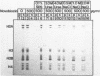
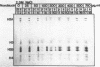
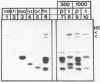
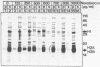
Abstract
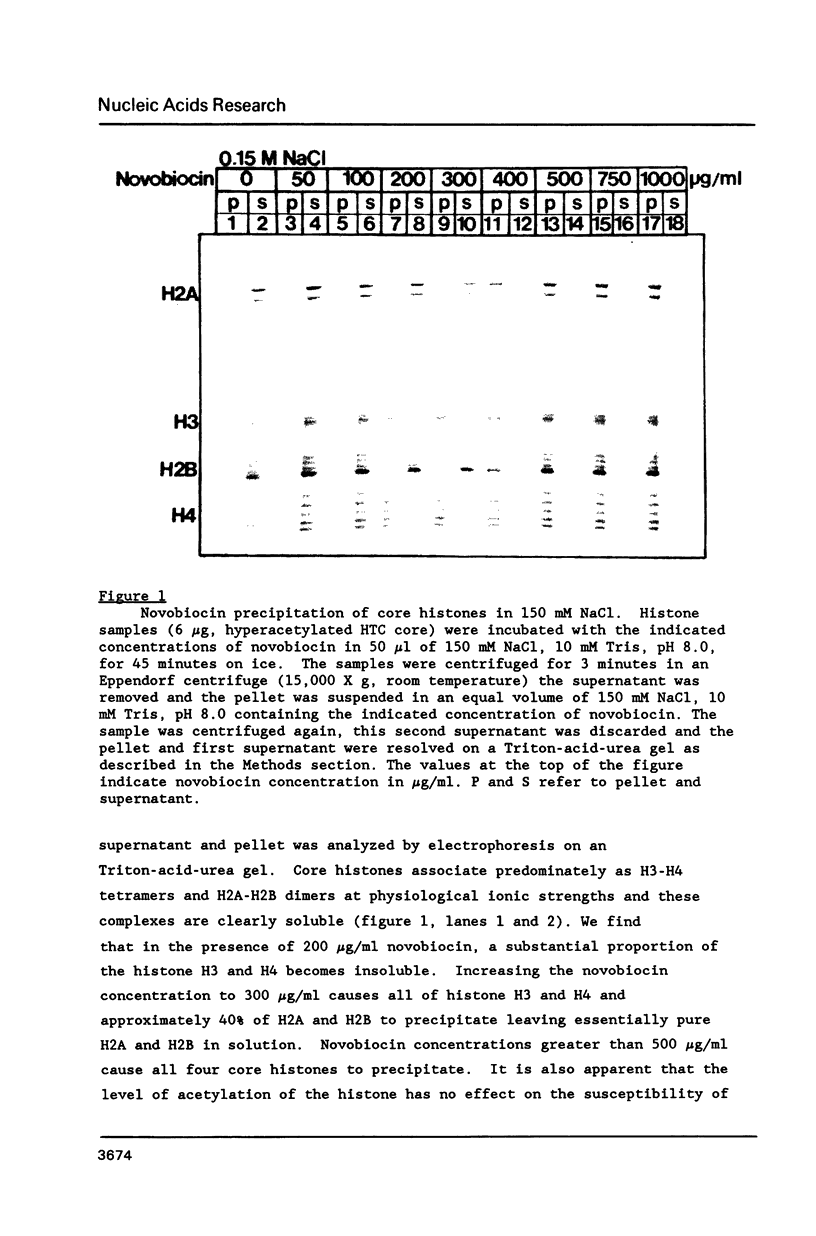
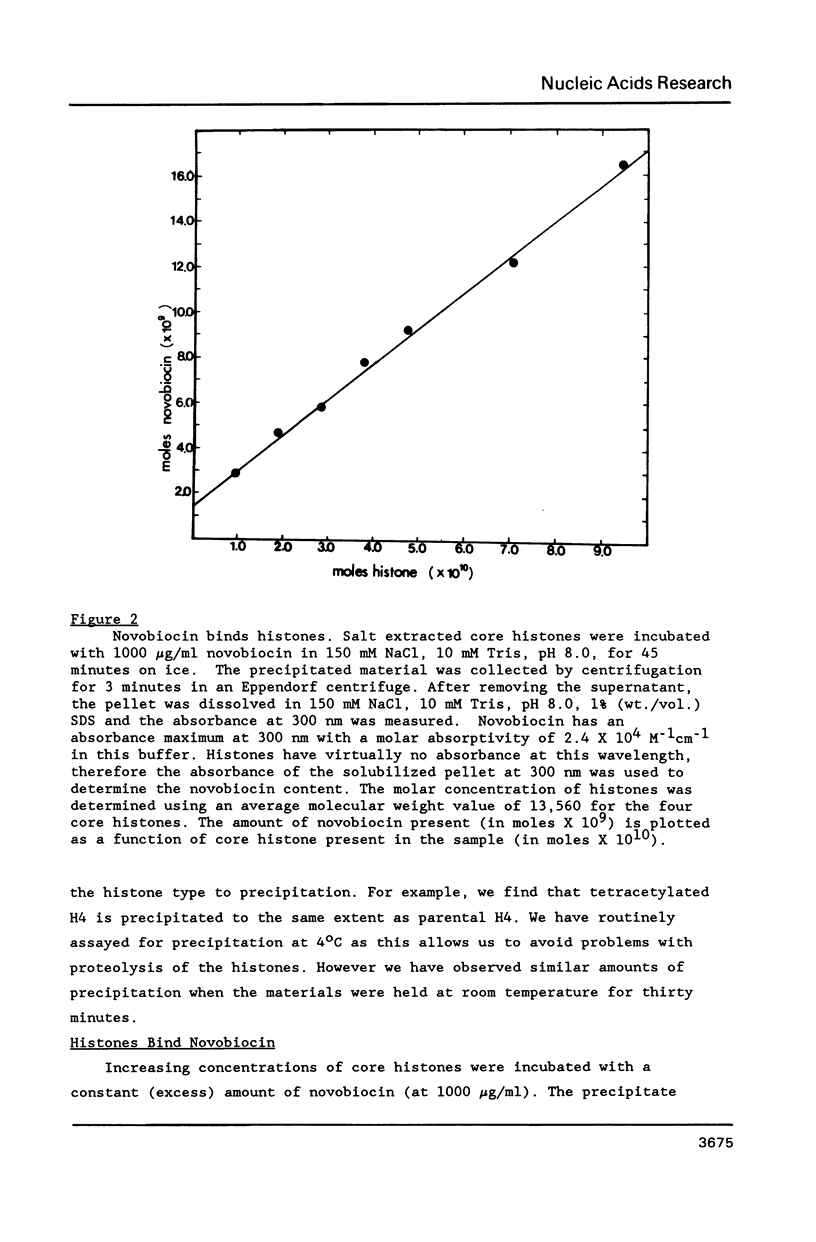
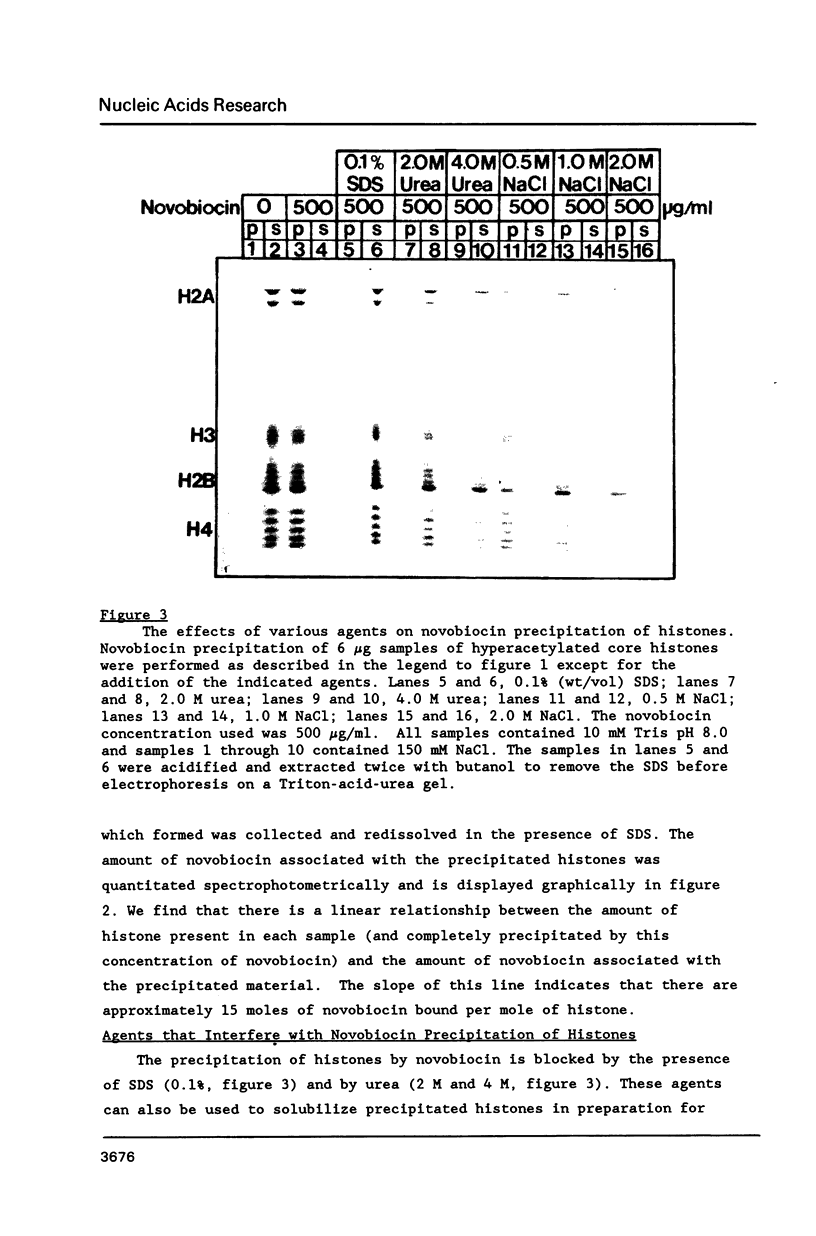
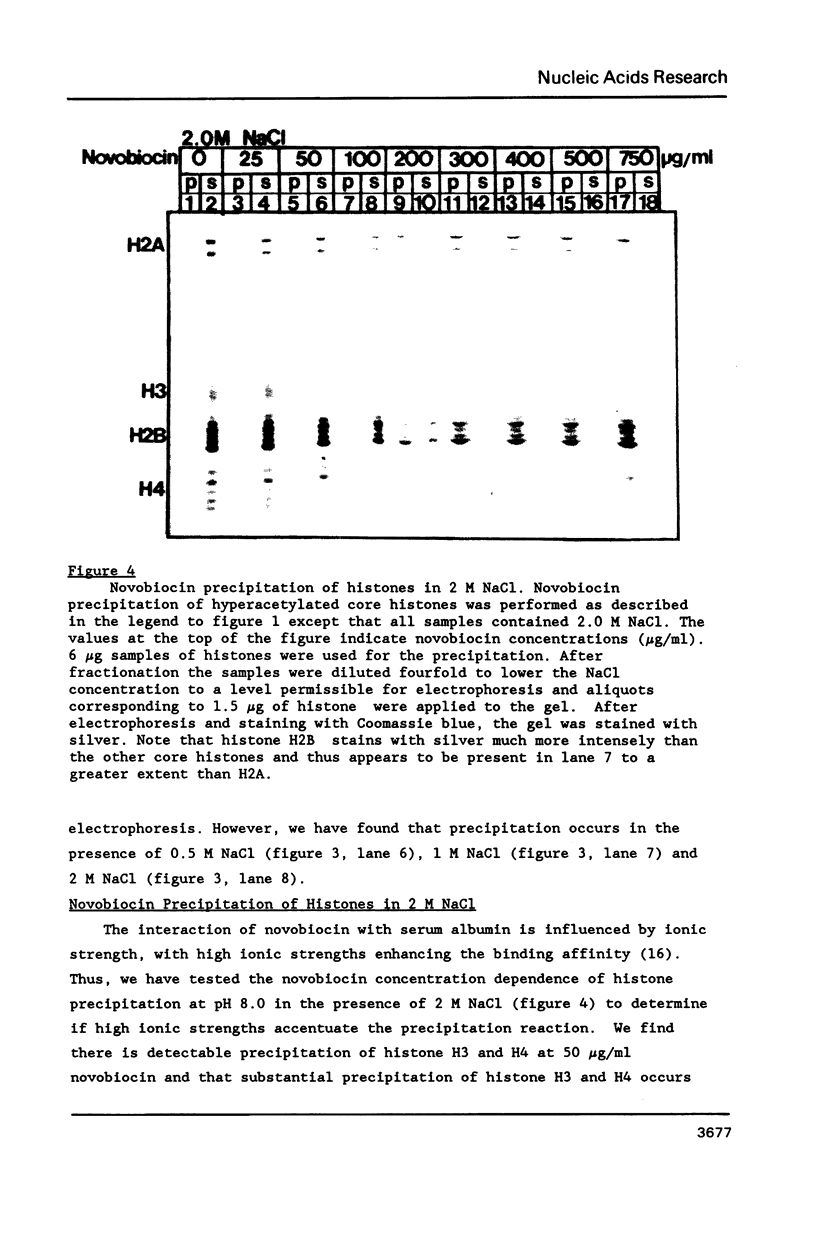
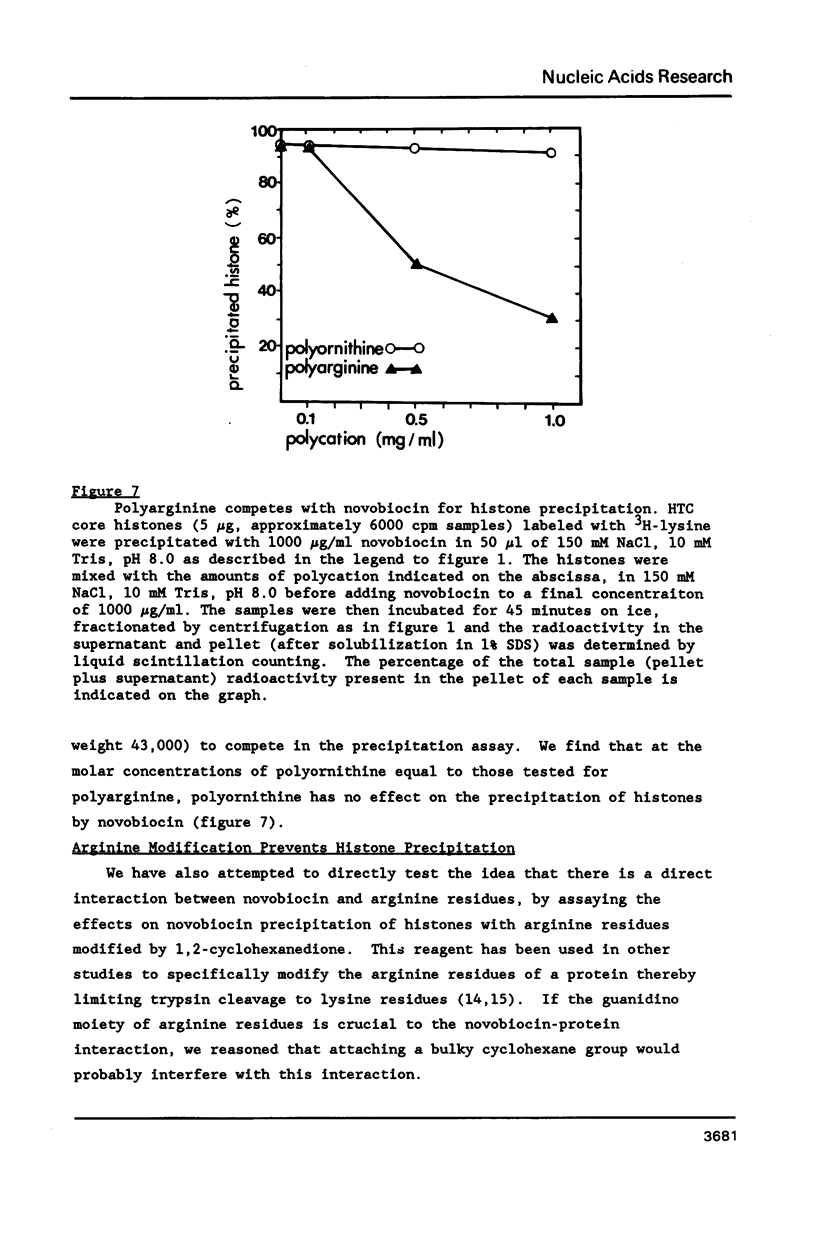
At concentrations normally used to inhibit eukaryotic type II topoisomerase activity (100-1000 micrograms/ml) novobiocin binds core histones. Approximately 15 moles of novobiocin bind per mole of histone resulting in histone precipitation from solution in either 0.15 M or 2 M NaCl. The interaction between novobiocin and proteins appears to involve arginine residues: histones H3 and H4 (13.5 and 14 mole percent arginine) are precipitated at lower novobiocin concentrations than histones H2A and H2B (9.5 and 6.5 mole percent arginine). Furthermore, polyarginine