were assessed for each animal: EAE incidence (clinical signs for >1 day), maximum EAE score (the highest clinical score observed during EAE), cumulative EAE score (the sum of daily clinical scores).
Quantitative Real-time PCR
Animals were euthanized using CO2 at selected time-points. Draining inguinal lymph nodes and cervical spinal cords were collected and transferred on to dry ice and kept at −70°C until isolation of mRNA using RNeasy mini kit (Qiagen, Hilden, Germany), including on-column DNA-digestion. Subsequent reverse transcription was performed using or iScript cDNA Synthesis Kit (BioRad Laboratories, Hercules, CA). All primers used for quantitative real-time PCR in the study are found in Table S1. Quantitative real-time PCR was performed using a BioRad iQ5 iCycler Detection System (BioRad Laboratories) with a three-step PCR protocol (95°C for 10 min, 40 or 50 cycles of 95°C for 15 sec, 60°C for 30 sec. and 72°C for 30 sec, followed by ramping from 55°C to 95°C at 1°C/minute for melt curve analysis), using SYBR green as fluorophore (BioRad Laboratories). Relative expression levels, corrected for amplification efficiency, were recorded in iQ5 v2.0 software (BioRad Laboratories) and are presented as fold change compared to the mean expression of the target in the experiment, with Gapdh as reference gene, using the formula 2−ΔΔCt.
Histopathological and IHC Analysis
On day 7, 8, 12 and 15 post immunization each 4–8 littermate controls and 4–8 DA.PVG-Eae18b animals were euthanized using CO2 and perfused via the left heart ventricle with PBS followed by 4% paraformaldehyde. Paraformaldehyde-fixed 3 to 5 mm thick paraffin embedded sections of the brain and spinal cord were dewaxed in xylol, rehydrated and then stained with Hematoxylin & eosin and Luxol fast blue to assess tissue inflammation and demyelination, respectively. The inflammatory index (I.I) and demyelination score (DM) were determined by the number and size of demyelinated lesions of each animal on an average of 12 complete brain and spinal cord cross-sections as previously described [5]. In adjacent serial sections IHC analysis was performed using antibodies against the following targets: α- CD68 (ED1, mouse anti-rat, AbD Serotec, 1∶1000), α-Iba1 (AIF1, mouse anti-rat, Millipore, 1∶1000), CD43 (W3/13 mouse anti-rat, Abcam, 1∶100), α-GFAP (astrocyte marker, mouse anti-rat, Abcam, 1∶200), α-CD3 (rabbit anti-human, cross-reactive to rat CD3; Dakopatts, Denmark), α-rat IgG (biotinylated, GE Healthcare, 1∶200), α-eotaxin (CCL11 (C-19) goat anti-rat, Santa Cruz BT, 1∶300), α-CCR3 (rabbit anti-rat, Abcam, 1∶300), α-Dysferlin (Ham1/7B6, mouse anti-rat, Abcam, prediluted), α-Occludin (rabbit anti-rat, Abcam 1∶100), α-CD4 (W3/25 Biotin, mouse anti-rat, Abcam, 1∶100), α-CD8α (Ox-8 mouse anti-rat, AbD Serotec, 1∶200), α-Ox6 (MHC class II, mouse anti-rat, AbD Serotec, 1∶150), α-CD11b (mouse anti-rat, BD Pharminogen, 1∶100), α-Ox62 (mouse anti-rat, AbD Serotec, 1∶100), α-CD45R (B220 mouse anti-rat, eBioscience, 1∶200) and EPX (H-75), Santa Cruz BT, 1∶100). Secondary antibodies used were biotinylated (Amersham Pharmacia Biotech, 1∶200) or alkaline phosphatase-conjugated (Dakopatts, Denmark, 1∶200).
