Abstract
The F-box protein, Ufo1, recruits Ho endonuclease to the SCFUfo1 complex for ubiquitylation. Both ubiquitylated Ho and Ufo1 are transferred by the UbL-UbA protein, Ddi1, to the 19S Regulatory Particle (RP) of the proteasome for degradation. The Ddi1-UbL domain binds Rpn1 of the 19S RP, the Ddi1-UbA domain binds ubiquitin chains on the degradation substrate. Here we used complex reconstitution in vitro to identify stages in the transfer of Ho and Ufo1 from the SCFUfo1 complex to the proteasome. We report SCFUfo1 complex at the proteasome formed in the presence of Ho. Subsequently Ddi1 is recruited to this complex by interaction between the Ddi1-UbL domain and Ufo1. The core of Ddi1 binds both Ufo1 and Rpn1; this interaction confers specificity of SCFUfo1 for Ddi1. The substrate-shield model predicts that Ho would protect Ufo1 from degradation and we find that Ddi1 binds Ho, Ufo1, and Rpn1 simultaneously forming a complex for transfer of Ho to the 19S RP. In contrast, in the absence of Ho, Rpn1 displaces Ufo1 from Ddi1 indicating a higher affinity of the Ddi1-UbL for the 19S RP. However, at high Rpn1 levels there is synergistic binding of Ufo1 to Ddi1 that is dependent on the Ddi1-UbA domain. Our interpretation is that in the absence of substrate, the Ddi1-UbL binds Rpn1 while the Ddi1-UbA binds ubiquitin chains on Ufo1. This would promote degradation of Ufo1 and disassembly of SCFUfo1 complexes.
Introduction
The Ubiquitin-proteasome system has a major role in regulation of cellular processes, in particular the cell cycle and many signaling pathways [1], [2]. Proteins targeted for degradation are conjugated to ubiquitin (Ub) by a cascade of enzymes, an E1 Ub activating- and E2 Ub conjugating enzyme, and an E3 Ub ligase responsible for substrate identification [3]. In some instances an E4 Ub chain elongating activity is also involved [4]. Ub chains comprising at least four K48-linked Ub molecules are recognized by the 19S Regulatory particle (RP) of the proteasome, either by an endogenous 19S RP subunit [5]–[7], or by a member of the UbL-UbA protein family. UbL-UbA proteins bind specific 19S RP subunits through their Ub-like (UbL) domain and K48-Ub chains on the substrate through their Ub-associated (UbA) domain. The yeast family of UbL-UbA proteins comprises Rad23, Dsk2, and Ddi1, and each family member participates in the degradation of a range of substrates either by itself, or as a Rad23-Dsk2 pair (reviewed in [8]).
UbL-UbA proteins are often referred to as shuttle proteins based on their recruitment of the ubiquitylated substrate from the E2-E3 complex and transfer to the 19S RP. This is supported particularly by the interaction between Rad23 and Dsk2 with the chain elongating E4, Ufd2, that occurs in the framework of a complex between Ufd2 and the AAA-ATPase ring hexamer, Cdc48 [9]. However, many E3s bind the 19S RP directly: these include Ubr1 and Ufd4 [10], Hul5 [11], Ufo1 [12], SCF (Skp1-Cullin1-F-box protein) and APC (Anaphase Promoting complex) [13], [14]. In the case of Ufd4, direct interaction between the E3 and the proteasome is essential for substrate degradation [15]. In some instances the UbL-UbA protein may be an essential stochiometric subunit of the E3 complex, as reported for KPC2 (Kip1 ubiquitylation-promoting complex 2) that regulates degradation of the p27 cell cycle inhibitor [16]. These reports raise the question whether other UbL-UbA proteins may also occur as intrinsic components of an E3-19S RP complex and if so whether it is possible to detect additional interactions between the core domain of the UbL-UbA protein and subunits of this complex. In the event of such interactions are they a prerequisite for interaction of the E3 complex with the 19S RP?
The SCF complex comprises a rigid cullin scaffold, in S. cerevisiae Cdc53, with the RING protein, Rbx1, attached to a C-terminal domain [17]. The RING domain serves as a landing pad for the Ub-charged E2, Cdc34 [18]. Substrate recruitment is executed by a series of different F-box proteins (FBPs), each of which binds a subset of targets many of which are recognized by phosphorylation [19]–[21]. FBPs have a F-box domain and a WD40- or LRR substrate-binding domain. The F-box domain binds the Skp1 adaptor that interacts with the N-terminal domain of Cdc53 [17], [22]–[25]. Exchange of FBPs within the SCF complex is achieved by auto-ubiquitylation of the FBP followed by degradation in the proteasome [26], [27]. A number of FBPs of SCF complexes and the related BTB/3-box domain receptor proteins [28] have been shown to occur as homo- or heterodimers. These include homodimers of yeast Cdc4 and of human Fbw7 FBP [29] and the heterodimeric S. pombe Pop1-Pop2 FBPs [30], [31]. FBP dimerization was shown to be required for dimerization of Cdc53 [32] and increasing experimental data support a model of a dimeric cullin-RING ligase complex. Indeed although monomeric FBPs bind their substrates and Skp1, substrate ubiquitylation was reported to require their dimerization [28], [32], [33].
Ddi1 is required for the final stages of proteasomal degradation of both Ho endonuclease [34] and of Ufo1, its cognate FBP [35]. Ubiquitylated Ho interacts with the UbA domain of Ddi1 via its ubiquitin chains and its transfer to the 19S RP requires the UbL domain of Ddi1 that interacts with the LRR domain of the 19S RP subunit, Rpn1 [36]. Ddi1 forms a homodimer mediated by residues in its core (residues 180–325) giving rise to an active aspartyl protease site [37], [38]. In ddi1Δ mutants ubiquitylated Ho endonuclease accumulates in the cytoplasm and is not transferred to the proteasome for degradation [34]. Ufo1 and its fungal orthologs are unique FBPs as they have four copies of the Ub interacting motif (UIMs) at their C-terminus in addition to the F-box and WD40 domains present in other FBPs [35], [39], [40]. The UIM is a simple α-helical ubiquitin binding domain [41] and the Ufo1-UIMs are separated by long linkers suggesting this is a flexible region. Turnover of Ufo1 is dependent on an interaction between its UIMs and the UbL domain of Ddi1 [35]. Furthermore the rpn1-D517A mutation that disrupts binding of Ddi1 to the proteasome stabilizes Ufo1 [36].
A protein fragment comprising the Ufo1-UIMs interacts with all three UbL-UbA proteins, Rad23, Dsk2, and Ddi1, however full-length (FL) Ufo1 interacts only with Ddi1 suggesting that the core residues are important for specificity [35]. UIMs have been shown to interact with Ub-charged E2s to promote monoubiquitylation of a different domain of their host protein [42]–[45]. Deletion of UFO1 has no obvious phenotype under normal growth conditions, however, a genomic UFO1 allele deleted for the UIMs is dominant lethal. Ectopic high level expression of UFO1 without the UIMs leads to stabilization of the protein and to cell cycle arrest at the end of G1. Substrates of other FBPs accumulate suggesting that Ddi1 is required for disassembly of SCFUfo1 complexes and recycling of the core complex subunits into alternative SCF complexes [35].
