Abstract
Transcription factors govern diverse aspects of cell growth and differentiation as major switches of gene expression. Etv5, a member of the E26 transformation-specific family of transcription factors, has many stories to share when it comes to reproduction. Etv5 deficient mice show complex infertility phenotypes both in males and females. In males, the infertility phenotype exhibited by Etv5 deficiency is sexually dimorphic, and it involves both somatic cells and germ cells. In Etv5-/- female mice, the problem is more complicated by hormonal involvement. This review synthesizes old and new information on this versatile transcription factor-from the inadvertent discovery of its role in the testes to its newly discovered role in maintaining spermatogonial stem cells.
Keywords: Etv5, ETS transcription factor, Testis
Introduction to the PEA3 subfamily of transcription factors
The E26 transformation-specific (ETS) genes encode a large family of transcription factors [1]. All ETS transcription factors have a DNA binding domain referred to as an ETS domain. These factors regulate transcription by binding to 10 bp elements in the promoters of target genes, known as ETS-binding sites (EBS, 5'-GGAA/T-3'). One EBS can be shared by different ETS factors. The location of the ETS domain and overall sequence homology classify ETS factors into several subfamilies [1,2]. More than 20 family members have been identified in mammals and are divided into subfamilies including ETS, ERG, ELG, TEL, and PEA3. ETS factors have been linked to diverse physiological and pathological processes, but no clear unifying theme has emerged.
The PEA3 subfamily is composed of three members: Etv1/ER81, Etv4/PEA3/E1AF, and Etv5/Erm (will be referred to as Etv1, Etv4, and Etv5 in this review) [3]. They share the highly conserved ETS domain and two transactivation domains. Each gene is located on a different chromosome [4], but all three genes share a common gene structure comprising 14 equivalently sized exons [5,6]. This suggests a close conservation among members of the PEA3 subfamily. Etv1, Etv4, and Etv5 induce activation of target genes via a consensus EBS on promoters [7]. Some of the known target genes of PEA3 transcription factors are follicle stimulating hormone receptor (FSH-R), cyclooxygenase-2 (COX-2), stromelysin, osteopontin, matrilysin, and urokinase plasminogen activator [7-12].
For more than a decade, studies have shown that members of the PEA3 subfamily are implicated in cell proliferation, differentiation, and tumorigenesis [1,3,13,14]. While these genes are expressed in various tissues, it is notable that more than one member always appears in a given scene of action. In the nervous system, Etv4 and Etv1 are differentially expressed in subsets of motor neurons and muscle sensory neurons. During organogenesis, PEA3 factors are differentially expressed in the mesenchyme and epithelium of the developing lung, salivary gland, gut, olfactory, and optic organs [15]. Etv5 and Etv4 are primarily expressed in the developing epithelium, and Etv1 is mostly confined to the surrounding mesenchyme [15]. Differential expression patterns suggest that they participate in epithelial-mesenchymal interactions, but the specific roles in organogenesis are not clear yet.
Roles for PEA3 transcription factors in reproduction
Each member of the PEA3 subfamily has been genetically inactivated in mice [16-18]. Etv1-/- mice are born without overt anatomical defects, but develop limb ataxia due to defective neural connections in subsets of the sensory and motor neurons [16]. The story of Etv5 in fundamental issues of development and reproduction began inadvertently when Etv4-/- mice and Etv5-/- mice exhibited certain deficits in male fertility narrated below.
1. The beginning: male sexual dysfunction in Etv4-/- mice
When the Etv4-/- mouse was first generated, researchers were hoping to observe phenotypes in organogenesis or in mammary oncogenesis [15]. But Etv4-/- mice were born at the expected Mendelian ratio and did not exhibit any noticeable anomaly [17]. The mice had one obvious problem: male infertility. Etv4-/- female mice reproduced normally, but mutant male mice could not sire any offspring. One of the major sites of Etv4 mRNA expression is the epididymis [19]. Thus, male reproductive organs of wildtype and Etv4-/- male mice were compared. Surprisingly, Etv4-/- male mice did not show any cellular defects in the reproductive organs. All stages of the germ cells as well as the supporting cells were observed [17]. Etv4-/- male mice could exhibit erections but they did not mate even with gonadotropin-stimulated female mice. Interestingly, while their sperms successfully fertilized eggs in vitro, Etv4-/- male mice did not copulate [17]. This result suggested that the male sexual dysfunction in Etv4-/- mice is attributed to defective sexual behavior rather than germ cell defects. There still remain many questions in regard to the mechanism of male sexual dysfunction in the absence of Etv4.