For IHC, paraffin sections of the spinal cord and inguinal lymph nodes were treated as previously described [34], deparaffinized in xylol and transferred to 90% ethanol. Endogenous peroxidase was blocked by incubation in methanol with 0.02% H2O2 (30 minutes, room temperature). Sections were then transferred to distilled water via a 90%, 70%, and 50% ethanol series. Antigen retrieval was performed with ethylenediamine tetraacetic acid buffer, pH 8.5, or citrate buffer pH 6.0 by warming for 60 minutes in a household steamer device (Braun, Germany). For detection of IgG sections were treated with 0.03% protease from Streptomyces griseus (Sigma) for 15 minutes at 37°C. Sections were incubated with 10% fetal calf serum in 0.1 mol/L PBS prior to incubation with primary antibody on 4°C, overnight. For double staining (co-staining) we used primary antibodies from two different species. After washing in PBS, sections were incubated with a mixture of one alkaline phosphatase-conjugated and one biotinylated secondary antibody in 10% fetal calf serum in PBS for 1 hour at room temperature. Finally, avidin peroxidase (1∶100, Sigma, Vienna, Austria) was applied for 1 hour at room temperature. Alkaline phosphatase label was visualized with Fast Blue B base or Fast Red TR Salt (Sigma) and avidin peroxidase with 3,3′diaminobenzidine-tetrahydrochloride (DAB, Sigma). Simple stainings were performed with biotin-avidin peroxidase detection system and visualized with DAB. Control sections were incubated in the absence of primary antibodies. Evaluation has been performed on at least 12 whole spinal cord cross sections. Imaging was performed on a Leica Polyvar 2 microscope.
Western Blot
Inguinal lymph nodes were harvested on day 7 p.i., frozen in liquid nitrogen and kept at −80°C. Tissue were homogenized in the lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl2, 1% Triton-X100, 0,5% deoxycholic acid and 0.1% SDS) supplemented with Complete Protease Inhibitor Cocktail (Roche). Tissue lysates were incubated on ice for 30 min followed by centrifugation at 16.000 g at 4°C for 20 min where after the lysates were subjected to SDS–PAGE on NuPage 12% Bis-Tris gels (Invitrogen) under reducing conditions. Separated proteins were blotted onto filters using the iBlot system (Invitrogen). Membranes were blocked for 1 h in Tris-buffered saline containing 0.1% Tween-20 (TBST) and 5% bovine serum albumin (BSA) followed by overnight incubation at 4 C with goat polyclonal anti-CCL11 antibody (Santa Cruz BT, 1∶ 1000 in TBST, 1% BSA), rabbit monoclonal CCR3 (Abcam, 1∶ 1000 in TBST, 1% BSA) or mouse monoclonal b-actin (1∶ 1000 in TBST, 1% BSA). Membranes were incubated with horseradish peroxidase conjugated antimouse IgG (dilution 1∶2000; Amersham Pharmacia Biotech.) for 1 h at room temperature and visualized by enhanced chemiluminescence (ECL, Amersham Pharmacia Biotech.). The optical density of the respective bands was digitized and analysed using the Biorad Geldoc XR (Biorad).
Elispot
Spleens were harvested on day 15 and 21 p.i. and placed in DMEM before being mechanically separated by passage through a mesh screen with the bolus of a syringe. Splenocytes from DA and DA.PVG-Eea18b (n = 4 for each condition) were used for IFN-γ Elispot analysis (R&D systems) in a 96 well plate. ConA (12.5 ng/well), MOG (50 ng/well) and MBP (50 ng/well) were used to stimulate 50.000 splenocytes/well in total volume of 100 µl. After 48 hours incubation IFN-γ producing cells were visualized by antibody staining according to the manufacturer’s protocol and the colony forming units (CFU) were visualized using an AID Elispot reader (AID, Germany).
ELISA
Cerebrospinal fluid (CSF) was harvested on day 7, 12 and 21 p.i., frozen in liquid nitrogen and stored at −70°C until used for ELISA. Samples and standards were incubated together with CCL11 conjugate according to the manufacturer’s protocol (Rat eosinophil chemotactic factor kit, Emelcabio, Netherlands).
Statistical Analysis
Mann-Whitney non-parametrical test was used for statistical analysis, if not stated otherwise. For all tests p-values <0.05 were considered significant. All statistical analyses were performed in GraphPad Prism versions 4 or 5 (GraphPad, San Diego, CA). Mean values with standard error are presented in graphs, if not stated otherwise.
Supporting Information
Description of flow cytometry, proliferation assay, determination of anti-MOG isotypes and CCL11 measurement in serum are found in Materials and Methods S1.