Here we used complex reconstitution in vitro to augment our in vivo data showing a role for Ddi1 in degradation of Ho and of its cognate FBP, Ufo1 [34], [35], [40], [46]. In particular we aimed to identify stages in the handover of Ho from the SCFUfo1 complex to the 19S RP and subsequent degradation of Ufo1. We delineate stages in the formation of SCFUfo1-Ho-Ddi1-19S RP complex. Domain analysis showing different modes of interaction of Ddi1 with Ufo1 and Rpn1 in the presence and absence of Ho support the “Substrate shield” model of protein degradation [47]. We present a model for sequential handover of Ho and Ufo1 to the proteasome.
Results
Ufo1 Forms Dimers Initiated by the UIMs
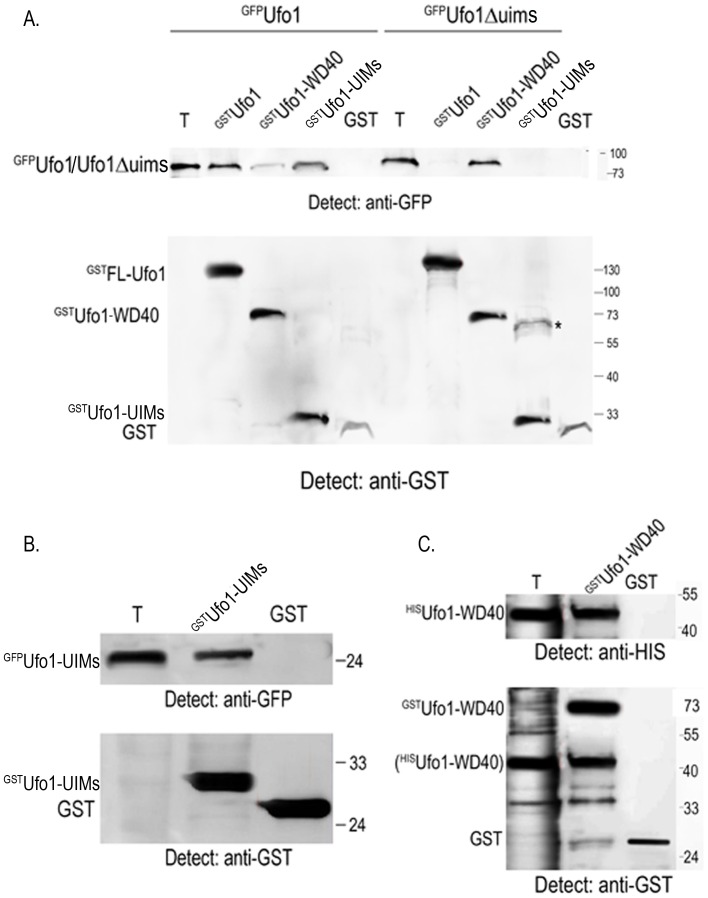
Given the importance of FBP dimerization for substrate ubiquitylation [28], [32], [33] we examined whether Ufo1 forms a dimer. Furthermore we aimed to determine which domain(s) of Ufo1 could have a role in dimerization. We incubated yeast extract from cells that produced full-length (FL), GFPFL-Ufo1, or Ufo1 truncated for the C-terminal UIMs, GFPUfo1Δuims, with GSH beads bound to recombinant GSTFL-Ufo1, GSTUfo1-WD40 domain, GSTUfo1-UIMs, or control GST (Figure S1). We observed a robust interaction of GFPFL-Ufo1 with GSTFL-Ufo1 and with GSTUfo1-UIMs whereas the interaction between GFPFL-Ufo1 and GSTUfo1-WD40 domain was extremely weak. GFPUfo1Δuims did not interact with GSTFL-Ufo1 or with GSTUfo1-UIMs. However, in contrast to GFPFL-Ufo1, truncated GFPUfo1Δuims interacted robustly with GSTUfo1-WD40 (Figure 1A).
Figure 1. Ufo1 forms a homodimer via its UIMs.
A. GSTFL-Ufo1, GSTUfo1-WD40 domain, GSTUfo1-UIMs or GST beads were incubated with yeast extract from cells expressing full-length pGAL-GFP-UFO1 or pGAL-GFP-UFO1Δuims. The bead fraction was analysed by Western blotting with anti-GFP and anti-GST antibodies. T is 10% of yeast extract with which the beads were incubated. *denotes contaminant band. B. Recombinant GSTUfo1-UIMs or control GST beads were incubated with yeast extract with GFPUfo1-UIMs and analysed as above. T is 10% of yeast extract as above. C. Recombinant GSTUfo1 WD40 domain protein or control GST on GSH beads were incubated with bacterial lysate from cells that expressed HISUfo1-WD40 and the bead fraction was analysed by Western blotting initially with anti-HIS and then with anti-GST antibodies. T is 10% of yeast extract as above. The brackets around HISUfo1-WD40 in the anti-GST Western blot indicate that these bands were observed after incubation with anti-HIS antibodies as shown in the upper part of the blot.
These results suggest both a positive and a negative role for the Ufo1-UIMs in Ufo1 dimerization. The positive role is indicated by the ability of FL-Ufo1 to dimerize with both FL-Ufo1 and with the isolated Ufo1-UIM fragment, whereas the negative role is indicated by the absence of dimerization between FL-Ufo1 and the Ufo1-WD40 domain fragment. This may indicate that the Ufo1-UIMs regulate access to the WD40 domain. To test directly whether the Ufo1-UIMs dimerize we incubated yeast extract with GFPUfo1-UIMs with recombinant GSTUfo1-UIMs on beads. We observed a robust interaction that was not found with the control GST beads indicating that isolated Ufo1-UIMs fragments dimerize (Figure 1B). The interaction between GFPUfo1Δuims and GSTUfo1-WD40 (Figure 1A) suggests that the Ufo1-WD40 domain by itself can dimerize. Indeed when we expressed the Ufo1-WD40 domain in bacteria with two different epitope tags we observed that GSTUfo1-WD40 bound to HISUfo1-WD40 (Figure 1C). Thus Ufo1 resembles other FBPs in forming dimers and both the unique Ufo1-UIMs and the Ufo1-WD40 domain participate in dimerization. Dimerization via the Ufo1-WD40 domains is supported by our previous finding of turnover of Ho in ufo1Δ mutants that produce plasmid-encoded Ufo1Δuims [35].
SCFUfo1 Complexes Interact with the 19S RP in vitro Only in the Presence of Substrate
Despite its nuclear role Ho must exit the nucleus to be degraded [46] and in ddi1Δ mutants stabilized Ho accumulates in the cytoplasm as an ubiquitylated conjugate [34]. SCFUfo1 complexes that have bound Ho may associate with the 19S RP as reported for SCFCdc4-Sic1 complexes [14], or alternatively Ddi1 could shuttle ubiquitylated Ho from a SCFUfo1-Ho complex to the proteasome. We therefore reconstituted SCFUfo1 complexes in vitro in the presence or absence of Ho. Recombinant GSTFL-Ufo1 and the GSTUfo1-WD40 domain proteins on GSH beads were incubated with yeast extract from cells that produced mycCdc53 and with the 19S RP complex tagged with Rpn11GFP. The experiment was performed both in the presence and the absence of GFPHo endonuclease. Experimental conditions are such that the 19S RP complex with a single tagged subunit remains intact in the yeast extract [5], [13], [14], [36], [51]. Both FL-Ufo1 and the Ufo1-WD40 domain on beads supported the formation of SCFUfo1-Ho-19S RP complexes and interacted with yeast mycCdc53, GFPHo, and with the tagged 19S RP complex. In addition endogenous Ddi1 was present as a major component of the GSTFL-Ufo1 and the GSTUfo1-WD40 domain bead fractions of complexes formed in the presence of Ho. In the absence of Ho, we found an interaction of GSTUfo1 with mycCdc53, but there was no interaction with Rpn11GFP. Ddi1 could still be detected in the GSTFL-Ufo1 and the GSTUfo1-WD40 domain bead fractions, although in a considerably diminished amount (Figure 2A). A similar result was observed using tagged Rpn1GFP (Figure S2). Rpn12 was present in the bead fraction indicating that 19S RP complexes and not just the tagged subunit were interacting with SCFUfo1 (Figure S3).