2. The development: sertoli-cell-only syndrome and anovulation in Etv5-/- mice
Like Etv4-/- mice, Etv5-/- mice showed the unexpected phenotype of male infertility [18]. Etv5-/- male mice initially carry a pool of spermatogonial stem cells (SSCs), but the pool does not seem to be reloaded after the first waves of spermatogenesis [18]. The loss of SSCs begins as early as 4 days after birth and continues until almost all of the stem cells disappear [20]. The loss of SSCs is largely due to defective self-renewal of SSCs, but apoptotic cell death also contributes [20]. However, Etv5 is dispensable for spermatogenesis, as normal sperms are produced from Etv5-/- mice as long as SSCs are available [18]. The problem with Etv5-/- male mice does not end here: they do not show interest in female mice nor are their sperms fertilization-competent. Natural breeding, artificial insemination, and in vitro fertilization were all unsuccessful in making female mice become pregnant by Etv5-/- mice [20]. These observations suggested that Etv5-/- male mice have more fundamental defects, possibly both in germ cell quality and sexual behavior.
Etv5-/- female mice are also infertile [21]. In the mouse ovary, Etv4 and Etv5 are always expressed together. They are both expressed in granulosa cells in immature and in adult ovaries [12], and also in germ cells in the immature mouse ovary [21]. At around 2 weeks of age, Etv5-/- ovaries show defects in tissue architecture, which is partly attributed to abnormal germ cell-somatic cell interactions and the reduced expression of tight junction protein Claudin-5 [21]. This is reminiscent of the phenotype observed in Etv5-/- testes, in which a defect in the blood-testis barrier is noted along with reduced expression of Claudin-5 [22].
Etv5-/- female mice show a complete ovulation failure. No vaginal plugs form after mating with stud male mice. Stimulation with gonadotropins is insufficient to encourage mating. However, one treatment can reverse all of this. When an injection of 17beta-estradiol (E2) is administered along with gonadotropins, Etv5-/- female mice ovulate, become plugged, and have fertilized embryos in their oviducts [21]. But only about a third progress up to the blastocyst stage [21]. Thus, oocytes developed in the absence of Etv5 cannot competently support full development of embryos. Partial rescue of fertility defects in Etv5-/- female mice by E2 suggests possible endocrine defects, but no clear answer has been given.
The infertility phenotype in Etv5-/- female mice is potentially biphasic: an early defect with oocyte competence and an endocrine defect manifested at the adult stage. Since both Etv4-/- male and Etv5-/- male mice show no interest in female mice, it is plausible that certain endocrine or behavior defects may be involved in these male mutants. So far, no endocrine deficit has been reported [17,18]. Taken together, a biphasic function of Etv5 is demonstrated at least in female mice: one function in relation to endowing the competence of germ cells during the prepubertal period, and the other in promoting sexual responsiveness in adults.
As stated above, both sperms and oocytes undoubtedly have reduced developmental competence in Etv5-/- mice [20,21]. The phenotype seems to be more severe in Etv5-/- male mice, which show complete failure of fertilization [20]. Does Etv5 play an expanded role in male germ cells? A series of recent papers seems to indicate so.
3. The turn: Etv5 and the self-renewal of SSCs
Etv5 was initially identified in Sertoli cells, and naturally its function in Sertoli cells was first investigated. Microarray analyses using primary Sertoli cells from wildtype and Etv5-/- mice showed that an array of chemokines are not produced from Etv5-/- Sertoli cells [18]. One of the identified chemokines, Chemokine (C-C motif) ligand 9 (CCL9), attracts progenitor spermatogonia to Sertoli cells [23]. Another study showed that Etv5 is also expressed in male germ cells during the early neonatal period [24]. Transplantation of 5-day-old SSCs to W/Wv recipient testes (c-kit mutant mice) fails to establish normal spermatogenesis, suggesting that Etv5 also plays a cell-autonomous function in germ cells as well as in Sertoli cells [24].
Glial cell-drived neurotrophic factor (GDNF) and fibroblast growth factor 2 (FGF2) are required for the self-renewal of SSCs in vitro [25,26], and Sertoli cells produce these factors. GDNF acts upon a tyrosine kinase receptor RET expressed in SSCs. GDNF-RET signaling is critical for the self-renewal of SSCs [27]. Independent studies have recently shown that Etv5 is a major downstream factor of both GDNF and FGF2 in male germ cells [28,29]. In SSCs, GDNF-induced Etv5 turns on expression of several known genes important for self-renewal of SSCs, including B-cell CLL/lymphoma 6 (Bcl6b) and LIM homeobox protein 1 (Lhx1) [28]. Furthermore, one of the highly enriched microRNAs in SSCs, the miR-21, is regulated by Etv5 [30]. These data provide strong evidence that multiple stimuli for the self-renewal of SSCs commonly utilize Etv5 as a regulator of the pathway. Thus a dual function of Etv5 in the mouse testis is established: one in regulating chemokine production in Sertoli cells and the other in maintaining the population of SSCs.