Supporting Information
Differental expression of chemokines in (A) inguinal lymph nodes and (B) spinal cord of parental DA and PVG.av1 rats. Expression of mRNA from naïve animals (4 animals/strain) and in tissues harvested day 5 (8 animals/strain), 7 (4 animals/strain) and 12 (8 animals/strain) post immunization. Error bars represent SEM, Mann-Whitney non-parametric test was used for statistical analysis. P-values <0.05 were considered significant and are indicated in the figure. nd: not detected.
(EPS)
DA and DA.PVG- Eae18b strains do not differ in proportions of main cell types (A) or subsets of T-cells (B). Cell from lymph nodes from 5 animals/strain were collected day 5, 7 and 12 post immunization and analyzed using flow cytometry. Error bars represent SEM, and Mann-Whitney non-parametric test was used for statistical analysis. No significant differences were found. n.a: not analyzed.
(EPS)
DA and DA.PVG- Eae18b strains do not differ in proportions of immune cell types. Analysis performed by flow cytometry on cells collected from the lymph nodes on day 7 (A) and 12 (B) after immunization, 5 to 8 animals in each group. Y-axis presents the percentage (%) of detected cells out of all gated viable cells; error bars show Standard Deviation and Mann-Whitney non-parametric test was used for statistical analysis. No significant differences were found.
(TIF)
In vitro proliferation reveals no differences between DA and DA.PVG- Eae18b rat strains. Proliferation assay performed on lymphocytes collected day 7 post immunization from 5 DA and 5 DA.PVG-Eae18b females. Cells were stimulated for 18 hours with MOG (19 µg/ml), ConA (0.5 µg/ml) or media alone. Error bars represent SEM, and Mann-Whitney non-parametric test was used for statistical analysis. No significant differences were found between strains.
(EPS)
DA and DA.PVG- Eae18b strains do not differ in secretion of specific antibodies. Anti-MOG specific antibody production of IgG1, IgG2b, IgG2c and total IgG isotypes measured using ELISA in sera collected day 12 post immunization from DA (12 females and 9 males), and DA.PVG-Eae18b (11 females and 8 males). Error bars represent SEM, and Mann-Whitney non-parametric test was used for statistical analysis. No significant differences were found.
(EPS)
DA and DA.PVG- Eae18b (Eae18b) rats have similar amounts of CCL11 in serum, at different time points after MOG immunization. Experiments repeated twice, 3–5 animals per group analyzed. Error bars represent SEM, Students t-test was used for statistical analysis. No significant differences were found.
(TIF)
Sequences of primers used for quantitative real-time PCR, designed using Primer Express software. The second pair of Ccl11 [Ccl11 (2)] primers was used for confirmation, excluding potential artifacts originating from annealing differences in the two strains.
(DOC)
Description of methods related to the supporting figures, including cytometry, proliferation assay, determination of anti-MOG isotypes and CCL11 measurement in serum.
(DOC)
Acknowledgments
We thank Sevasti Flytzani and Susanne Laukoter for assistance with ELISA and imaging, respectively. We also thank Marianne Leisser, Angela Kury and Thomas Pekar for technical assistance.
Footnotes
Competing Interests: The authors have read the journal’s policy and have the following conflicts: Milena Zeitelhofer-Adzemovic was partly funded by a research grant from Biogen Idec, Inc. This does not alter the authors’ adherence to all the PLoS ONE policies on sharing data and materials.