Figure 2. SCFUfo1-Ho-19S RP complex formation in vitro.
A. GSTUfo1, GSTUfo1-WD40 domain, or control GST beads were incubated with yeast extract from cells with tagged genomic RPN11-GFP that were transformed with pGAL-MYC-CDC53 either with pGAL-GFP-HO or alone. The bead fraction was analysed by Western blotting with anti-GFP antibodies to detect Rpn11GFP and GFPHo, with anti-myc antibodies to detect mycCdc53, and with anti-Ddi1 and anti-GST antibodies. T is 10% of total yeast extract with which the beads were incubated (Lanes 1 and 2). Lane 3: GSTUfo1 beads incubated with mycCdc53, Rpn11GFP and GFPHo; Lane 4: GSTUfo1 WD40 domain incubated with mycCdc53, Rpn11GFP and GFPHo; Lane 5: control GST beads incubated with these yeast extracts; Lane 6: GSTUfo1 beads incubated with mycCdc53 and Rpn11GFP; Lane 7: GSTUfo1 WD40 domain incubated with mycCdc53 and Rpn11GFP; Lane 8: control GST beads incubated with these yeast extracts. B. GSTUfo1, GSTUfo1-WD40, or control GST beads were incubated with yeast extract from ddi1Δ mutant cells that expressed pGAL-MYC-CDC53, pGFP-RPN11, with or without pGAL-GFP-HO. The bead fractions were analysed by Western blotting with anti-myc, anti-GFP, anti-Ddi1, and anti-GST antibodies as in A. T is 10% of total yeast extract with which the beads were incubated (Lanes 1-3). Lane 4: GSTUfo1 beads incubated with mycCdc53, Rpn11GFP and GFPHo; Lane 5: GSTUfo1 WD40 domain incubated with mycCdc53, Rpn11GFP and GFPHo; Lane 6: control GST beads incubated with these yeast extracts; Lane 7: GSTUfo1 beads incubated with mycCdc53 and Rpn11GFP; Lane 8: GSTUfo1 WD40 domain incubated with mycCdc53 and Rpn11GFP; Lane 9: control GST beads incubated with these yeast extracts.
Ddi1 is involved in the final stages of transfer of Ho and of Ufo1 to the 19S RP and could be recruited to the SCFUfo1-Ho-19S RP complex after its assembly. We therefore repeated the above experiment using extracts of transformed ddi1Δ mutants. As in w.t. cells, Ho was crucial for formation of complex between SCFUfo1 and the19S RP, however, there was no requirement for Ddi1 for formation of the SCFUfo1-Ho-19S RP complex (Figure 2B). These results taken together and supported by our in vivo data that show that both Ho and Ufo1 accumulate as ubiquitylated conjugates in ddi1Δ mutants suggest that in vivo Ddi1 is recruited to the SCFUfo1-Ho-19S RP complex after its assembly.
SCFUfo1-Ho-Ddi1-19S RP Complexes can be Reconstituted in vitro with Immobilized GSTDdi1 or GSTRpn1
Reconstitution of SCFUfo1-Ho-19S RP complexes in vitro in the above experiments was achieved with GSTFL-Ufo1 or the GSTUfo1-WD40 domain on beads. To determine whether complex reconstitution is also possible with immobilized GSTDdi1 or GSTRpn1, the 19S RP subunit bound by Ddi1 [36], [52], we incubated GSTDdi1 or control GST on GSH beads with yeast extract from cells that produced mycCdc53 and GFPUfo1. We observed a robust interaction of both proteins with GSTDdi1 that was not observed with the GST control beads (Figure 3A). Similarly GSTRpn1 on beads could reconstitute SCFUfo1-GFPHo-GSTRpn1 complexes that included endogenous Ddi1 present in the yeast extract (Figure 3B). No complexes were formed with the control GST beads. Thus it is possible to reconstitute complexes in vitro irrespective of which component is immobilized. Recombinant HISUfo1 interacted extremely weakly with irrelevant control GSTRpn10 beads, however we did observe an interaction of GFPHo and of mycCdc53 with this 19S RP subunit.
Figure 3. Immobilized Ddi1 and Rpn1 reconstitute SCFUfo1 complexes in vitro.
A. GSTDdi1 or control GST on GSH beads were incubated with yeast extract from cells that produced mycCdc53 and GFPUfo1. Analysis was by Western blotting with anti-myc, anti-GFP, and anti-GST antibodies. T represents 10% of the yeast extract with which the beads were incubated. B. GSTRpn1, GSTRpn10, or GST beads were incubated with yeast extract from cells that produced mycCdc53 and GFPHo mixed with bacterial lysate with recombinant HISUfo1. The bead fraction was analysed by Western blotting with anti-myc, anti-HIS, anti-GFP, anti-Ddi1, and anti-GST antibodies. T represents 10% of the yeast extract incubated with the beads. C. GSTUfo1 WD40 domain, GSTCdc53, or control GST on GSH beads were incubated with bacterial lysate from cells that produced recombinant HISΔΔDdi1. The bead fraction was analysed by Western blotting with anti-HIS and with anti-GST antibodies as indicated. T is 10% of the HISΔΔDdi1 bacterial lysate incubated with the beads. D. The HISΔΔDdi1 bacterial lysate was incubated with GSTRpn1, GSTRpn10, or GST beads and analysed as above.
The Core of Ddi1 Binds Cdc53, the Ufo1-WD40 Domain, and Rpn1
The Ufo1-UIMs fragment in isolation interacts with all three UbL-UbA proteins, Rad23, Dsk2, and Ddi1, however, FL-Ufo1 discriminates between them [35]. This suggests that the initial interaction between Ufo1 and Ddi1 occurs via interaction of its UIMs with the Ddi1-UbL domain and that specificity of UbL-UbA protein may be conferred by further interactions between Ufo1 and the core of Ddi1. We subcloned HISΔΔDdi1 without the UbL and UbA domains comprising residues 180–325. Indeed both the GSTUfo1-WD40 domain and GSTCdc53 bound core HISΔΔDdi1 (Figure 3C). Ddi1 binds the LRR domain of the Rpn1 subunit of the 19S RP [36], [52] via its UbL domain and here we found that the core HISΔΔDdi1 fragment bound GSTRpn1 robustly but showed only extremely weak binding to control GSTRpn10 (Figure 3D). Thus after the initial interaction between the Ufo1-UIMs and the Ddi1-UbL these additional interactions with the Ddi1 core could secure Ddi1 within the SCFUfo1-Ho-Ddi1-19S RP complex. They could also allow flexibility to the Ddi1-UbL allowing it to switch to binding Rpn1 for substrate or FBP transfer.