4. The conclusion: functional redundancy and sexual dimorphism
In several tissues and organs in mice, Etv4 and Etv5 exhibit overlapping expression patterns [31]. In the mouse kidney, GDNF is a critical morphogen driving renal branching morphogenesis. Etv5 again acts downstream of GDNF during budding of the ureteric buds [32], as in SSCs. However, a redundancy of gene function by Etv4 is noted in the kidney. A single targeting of either Etv4 or Etv5 is not sufficient to cause severe kidney defects, but Etv4-/-/Etv5-/- double knockout mice do not develop kidneys [32]. Whether Etv4 and Etv5 play any redundant functions in the testes is not clear, but considering the somewhat restricted reproductive phenotypes in Etv4-/- mice, Etv5 seems to be a chief PEA3 transcription factor with unique functions in this domain. Etv4 is also expressed in SSCs of the mouse testis (unpublished observation, Eo and Lim), but it does not seem to play a critical function in SSC self-renewal because Etv4-/- male mice do produce sperms [17].
Overall, the sexual dimorphism of Etv5 deficiency is evident. In the ovary, Etv5 and Etv4 always exhibit a similar expression pattern [12, 21]. Thus, partial failure of preimplantation embryo development in Etv5-/- mice may be due to a compensatory mechanism endowed by Etv4. This hypothesis is yet to be proven with compound knockout mice. In Etv5-/- male mice, the phenotype is close to complete failure of fertility, a much stronger phenotype than in Etv5-/- female mice. This might be partially explained by the facts that the self-renewal of SSCs is heavily dependent upon GDNF and that Etv5 is directly downstream from it. While the relationship between GDNF and Etv5 has not been directly investigated in the ovary, GDNF is known to be one of the intraovarian factors, and is shown to enhance oocyte maturation and embryonic development [33]. Thus, it is possible that some phenotypes observed in Etv5-/- female mice as stated above are what the deficiency of GDNF might partially manifest.
Conclusion
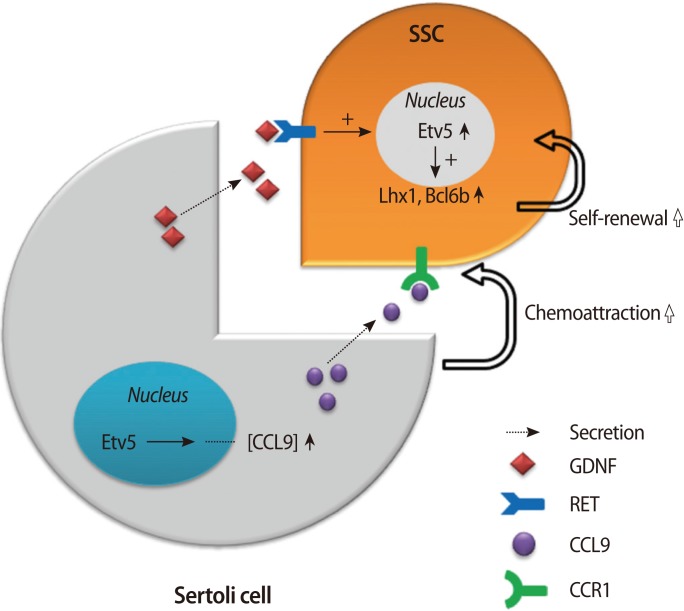
Etv5 is induced by GDNF or other secretory factors depending on the cell type. During the ureteric bud morphogenesis in the kidney, the GDNF-RET signaling pathway turns on multiple cytoplasmic kinases, and they contribute to the induction of Etv4 and Etv5 [31]. Sertoli cells also produce GDNF and other factors, which act upon the receptors on SSCs. Etv5 in SSCs is turned on and induces other genes critical for self-renewal of SSCs. Thus, GDNF is a paracrine activator of Etv5 in both cases. In Sertoli cells, Etv5 also contributes to the production of chemokines that are utilized for the chemoattraction of germ cells, and this exemplifies a cell-autonomous function of Etv5. The molecular pathway governed by Etv5 in the testes is diagrammed in Figure 1.
Figure 1.
Dual molecular pathway governed by Etv5 in the testes. Sertoli cells produce GDNF and other factors that act upon receptors on SSCs. Etv5 in SSCs is turned on and induces genes such as Lh×1 and Bcl6b that are critical for self-renewal of SSCs. In Sertoli cells, Etv5 also contributes to the production of CCL9, which is utilized for the chemoattraction of germ cells, and this exemplifies a cell-autonomous function of Etv5. GDNF, glial cell-derived neurotrophic factor; SSCs, spermatogonial stem cells; CCL9, chemokine (C-C motif) ligand 9; CCR1, C-C chemokine receptor type 1; Lh×1, LIM homeobox protein 1; Bcl6b, B-cell CLL/lymphoma 6.