Funding: This study was supported by grants from Biogen Idec, the Wenner-Gren Foundation, the Swedish Research Council, Ragnar Söderberg’s Foundation, Bibbi and Niels Jensen’s Foundation, Montel William’s Foundation, Åke-Wibergs Stiftelse, the Swedish Association of Persons with Neurological Disabilities, Swedish Brain Foundation, the Multiple Sclerosis Society of Canada and the EU 6TH Framework EURATools (LSHG-CT-2005-019015) and Neuropromise (LSHM-CT-2005-018637). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.De Jager PL, Jia X, Wang J, de Bakker PI, Ottoboni L, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Jager PL, Baecher-Allan C, Maier LM, Arthur AT, Ottoboni L, et al. The role of the CD58 locus in multiple sclerosis. Proc Natl Acad Sci U S A. 2009;106:5264–5269. doi: 10.1073/pnas.0813310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawcer S. The complex genetics of multiple sclerosis: pitfalls and prospects. Brain. 2008;131:3118–3131. doi: 10.1093/brain/awn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Storch MK, Stefferl A, Brehm U, Weissert R, Wallström E, et al. Autoimmunity to Myelin Oligodendrocyte Glycoprotein in Rats Mimics the Spectrum of Multiple Sclerosis Pathology Brain Pathol. 1998;8:681–694. doi: 10.1111/j.1750-3639.1998.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Connor RA, Prendergast CT, Sabatos CA, Lau CW, Leech MD, et al. Cutting edge: Th1 cells facilitate the entry of Th17 cells to the central nervous system during experimental autoimmune encephalomyelitis. J Immunol. 2008;181:3750–3754. doi: 10.4049/jimmunol.181.6.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stromnes IM, Cerretti LM, Liggitt D, Harris RA, Goverman JM. Differential regulation of central nervous system autoimmunity by T(H)1 and T(H)17 cells. Nat Med. 2008;14:337–342. doi: 10.1038/nm1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thessen Hedreul M, Gillett A, Olsson T, Jagodic M, Harris RA. Characterization of Multiple Sclerosis candidate gene expression kinetics in rat experimental autoimmune encephalomyelitis. Journal of Neuroimmunology. 2009. doi: 10.1016/j.jneuroim.2009.02.010. [DOI] [PubMed]
- 9.Dahlman I, Wallstrom E, Weissert R, Storch M, Kornek B, et al. Linkage analysis of myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis in the rat identifies a locus controlling demyelination on chromosome 18. Hum Mol Genet. 1999;8:2183–2190. doi: 10.1093/hmg/8.12.2183. [DOI] [PubMed] [Google Scholar]
- 10.Jagodic M, Kornek B, Weissert R, Lassmann H, Olsson T, et al. Congenic mapping confirms a locus on rat chromosome 10 conferring strong protection against myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis. Immunogenetics. 2001;53:410–415. doi: 10.1007/s002510100342. [DOI] [PubMed] [Google Scholar]
- 11.Teuscher C, Butterfield RJ, Ma RZ, Zachary JF, Doerge RW, et al. Sequence polymorphisms in the chemokines Scya1 (TCA-3), Scya2 (monocyte chemoattractant protein (MCP)-1), and Scya12 (MCP-5) are candidates for eae7, a locus controlling susceptibility to monophasic remitting/nonrelapsing experimental allergic encephalomyelitis. J Immunol. 1999;163:2262–2266. [PubMed] [Google Scholar]
- 12.Jagodic M, Becanovic K, Sheng JR, Wu X, Bäckdahl L, et al. An advanced intercross line resolves Eae18 into two narrow quantitative trait loci syntenic to multiple sclerosis candidate loci. J Immunol. 2004;173:1366–1373. doi: 10.4049/jimmunol.173.2.1366. [DOI] [PubMed] [Google Scholar]
- 13.Ockinger J, Stridh P, Beyeen AD, Lundmark F, Seddighzadeh M, et al. Genetic variants of CC chemokine genes in experimental autoimmune encephalomyelitis, multiple sclerosis and rheumatoid arthritis. Genes Immun. 2009;11:142–154. doi: 10.1038/gene.2009.82. [DOI] [PubMed] [Google Scholar]
- 14.Bromley ST, Illas F, Mas-Torrent M. Dependence of charge transfer reorganization energy on carrier localisation in organic molecular crystals. Phys Chem Chem Phys. 2008;10:121–127. doi: 10.1039/b713688a. [DOI] [PubMed] [Google Scholar]