SCFUfo1-Ddi1-19S RP Complex Subunits Immunoprecipitate Together in the Presence of Ho
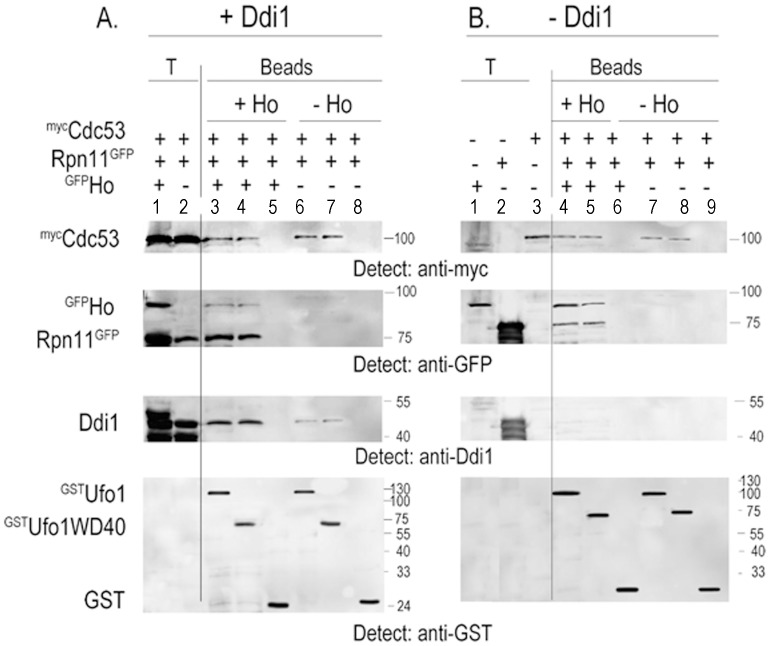
The above experiments demonstrate that in the presence of Ho a SCFUfo1-Ho-Ddi1-19S RP complex is formed in vitro. To verify that this is indeed a complex we prepared a reaction mix comprising yeast extract with mycCdc53, with or without GFPHo, and bacterial lysate with GSTUfo1 and HISRpn1, and immunoprecipitated each tagged protein separately. In the presence of Ho, immunoprecipitation of mycCdc53, of GFPHo, of GSTUfo1 or of HISRpn1 led to reciprocal coimmunoprecipitation of the other three proteins and of Ddi1 present in the yeast extract. In the absence of Ho, immunoprecipitation of mycCdc53, GSTUfo1 or HISRpn1 led to coimmunoprecipitation of endogenous Ddi1 from the yeast extract, but not of any of the other proteins of the complex formed in the presence of substrate. This result indicates that in the presence of Ho a bona fide complex is formed between SCFUfo1-Ho-Ddi1 and Rpn1. This complex does not form in the absence of Ho (Figure 4).
Figure 4. Cdc53, Ufo1, Rpn1 Ho and Ddi1 form a complex.
Bacterial lysates from cells that produced HISRpn1 or GSTUfo1 were mixed with yeast extracts with mycCdc53 with or without GFPHo and divided into equal aliquots. Each aliquot was immunoprecipitated with a different antibody: anti-myc, anti-GFP, anti-GST or anti-HIS, in the presence of Protein A beads. The bead fractions were analysed by Western blotting with each antibody. + Ho: Lanes 1–3: Lane 1. 10% of the total (T) extract from yeast that produced mycCdc53, GFPHo and endogenous Ddi1; Lane 2. Bacterial lysate with GSTUfo1; Lane 3. HISRpn1 bacterial lysate; Lane 4. anti-myc immunoprecipitation; Lane 5. anti-GFP immunoprecipitation; Lane 6. anti-GST immunoprecipitation; Lane 7. anti-HIS immunoprecipitation. – Ho: Lane 8. 10% of the yeast extract with mycCdc53 and endogenous Ddi1 and bacterial lysate with GSTUfo1, Lane 9. 10% of bacterial lysate with HISRpn1. Lane 10. anti-myc immunoprecipitation; Lane 11. anti-GST immunoprecipitation; Lane 12. anti-HIS immunoprecipitation;The IP lanes are headed by the antibodies used for immunoprecipitation of each protein.
Ufo1 and Rpn1 Bind Ddi1 in Both a Competitive and a Synergistic Manner
(a) Competitive interaction: GSTRpn1 abrogates binding of GFPUfo1 to HISDdi1
The Ddi1-UbL domain binds both the Ufo1-UIMs and Rpn1 [35], [52], however, interaction between Ddi1 and Rpn1 is essential for turnover of Ufo1 [36]. Both Ufo1 and Rpn1 bind the core of Ddi1 (Figure 3C and 3D) and this interaction may facilitate the switch of the Ddi1-UbL domain from the Ufo1-UIMs to Rpn1 for transfer of Ho or Ufo1 to the 19S RP. We therefore examined whether there is competition between Ufo1 and Rpn1 for interaction with Ddi1. Each protein incubated separately with Ddi1 beads was present in the HISDdi1 bead fraction (Figure 5A, Lanes 4–6). However, Rpn1 displaced Ufo1 from Ddi1 when both GSTUfo1 and GSTRpn1 were incubated together with the HISDdi1 beads (Lane 7). In contrast addition of yeast extract with ubiquitylated GFPHo to the reaction mix with either GSTUfo1 or GSTRpn1 did not affect the binding of either protein to HISDdi1 (Lanes 8 and 9). Furthermore, Ho in the reaction mix comprising Ufo1, Rpn1, and Ddi1, abrogated the competition between Ufo1 and Rpn1 and all three proteins bound the HISDdi1 beads (Lane 10) and Figure 2. Thus Ho protects Ufo1 from displacement from Ddi1 by Rpn1. In this complex the Ddi1-UbL would bind Rpn1, Ufo1 would be bound via its WD40 domain to Ho and to the Ddi1 core, and further interactions would occur between the Ddi1-UbA and the Ub chains on Ho. This is the complex we predict to underlie transfer of ubiquitylated Ho to the 19S RP (Figure 6).
Figure 5. Rpn1 and Ufo1 exhibit synergistic binding to Ddi1.
A. Bacterial lysate with GSTRpn1 or GSTUfo1 and yeast extract with GFPHo were incubated with HISDdi1 beads alone (Lanes 4–6), in pairs Lanes 7–9), or all three together (Lane 10). The bead fractions were analysed by Western blotting with anti-GST, anti-GFP, anti-HIS and anti-ubiquitin antibodies. Lanes 1–3 (T) show 10% of the lysate/extract for bead incubation. B. Yeast extract with GFPUfo1 was incubated with HISDdi1 and HISΔΔDdi1 beads in the absence or the presence of increasing amounts of HISRpn1. The bead fractions were analysed by Western blotting with anti-GFP, anti-GST antibodies and anti-HIS. Lanes 1 and 2 (T) show 10% of the lysate/extract with which the beads were incubated. C. Bacterial lysate with GSTUfo1 was mixed with increasing amounts of lysate with GSTRpn1 or GSTRpn10 and incubated with nickel beads with HISDdi1. The Western blots were analysed with anti-GST and anti-HIS antibodies. D. Bacterial lysate with HISRpn1 was incubated with GSTUfo1, the GSTUfo1-WD40 domain, the GSTUfo1-UIMs or control GST beads. The Western blots were analysed with anti-HIS and anti-GST antibodies. T indicates 10% of the lysate with which the beads were incubated. E. Recombinant HISRpn1 made in bacteria and GFPUfo1 from yeast extract were incubated alone or together with GSH beads bound to GSTDdi1, GSTDdi1ΔUbL, or GSTDdi1ΔUbA produced in bacteria. The bead fractions were analysed by Western blotting with anti-HIS and anti-GFP antibodies to show proteins that bound the GSH beads. The latter were detected with anti-GST antibodies. F. As above except that GFPUfo1Δuims was used instead of FL-Ufo1. T denotes 10% of the yeast extract incubated with the beads.