but not polyornithine competes for novobiocin in histone precipitation. Moreover, histones with arginine residues modified with 1,2-cyclohexanedione are soluble in 1000 micrograms/ml novobiocin. Because novobiocin can remove histones from solution as well as inhibit topoisomerase activity, and because both of these events can alter DNA topology, novobiocin should be used with caution in experiments designed to implicate topoisomerase activity in chromatin dynamics.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brand J. G., Toribara T. Y. Equilibrium dialysis and circular dichroic analysis of the novobiocin-bovine serum albumin complex. Arch Biochem Biophys. 1976 Jun;174(2):541–545. doi: 10.1016/0003-9861(76)90381-7. [DOI] [PubMed] [Google Scholar]
- Colwill R. W., Sheinin R. ts A1S9 locus in mouse L cells may encode a novobiocin binding protein that is required for DNA topoisomerase II activity. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4644–4648. doi: 10.1073/pnas.80.15.4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Chalkley R. Hyperacetylated histones facilitate chromatin assembly in vitro. Nucleic Acids Res. 1985 Jan 25;13(2):401–414. doi: 10.1093/nar/13.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Itoh T., Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glikin G. C., Ruberti I., Worcel A. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell. 1984 May;37(1):33–41. doi: 10.1016/0092-8674(84)90298-8. [DOI] [PubMed] [Google Scholar]
- Han S., Udvardy A., Schedl P. Novobiocin blocks the Drosophila heat shock response. J Mol Biol. 1985 May 5;183(1):13–29. doi: 10.1016/0022-2836(85)90277-3. [DOI] [PubMed] [Google Scholar]
- Jackson V. Studies on histone organization in the nucleosome using formaldehyde as a reversible cross-linking agent. Cell. 1978 Nov;15(3):945–954. doi: 10.1016/0092-8674(78)90278-7. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., O'Dea M. H., Gellert M. DNA gyrase: subunit structure and ATPase activity of the purified enzyme. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5960–5963. doi: 10.1073/pnas.75.12.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patthy L., Smith E. L. Identification of functional arginine residues in ribonuclease A and lysozyme. J Biol Chem. 1975 Jan 25;250(2):565–569. [PubMed] [Google Scholar]
- Patthy L., Smith E. L. Reversible modification of arginine residues. Application to sequence studies by restriction of tryptic hydrolysis to lysine residues. J Biol Chem. 1975 Jan 25;250(2):557–564. [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Chromatin assembly in Xenopus oocytes: in vivo studies. Cell. 1984 May;37(1):21–32. doi: 10.1016/0092-8674(84)90297-6. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L., Orr E. DNA gyrase: affinity chromatography on novobiocin-Sepharose and catalytic properties. Nucleic Acids Res. 1981 Aug 11;9(15):3589–3603. doi: 10.1093/nar/9.15.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Higgins N. P., Brown P. O., Peebles C. L., Cozzarelli N. R. Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4838–4842. doi: 10.1073/pnas.75.10.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R. J., Mosig G. An ATP-dependent supercoiling topoisomerase of Chlamydomonas reinhardtii affects accumulation of specific chloroplast transcripts. Nucleic Acids Res. 1985 Feb 11;13(3):873–891. doi: 10.1093/nar/13.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeponteau B., Lundell M., Martinson H. Torsional stress promotes the DNAase I sensitivity of active genes. Cell. 1984 Dec;39(3 Pt 2):469–478. doi: 10.1016/0092-8674(84)90454-9. [DOI] [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]