- 15.Daugherty BL, Siciliano SJ, DeMartino JA, Malkowitz L, Sirotina A, et al. Cloning, expression, and characterization of the human eosinophil eotaxin receptor. J Exp Med. 1996;183:2349–2354. doi: 10.1084/jem.183.5.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, et al. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Correale J, Fiol M. Activation of humoral immunity and eosinophils in neuromyelitis optica. Neurology. 2004;63:2363–2370. doi: 10.1212/01.wnl.0000148481.80152.bf. [DOI] [PubMed] [Google Scholar]
- 18.Hochmeister S, Grundtner R, Bauer J, Engelhardt B, Lyck R, et al. Dysferlin is a new marker for leaky brain blood vessels in multiple sclerosis. J Neuropathol Exp Neurol. 2006;65:855–865. doi: 10.1097/01.jnen.0000235119.52311.16. [DOI] [PubMed] [Google Scholar]
- 19.Kniesel U, Wolburg H. Tight junctions of the blood-brain barrier. Cell Mol Neurobiol. 2000;20:57–76. doi: 10.1023/A:1006995910836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vyshkina T, Kalman B. Haplotypes within genes of beta-chemokines in 17q11 are associated with multiple sclerosis: a second phase study. Human Genetics. 2005;V118:67. doi: 10.1007/s00439-005-0003-2. [DOI] [PubMed] [Google Scholar]
- 21.Vyshkina T, Shugart YY, Birnbaum G, Leist TP, Kalman B. Association of haplotypes in the [beta]-chemokine locus with multiple sclerosis. Eur J Hum Genet. 2004;13:240. doi: 10.1038/sj.ejhg.5201295. [DOI] [PubMed] [Google Scholar]
- 22.Bugeja MJ, Booth D, Bennetts B, Heard R, Rubio J, et al. An investigation of polymorphisms in the 17q11.2–12 CC chemokine gene cluster for association with multiple sclerosis in Australians. BMC Med Genet. 2006;7:64. doi: 10.1186/1471-2350-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitaura M, Nakajima T, Imai T, Harada S, Combadiere C, et al. Molecular cloning of human eotaxin, an eosinophil-selective CC chemokine, and identification of a specific eosinophil eotaxin receptor, CC chemokine receptor 3. J Biol Chem. 1996;271:7725–7730. doi: 10.1074/jbc.271.13.7725. [DOI] [PubMed] [Google Scholar]
- 24.Ponath PD, Qin S, Post TW, Wang J, Wu L, et al. Molecular cloning and characterization of a human eotaxin receptor expressed selectively on eosinophils. J Exp Med. 1996;183:2437–2448. doi: 10.1084/jem.183.6.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao J-L, Sen AI, Kitaura M, Yoshie O, Rothenberg ME, et al. Identification of a Mouse Eosinophil Receptor for the CC Chemokine Eotaxin. Biochemical and Biophysical Research Communications. 1996;223:679–684. doi: 10.1006/bbrc.1996.0955. [DOI] [PubMed] [Google Scholar]
- 26.Beaulieu S, Robbiani DF, Du X, Rodrigues E, Ignatius R, et al. Expression of a functional eotaxin (CC chemokine ligand 11) receptor CCR3 by human dendritic cells. J Immunol. 2002;169:2925–2936. doi: 10.4049/jimmunol.169.6.2925. [DOI] [PubMed] [Google Scholar]
- 27.Sallusto F, Mackay CR, Lanzavecchia A. Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells. Science. 1997;277:2005–2007. doi: 10.1126/science.277.5334.2005. [DOI] [PubMed] [Google Scholar]
- 28.Stevenson NJ, Addley MR, Ryan EJ, Boyd CR, Carroll HP, et al. CCL11 blocks IL-4 and GM-CSF signaling in hematopoietic cells and hinders dendritic cell differentiation via suppressor of cytokine signaling expression. J Leukoc Biol. 2009;85:289–297. doi: 10.1189/jlb.0708394. [DOI] [PubMed] [Google Scholar]
- 29.Kroenke MA, Carlson TJ, Andjelkovic AV, Segal BM. IL-12- and IL-23-modulated T cells induce distinct types of EAE based on histology, CNS chemokine profile, and response to cytokine inhibition. J Exp Med. 2008;205:1535–1541. doi: 10.1084/jem.20080159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thakker P, Leach MW, Kuang W, Benoit SE, Leonard JP, et al. IL-23 is critical in the induction but not in the effector phase of experimental autoimmune encephalomyelitis. J Immunol. 2007;178:2589–2598. doi: 10.4049/jimmunol.178.4.2589. [DOI] [PubMed] [Google Scholar]