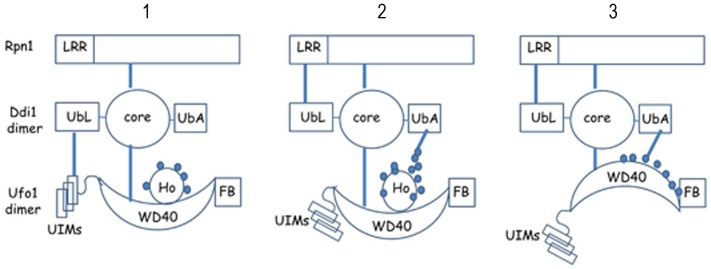
Figure 6. Model for sequential interactions of Ho, Ufo1, and Rpn1 with Ddi1.
Panel 1. Active SCFUfo1-Ho complexed with the 19S RP recruits Ddi1 by interaction of the Ufo1-UIMs with the Ddi1-UbL domain ([35] and Figures 2, 4 and 5F). Subsequently the core of Ddi1 binds the Ufo1-WD40 domain and Rpn1 (Figure 3C and D). Both Ufo1 (Figure 1) and Ddi1 [38] form dimers but are drawn here as monomers for clarity. Panel 2. The Ddi1-UbA domain interacts with ubiquitin chains on Ho and the Ddi1-UbL binds Rpn1 for transfer of ubiquitylated Ho to the 19S RP [34]. At this stage Ho, Ufo1, and Rpn1 bind Ddi1 simultaneously (Figure 5A). Panel 3. After degradation of Ho, Ufo1 can no longer bind Ddi1 in the presence of Rpn1 (competitive interaction, Figure 5B and C). However, at high levels of Rpn1 there is synergistic binding that is supported to a small extent by the Ddi1 core (Figure 5B) and is totally dependent on the Ddi1-UbA domain (Figure E). Based on the higher affinity of the Ddi1-UbL for Rpn1 seen in the competitive interaction we propose that at this stage the Ddi1-UbL binds Rpn1 and the Ddi1-UbA binds ubiquitin chains on Ufo1. This would lead to degradation of Ufo1 [36].
(b) Synergistic interaction: GSTRpn1 and GFPUfo1 bind HISDdi1 in a tertiary complex that requires the Ddi1 UbA domain and does not involve the Ddi1 UbL domain
The competitive interaction between Ufo1 and Rpn1 may occur during handover of the FBP to the 19S RP after degradation of Ho. To explore this hypothesis we examined whether exclusion of GSTUfo1 from binding to HISDdi1 by GSTRpn1 is concentration dependent. We calibrated the system by determining an amount for each lysate/extract that would give detectable binding of protein to the Ddi1 beads (x1, Figure 5B, Lanes 3 and 4). Then keeping the amount of GFPUfo1 extract constant in a fixed reaction volume we increased the amount of GSTRpn1 lysate two- and threefold. In this experiment we used ubiquitylated GFPUfo1 produced in yeast [35]. GSTRpn1 at x1 and x2 in the reaction mix gave a similar amount bound to the Ddi1 beads. Both these GSTRpn1 concentrations abrogated binding of GFPUfo1 to Ddi1 (Figure 5B, Lanes 5 and 6 and as observed in Figure 5A, Lane 7). However, x3 the amount of GSTRpn1 lysate induced synergistic binding of GSTRpn1 and GFPUfo1 to the HISDdi1 beads. A similar although considerably weaker signal was obtained when core HISΔΔDdi1 beads were used. In contrast binding of GSTRpn10 to the HISDdi1 beads was not affected by GSTUfo1 nor was any synergistic effect observed between them in binding to Ddi1 (Figure 5C). In contrast to Ddi1 [36] there is no direct binding between Ufo1 and Rpn1 (Figure 5D).
The competition between Ufo1 and Rpn1 for binding Ddi1 may involve the Ddi1-UbL which binds both proteins (above). To address this question we repeated the synergistic binding experiment described in Figure 5B but this time in addition to GSTFL-Ddi1 beads we used Ddi1 that lacked either the UbL or UbA domain: GSTDdi1ΔUbL, and GSTDdi1ΔUbA, respectively (Figure 5E, Lanes 1–3). Ddi1ΔUbL exhibited severely reduced binding to Rpn1 and did not bind Ufo1 when each protein was incubated separately with the beads. In contrast, Ddi1ΔUbL bound both Rpn1 and Ufo1 synergistically when both were present in the reaction mix. This suggests a role for the Ddi1-UbA in the synergistic binding of Rpn1 and Ufo1 to Ddi1. Surprisingly although Rpn1 binds the Ddi1-UbL, when we incubated HISRpn1 with GSTDdi1ΔUbA beads it interacted less strongly than with GSTFL-Ddi1 beads (Figure 5E, compare Lane 1 with Lane 4). GFPUfo1 bound GSTDdi1ΔUbA beads and there was an extremely weak synergistic binding of Rpn1 and Ufo1 to GSTDdi1ΔUbA beads when both were present in the reaction mix. Our previous in vivo experiments indicated that Ufo1 and Ddi1 interact via the Ufo1-UIMs and the Ddi1-UbL [35]. We therefore substituted GFPUfo1Δuims for GFPFL-Ufo1. Indeed GFPUfo1Δuims did not interact with GSTFL-Ddi1, GSTDdi1ΔUbL or GSTDdi1ΔUbA beads both in the presence or the absence of Rpn1 (Figure 5F).
Discussion
Complex reconstitution in vitro indicated that SCFUfo1 complexes that contain their substrate, Ho, are associated with the 19S RP. These complexes can assemble in the absence of Ddi1, however, in experiments with extracts from w.t. cells Ddi1 is found in association with the SCFUfo1-Ho-19S RP complex. Our interpretation is that Ddi1 is recruited to preformed SCFUfo1-Ho-19S RP complex. Based on our previous experiments in vivo we propose that Ddi1 enters the SCFUfo1-Ho-19S RP complex via initial interaction between the Ufo1-UIMs and the Ddi1-UbL ([35] and Figure 5F). Subsequent interaction between the Ufo1-WD40 and the core of Ddi1 detected here could explain the specificity of the interaction of SCFUfo1 for Ddi1 [35]. The recruitment of Ddi1 after formation of the SCFUfo1-Ho-19S RP complex supports our in vivo results that suggested Ddi1 is required for disassembly of SCFUfo1 complexes after substrate degradation. This hypothesis is based on accumulation of ubiquitylated Ho in the cytoplasm of ddi1Δ mutants [34], stabilization of full-length Ufo1 in ddi1Δ mutants, cell cycle arrest at the G1-S interphase by overexpression of UFO1Δuims in wild type cells or of full-length UFO1 in ddi1Δ mutants, and by the accumulation of Cln2, a substrate of the FBP, Grr1 [21], in cells with a high level of Ufo1Δuims [35].