- 31.Wainwright DA, Xin J, Mesnard NA, Beahrs TR, Politis CM, et al. Exacerbation of facial motoneuron loss after facial nerve axotomy in CCR3-deficient mice. ASN Neuro. 2009;1:e00024. doi: 10.1042/AN20090017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darvasi A. Experimental strategies for the genetic dissection of complex traits in animal models. Nat Genet. 1998;18:19. doi: 10.1038/ng0198-19. [DOI] [PubMed] [Google Scholar]
- 33.Amor S, Groome N, Linington C, Morris MM, Dornmair K, et al. Identification of epitopes of myelin oligodendrocyte glycoprotein for the induction of experimental allergic encephalomyelitis in SJL and Biozzi AB/H mice. J Immunol. 1994;153:4349–4356. [PubMed] [Google Scholar]
- 34.Bradl M, Bauer J, Flugel A, Wekerle H, Lassmann H. Complementary contribution of CD4 and CD8 T lymphocytes to T-cell infiltration of the intact and the degenerative spinal cord. Am J Pathol. 2005;166:1441–1450. doi: 10.1016/S0002-9440(10)62361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differental expression of chemokines in (A) inguinal lymph nodes and (B) spinal cord of parental DA and PVG.av1 rats. Expression of mRNA from naïve animals (4 animals/strain) and in tissues harvested day 5 (8 animals/strain), 7 (4 animals/strain) and 12 (8 animals/strain) post immunization. Error bars represent SEM, Mann-Whitney non-parametric test was used for statistical analysis. P-values <0.05 were considered significant and are indicated in the figure. nd: not detected.
(EPS)
DA and DA.PVG- Eae18b strains do not differ in proportions of main cell types (A) or subsets of T-cells (B). Cell from lymph nodes from 5 animals/strain were collected day 5, 7 and 12 post immunization and analyzed using flow cytometry. Error bars represent SEM, and Mann-Whitney non-parametric test was used for statistical analysis. No significant differences were found. n.a: not analyzed.
(EPS)
DA and DA.PVG- Eae18b strains do not differ in proportions of immune cell types. Analysis performed by flow cytometry on cells collected from the lymph nodes on day 7 (A) and 12 (B) after immunization, 5 to 8 animals in each group. Y-axis presents the percentage (%) of detected cells out of all gated viable cells; error bars show Standard Deviation and Mann-Whitney non-parametric test was used for statistical analysis. No significant differences were found.
(TIF)
In vitro proliferation reveals no differences between DA and DA.PVG- Eae18b rat strains. Proliferation assay performed on lymphocytes collected day 7 post immunization from 5 DA and 5 DA.PVG-Eae18b females. Cells were stimulated for 18 hours with MOG (19 µg/ml), ConA (0.5 µg/ml) or media alone. Error bars represent SEM, and Mann-Whitney non-parametric test was used for statistical analysis. No significant differences were found between strains.
(EPS)
DA and DA.PVG- Eae18b strains do not differ in secretion of specific antibodies. Anti-MOG specific antibody production of IgG1, IgG2b, IgG2c and total IgG isotypes measured using ELISA in sera collected day 12 post immunization from DA (12 females and 9 males), and DA.PVG-Eae18b (11 females and 8 males). Error bars represent SEM, and Mann-Whitney non-parametric test was used for statistical analysis. No significant differences were found.
(EPS)
DA and DA.PVG- Eae18b (Eae18b) rats have similar amounts of CCL11 in serum, at different time points after MOG immunization. Experiments repeated twice, 3–5 animals per group analyzed. Error bars represent SEM, Students t-test was used for statistical analysis. No significant differences were found.
(TIF)
Sequences of primers used for quantitative real-time PCR, designed using Primer Express software. The second pair of Ccl11 [Ccl11 (2)] primers was used for confirmation, excluding potential artifacts originating from annealing differences in the two strains.
(DOC)
Description of methods related to the supporting figures, including cytometry, proliferation assay, determination of anti-MOG isotypes and CCL11 measurement in serum.
(DOC)