The Ufo1-UIMs promote dimerization of Ufo1 and are crucial for all interactions of Ufo1 with Ddi1. They may fulfill two roles in dimerization: one is physical interaction between the UIMs of two Ufo1 molecules to initiate dimerization. The other is regulation of access to the Ufo1-WD40 domain as full-length Ufo1 did not dimerize with an Ufo1-WD40 domain fragment. Thus dimerization may start at the C-terminal UIMs and proceed to include the Ufo1-WD40 domains. We previously reported that SCF complexes from cells that produced Ufo1Δuims are capable of degrading Ho [35]. Given that dimerization of FBPs has been shown to be a prerequisite for substrate ubiquitylation in some instances [28], [32], [33], our current results support an interpretation that in the absence of its UIMs the Ufo1-WD40 domains of each monomer are able to interact with one another in vivo. The Ufo1-WD40 domain alone is sufficient for formation of complexes that include GFPHo, the 19S RP, and Ddi1 and indeed in our yeast two-hybrid experiments we reported an interaction between Cdc53 and the Ufo1-WD40 domain [35]. This is unusual as the solved SCF structures do not display interaction between the cullin and the WD40 domain of the FBP [23], [25] or with the related BTB/3-box domain receptor protein [28]. The Ufo1 WD40 sequence has a rather degenerate β-propeller sequence and a full analysis of this unusual interaction awaits solution of the 3D structure of Ufo1. A dimerization sequence has been identified in the N-terminal region of certain FBPs [26], [27] and it is conceivable that there is one in Ufo1 too that could serve for dimerization in the absence of the Ufo1 UIMs.
The Ddi1-UbL – Ufo1-UIMs interaction is essential for recruitment of Ddi1 to the SCFUfo1-Ho-19S RP complex ([35] and Figure 5F). However, degradation of the ubiquitylated substrate requires transfer of the Ddi1-UbL from its interaction with the Ufo1-UIMs to Rpn1 [36]. In the presence of Ho a complex is formed that includes Ufo1, Rpn1, and Ddi1. Binding of Ho to Ddi1 is mediated by interaction of its ubiquitin chains with the Ddi1-UbA domain [34]. We propose that interaction of the Ddi1-UbA with a critical amount of Ub chains on Ho could lead to switching of the Ddi1-UbL domain from the Ufo1-UIMs to Rpn1 for transfer of Ho to the 19S RP. Transfer of the Ddi1-UbL without disruption of the complex between these proteins would be supported further by concurrent binding of the Ddi1 core to both Ufo1 and Rpn1 and by interactions of Ho with both the Ufo1-WD40 domain [40] and with the Ddi1-UbA domain via its Ub chains ([34] and Figure 6).
The “substrate shield” model proposes that the substrate protects the FBP from degradation [47]. In the reaction lacking Ho (comparable to an in vivo situation after substrate degradation but prior to SCFUfo1 complex disassembly) we observed two different modes of interaction of Ufo1 and Rpn1 with Ddi1: (a) competitive - Rpn1 excludes Ufo1 from binding to Ddi1; (b) synergistic - high levels of Rpn1 formed a tertiary complex between Ufo1, Ddi1, and Rpn1. The competitive interaction indicates that the Ddi1-UbL has higher affinity for Rpn1 than for the Ufo1-UIMs. The dependence of synergistic binding of Ufo1 and Rpn1 on the Ddi1-UbA domain suggests that Ub chains on Ufo1 are involved. The higher ratio of Rpn1 to Ufo1 in the in vitro Ddi1 synergistic binding experiment could parallel molecular crowding within the SCFUfo1-19S RP complex. Thus in the absence of Ho our data support a complex in which the Ddi1-UbL is bound to Rpn1 while the Ddi1-UbA domain binds Ub chains on Ufo1. This model for sequential transfer of Ho and of Ufo1 to the 19S RP is presented in Figure 6.
Materials and Methods
Yeast Strains
Wild type BY4741 (MAT a ; his3Δ1; leu2Δ0; met15 Δ0; ura3Δ0), and ddi1Δ (MATα, his3 Δ1, leu2 Δ0, lys2 Δ0, ura3 Δ00, YER143w::kanMX4) were purchased from Euroscarf. Strains with genomic RPN1-GFP and RPN11-GFP are from the library of [48].
Bacterial Strains
Rosetta bacteria (Novagen) (F–, ompT, hsdSB(rB-, mB-), dcm, gal, lacY1, pRARE (argU, argW, ileX, glyT, leuW, proL) (CmR) were used for most recombinant protein expression. His-tagged recombinant proteins were expressed in BL21 (Promega) (F–, ompT, hsdSB(rB-, mB-), dcm, gal, λ(DE3), pLysS (Cmr)) or M15 (Qiagen) (NaIS, strS, rifS, thi-, lac-, ara+, gal+, mtl-,F-, recA+, uvr+, lon+(Kmr)) bacteria.XL1 MRF1 (StrataGene) (Nalr) gyrA96 end A1 Δβ(lacz)M15/recA1 laclq proA+B+ lacF’::Tn10 relA1 supE44 thi hsdR17(rk-mk+) was used for plasmid amplification.
Yeast Plasmids
YCPGAL-GFP (GFP-UFO1, GFP- ΔUIMs, GFP-HO) are described in [35]; pMT2989 for expression of MYC-CDC53 from the GAL promoter was obtained from M. Tyers [21]. pYE-RPN11-GFP in which expression of RPN11-GFP is from the ADH1 promoter was obtained from M. Glickman [49].
Growth media and yeast transformation by LiOAc are as in [50].
Bacterial Plasmids
pHB2-GST-CDC53, pGST-DDI1, pGST-DDI1-ΔUBL, pGST-DDI1-ΔUBA, pHIS-DDI1, pHIS-ΔΔDDI1, pGST-RPN1, and pGST-RPN10 (gift of Dorota Skowyra); pGEX-5X-1 (Amersham Biosciences) was used to construct pGST-UFO1 by amplifying the UFO1 gene from genomic DNA using primer pair Ufo1F (GAATTCATGGAGCGGCCTGGCTTGGTATT) and Ufo1R (CTCGAGTCAATTGATTTCACTCAATGACAACG). pGST-UIMs was constructed using Ufo1UIMsF ( GAATTCAAAACGACATTCAGTTGAGAATTGCA ) and Ufo1R. pGST-WD40 employed primer pair WD40GstF (GAATTCAATATTAATGCTGCAGTG) and WD40GstR (CTCGAGGTTTTCTTCATCGGTGTC). pCDFduet1- HIS-UFO1 was constructed by amplifying the UFO1 ORF with primer pair Ufo1HisF (GGATTCATGG AGCGGCCTGGCTTGGTATT) and Ufo1HisR (CTCGAGTCAATTGATTTCACTCAATG ACAACG). pCDFduet1- HIS-WD40 was constructed by amplifying the WD40 domain of UFO1 with primer pair WD40HisF (GGCGCGCCAATATTAATGCTGCAGTG) and WD40HisR (CTCGAGGTTTTCTTCATCGGTGTC).
Immunoprecipitation and immunoblotting were performed as described in [5], [40]. Briefly, proteins were induced from the GAL promoter by overnight growth in minimal medium with 2% galactose. Next morning the culture was diluted 1∶3 and grown for a further 1.5 hours. 50 mls of logarithmic culture served as the source of a 300 µl extract with 80 µg/µl protein. 200 µl were taken for immunoprecipitation (IP) with the appropriate antibody and the immunoprecipitate was run in a single lane for Western blotting (WB).
Antibodies
Mouse anti-GFP (Roche Applied Science), mouse 9E10 anti-myc (Enzo), and mouse anti-HIS (Sigma) antibodies were used at a dilution of 1∶250 for IP and at 1∶1,000 for WB; mouse anti-GST (Santa Cruz Biotechnology) antibodies were diluted 1∶1,000 for IP and 1∶2,000 for WB, rabbit anti-Ddi1 (gift from Jeffrey Gerst) was used at 1∶5,000 for WB. Goat anti-mouse and anti-rabbit antisera, used at 1∶1,000 were from Santa Cruz Biotechnology. Protein A-sepharose was purchased from Amersham and used at 50%; 30 µl were added to each sample.
TCA precipitation proteins were precipitated from 300 µl cell extract by adding TCA to 10% with 10 minutes incubation on ice. The pellet was centrifuged at 12,000 g for 10 minutes and five volumes of cold acetone were added. The protein pellets were harvested and dried. For WB analysis the pellets were dissolved in 30 µl of sample buffer and 5 µl of each fraction was separated by SDS-PAGE.
Expression of GST and HIS Fusion Proteins in Rosetta Bacteria
Bacteria were transformed by electroporation and the colonies selected on LB-agar plates with ampicillin and kanamycin, each at 100 µg/ml, and chloramphenicol at 34µg/ml. A single colony was grown in 1 liter of LB (with ampicillin and chloramphenicol) to an OD600 of 0.6–0.8 (3–5 hours) with vigorous agitation at 37°C. IPTG was added to 0.4 mM to induce expression and the culture was incubated overnight at 20°C. The cells were harvested by centrifugation at 4°C for 10 min at 6,000 rpm. The cell pellet was washed with 20 ml of ice-cold PBS and resuspended in 3 ml yeast extract buffer (50 mM Tris-Cl pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% NP40, 1∶25 Protease Inhibitor cocktail (Roche)). The cell suspension was disrupted with an ultra-sound sonicator on ice using 6 cycles each of 10 seconds and clarified by centrifugation for 10 min at 4,000 rpm at 4°C. The supernatants with the GST-fusion proteins were incubated with Glutathione-sepharose 4B (GSH) beads (Amersham Biosciences) prewashed in yeast extract buffer with 1% Triton-X100; HIS-fusion proteins were incubated with washed Ni-sepharose (Clontech) for 1.5 hr at 4°C. The bead fractions were washed 5 times in extract buffer with 2.5% Triton-X100. The GST- and HIS-fusion proteins on beads were stored at -20°C after addition of glycerol to 5%.
GST in vitro Binding Assay
Yeast cells were grown overnight to late log phase (OD600 = 0.8) in 2% galactose medium for the GAL-regulated constructs, or in YePD. The cells were harvested by centrifugation at room temperature for 5 minutes at 4,000 rpm, washed in 50 ml TE and resuspended in 600 µl extract buffer. 0.5–0.6 mg of glass beads were added and the cells were broken by vigorous vortexing for 25 minutes at 4°C. The extract was clarified by centrifugation at 12,000 g for 20 minutes at 4°C and protein concentration was measured with the Bio-Rad protein reagent. 5–10 mg of protein extract were taken for each GST pull-down in a total volume of 350–400 µl extract buffer. 30–50 µl of 50% Glutathione Sepharose 4B beads coupled to GST fusion protein were added to each sample and incubated at 4°C for 1–2 hours with very mild shaking. The samples were washed 6 times with extract buffer with 2.5% Triton X100. The pellet was resuspended in 30–50 µl sample buffer x2, boiled for 5 minutes and centrifuged for 3 minutes at high speed to remove insoluble material. The supernatant was separated on a 12% polyacrylamide SDS gel with protein size standards followed by WB analysis.
HIS-tagged Protein in vitro Binding Assay
As above, but with 30–50 µl of 50% Ni Sepharose beads coupled to the HIS fusion protein added to each sample and incubated at 4°C for 1–2 hours with very mild shaking. The samples were washed 6 times with extract buffer with 2.5% Triton X100 and 100 mM Imidazole. The pellet was resuspended in 30–50 µl sample buffer x2, boiled for 5 minutes and centrifuged for 3 minutes at high speed to remove insoluble material. The supernatant was separated on a 12% polyacrylamide SDS gel with protein size standards followed by WB analysis.
Supporting Information
Domains of Ufo1 and Ddi1 used in experiments. The protein fragments used in the experiments depicted in the Figures are shown.
(TIF)
Formation of SCFUfo1-Ho-19S RP complex with yeast extract from RPN1-GFP cells. GSTUfo1 or control GST beads were incubated with yeast extract from cells with tagged genomic RPN1-GFP that were cotransformed with pGFP-HO and with pMYC-CDC53. The bead fraction was analysed by Western blotting with anti-GFP antibodies to detect Rpn1 and Ho, with anti-myc antibodies to detect Cdc53, and with anti-Ddi1 antibodies.
(TIF)
Rpn12 is present in the GSTUfo1 bead fraction. A further experiment in which GSTUfo1 and GST beads were incubated with yeast extract in the presence of GFPHo as in Figures 2 and S2. Here the Western blot employed antibodies made to GSTRpn12. The presence of Rpn12 is an indication that the 19S RP is intact.
(TIF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by FP7 EU contract PITN-GA-2008-215148 and the Israel Cancer Association. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kornitzer D, Ciechanover A. Modes of regulation of ubiquitin-mediated protein degradation. J Cell Physiol. 2000;182:1–11. doi: 10.1002/(SICI)1097-4652(200001)182:1<1::AID-JCP1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 2.Kirkin V, Dikic I. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr Opin Cell Biol. 2007;19:199–205. doi: 10.1016/j.ceb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 4.Koegl M, Hoppe T, Schlenker S, Ulrich H, Mayer T, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 5.Voloshin O, Gocheva Y, Gutnick M, Movshovich N, Bakhrat A, et al. Tubulin chaperone E binds microtubules and proteasomes and protects against misfolded protein stress Cell and Molec Life Sci. 2010;67:2025–2038. doi: 10.1007/s00018-010-0308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isasa M, Katz EJ, Kim W, Yugo V, Gonzalez S, et al. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol Cell. 38:733–745. doi: 10.1016/j.molcel.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hänzelmann P, Stingele J, Hofmann K, Schindelin H, Raasi S. The yeast E4 ubiquitin ligase Ufd2 interacts with the ubiquitin-like domains of Rad23 and Dsk2 via a novel and distinct ubiquitin-like binding domain. J Biol Chem. 2010;285:20390–20398. doi: 10.1074/jbc.M110.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie Y, Varshavsky A. Physical association of ubiquitin ligases and the 26S proteasome. Proc Natl Acad Sci U S A. 2000;97:2497–2502. doi: 10.1073/pnas.060025497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 12.Baranes-Bachar K, Khalaila I, Ivantsiv Y, Lavut A, Voloshin O, et al. New interacting partners of the F-box protein Ufo1 of yeast. Yeast. 2008;25:733–743. doi: 10.1002/yea.1615. [DOI] [PubMed] [Google Scholar]
- 13.Verma R, Chen S, Feldman R, Schieltz D, Yates J, et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babbitt SE, Kiss A, Deffenbaugh AE, Chang YH, Bailly E, et al. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell. 2005;121:553–565. doi: 10.1016/j.cell.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 15.Xie Y, Varshavsky A. UFD4 lacking the proteasome-binding region catalyses ubiquitination but is impaired in proteolysis. Nat Cell Biol. 2002;4:1003–1007. doi: 10.1038/ncb889. [DOI] [PubMed] [Google Scholar]
- 16.Hara T, Kamura T, Kotoshiba S, Takahashi H, Fujiwara K, et al. Role of the UBL-UBA Protein KPC2 in Degradation of p27 at G1 Phase of the Cell Cycle. Mol Cell Biol. 2005;25:9292–9303. doi: 10.1128/MCB.25.21.9292-9303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 18.Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 19.Bai C, Sen P, Hofmann K, Ma L, Goebl M, et al. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 20.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 21.Patton E, Willems A, Sa D, Kuras L, Thomas D, et al. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, et al. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol Cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 24.Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, et al. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 25.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, et al. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 26.Mathias N, Johnson S, Byers B, Goebl M. The abundance of cell cycle regulatory protein Cdc4p is controlled by interactions between its F box and Skp1p. Mol Cell Biol. 1999;19:1759–1767. doi: 10.1128/mcb.19.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galan JM, Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc Natl Acad Sci U S A. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 30.Wolf DA, McKeon F, Jackson PK. F-box/WD-repeat proteins Pop1p and Sud1p/Pop2p form complexes that bind and direct the proteolysis of cdc18p. Curr Biol. 1999;9:373–376. doi: 10.1016/s0960-9822(99)80165-1. [DOI] [PubMed] [Google Scholar]
- 31.Seibert V, Prohl C, Schoultz I, Rhee E, Lopez R, et al. Combinatorial diversity of fission yeast SCF ubiquitin ligases by homo- and heterooligomeric assemblies of the F-box proteins Pop1p and Pop2p. BMC Biochem. 2002;3:22. doi: 10.1186/1471-2091-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, et al. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 33.Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplun L, Tzirkin R, Bakhrat A, Shabek N, Ivantsiv Y, et al. The DNA damage-inducible UbL-UbA protein Ddi1 participates in Mec1-mediated degradation of Ho endonuclease. Mol Cell Biol. 2005;25:5355–5362. doi: 10.1128/MCB.25.13.5355-5362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivantsiv Y, Kaplun L, Tzirkin-Goldin R, Shabek N, Raveh D. Turnover of SCFUfo1 complexes requires the UbL-UbA motif protein, Ddi1. Mol Cell Biol. 2006;26:1579–1588. doi: 10.1128/MCB.26.5.1579-1588.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez T, Kolawa N, Gee M, Sweredoski M, Deshaies R. Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 2011;9:33. doi: 10.1186/1741-7007-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krylov DM, Koonin EV. A novel family of predicted retroviral-like aspartyl proteases with a possible key role in eukaryotic cell cycle control. Curr Biol. 2001;11:R584–587. doi: 10.1016/s0960-9822(01)00357-8. [DOI] [PubMed] [Google Scholar]
- 38.Sirkis R, Gerst JE, Fass D. Ddi1, a eukaryotic protein with the retroviral protease fold. J Mol Biol. 2006;364:376–387. doi: 10.1016/j.jmb.2006.08.086. [DOI] [PubMed] [Google Scholar]
- 39.Hofmann K, Falquet L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci. 2001;26:347–350. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- 40.Kaplun L, Ivantsiv Y, Bakhrat A, Raveh D. DNA damage response-mediated degradation of Ho endonuclease via the ubiquitin system involves its nuclear export. J Biol Chem. 2003;278:48727–48734. doi: 10.1074/jbc.M308671200. [DOI] [PubMed] [Google Scholar]
- 41.Hirano S, Kawasaki M, Ura H, Kato R, Raiborg C, et al. Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nat Struct Mol Biol. 2006;13:272–277. doi: 10.1038/nsmb1051. [DOI] [PubMed] [Google Scholar]
- 42.Oldham CE, Mohney RP, Miller SL, Hanes RN, O’Bryan JP. The ubiquitin-interacting motifs target the endocytic adaptor protein epsin for ubiquitination. Curr Biol. 2002;12:1112–1116. doi: 10.1016/s0960-9822(02)00900-4. [DOI] [PubMed] [Google Scholar]
- 43.Katz M, Shtiegman K, Tal-Or P, Yakir L, Mosesson Y, et al. Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic. 2002;3:740–751. doi: 10.1034/j.1600-0854.2002.31006.x. [DOI] [PubMed] [Google Scholar]
- 44.Regan-Klapisz E, Sorokina I, Voortman J, de Keizer P, Roovers RC, et al. Ubiquilin recruits Eps15 into ubiquitin-rich cytoplasmic aggregates via a UIM-UBL interaction. J Cell Sci. 2005;118:4437–4450. doi: 10.1242/jcs.02571. [DOI] [PubMed] [Google Scholar]
- 45.Hoeller D, Hecker CM, Wagner S, Rogov V, Dotsch V, et al. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell. 2007;26:891–898. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 46.Kaplun L, Ivantsiv Y, Kornitzer D, Raveh D. Functions of the DNA damage response pathway target Ho endonuclease of yeast for degradation via the ubiquitin-26S proteasome system. Proc Natl Acad Sci U S A. 2000;97:10077–10082. doi: 10.1073/pnas.97.18.10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 48.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 49.Rinaldi T, Pick E, Gambadoro A, Zilli S, Maytal-Kivity V, et al. Participation of the proteasomal lid subunit Rpn11 in mitochondrial morphology and function is mapped to a distinct C-terminal domain. Biochem J. 2004;381:275–285. doi: 10.1042/BJ20040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. CSHL Press, NY. 1997.
- 51.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 52.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Domains of Ufo1 and Ddi1 used in experiments. The protein fragments used in the experiments depicted in the Figures are shown.
(TIF)
Formation of SCFUfo1-Ho-19S RP complex with yeast extract from RPN1-GFP cells. GSTUfo1 or control GST beads were incubated with yeast extract from cells with tagged genomic RPN1-GFP that were cotransformed with pGFP-HO and with pMYC-CDC53. The bead fraction was analysed by Western blotting with anti-GFP antibodies to detect Rpn1 and Ho, with anti-myc antibodies to detect Cdc53, and with anti-Ddi1 antibodies.
(TIF)
Rpn12 is present in the GSTUfo1 bead fraction. A further experiment in which GSTUfo1 and GST beads were incubated with yeast extract in the presence of GFPHo as in Figures 2 and S2. Here the Western blot employed antibodies made to GSTRpn12. The presence of Rpn12 is an indication that the 19S RP is intact.
(TIF)