Questions still remain with respect to the role of Etv5 in the ovaries. Is Etv5 also needed for the maintenance of the ovarian germ cell reserves? Does it take part in the endocrine axis that drives ovulation and sexual responsiveness in female mice? Along with this, compromised developmental competence of germ cells in both Etv5-/- female and male mice suggests an interesting possibility regarding the nature of Etv5. It might be a potential "stem-ness" factor that is needed for the maintenance of developmental potency. Further studies are needed to tackle these challenging issues.
Footnotes
This study was written as part of Konkuk University's research support program for its faculty during a sabbatical leave in 2012.
No potential conflict of interest relevant to this article was reported.
References
- 1.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 2.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 3.Oh S, Shin S, Janknecht R. ETV1,4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta. 2012;1826:1–12. doi: 10.1016/j.bbcan.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeon IS, Shapiro DN. Phylogenetically interrelated ETS genes, ETV1, ERM and E1A-F locate on different chromosomes. J Korean Med Sci. 1998;13:355–360. doi: 10.3346/jkms.1998.13.4.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monté D, Baert JL, Defossez PA, de Launoit Y, Stéhelin D. Molecular cloning and characterization of human ERM, a new member of the Ets family closely related to mouse PEA3 and ER81 transcription factors. Oncogene. 1994;9:1397–1406. [PubMed] [Google Scholar]
- 6.Coutte L, Monté D, Imai K, Pouilly L, Dewitte F, Vidaud M, et al. Characterization of the human and mouse ETV1/ER81 transcription factor genes: role of the two alternatively spliced isoforms in the human. Oncogene. 1999;18:6278–6286. doi: 10.1038/sj.onc.1203020. [DOI] [PubMed] [Google Scholar]
- 7.Howe LR, Crawford HC, Subbaramaiah K, Hassell JA, Dannenberg AJ, Brown AM. PEA3 is up-regulated in response to Wnt1 and activates the expression of cyclooxygenase-2. J Biol Chem. 2001;276:20108–20115. doi: 10.1074/jbc.M010692200. [DOI] [PubMed] [Google Scholar]
- 8.Rorth P, Nerlov C, Blasi F, Johnsen M. Transcription factor PEA3 participates in the induction of urokinase plasminogen activator transcription in murine keratinocytes stimulated with epidermal growth factor or phorbol-ester. Nucleic Acids Res. 1990;18:5009–5017. doi: 10.1093/nar/18.17.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford HC, Fingleton B, Gustavson MD, Kurpios N, Wagenaar RA, Hassell JA, et al. The PEA3 subfamily of Ets transcription factors synergizes with beta-catenin-LEF-1 to activate matrilysin transcription in intestinal tumors. Mol Cell Biol. 2001;21:1370–1383. doi: 10.1128/MCB.21.4.1370-1383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levallet J, Koskimies P, Rahman N, Huhtaniemi I. The promoter of murine follicle-stimulating hormone receptor: functional characterization and regulation by transcription factor steroidogenic factor 1. Mol Endocrinol. 2001;15:80–92. doi: 10.1210/mend.15.1.0583. [DOI] [PubMed] [Google Scholar]
- 11.El-Tanani M, Platt-Higgins A, Rudland PS, Campbell FC. Ets gene PEA3 cooperates with beta-catenin-Lef-1 and c-Jun in regulation of osteopontin transcription. J Biol Chem. 2004;279:20794–20806. doi: 10.1074/jbc.M311131200. [DOI] [PubMed] [Google Scholar]
- 12.Eo J, Han K, K MM, Song H, Lim HJ. Etv5, an ETS transcription factor, is expressed in granulosa and cumulus cells and serves as a transcriptional regulator of the cyclooxygenase-2. J Endocrinol. 2008;198:281–290. doi: 10.1677/JOE-08-0142. [DOI] [PubMed] [Google Scholar]
- 13.Bartel FO, Higuchi T, Spyropoulos DD. Mouse models in the study of the Ets family of transcription factors. Oncogene. 2000;19:6443–6454. doi: 10.1038/sj.onc.1204038. [DOI] [PubMed] [Google Scholar]
- 14.Maroulakou IG, Bowe DB. Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene. 2000;19:6432–6442. doi: 10.1038/sj.onc.1204039. [DOI] [PubMed] [Google Scholar]
- 15.Chotteau-Lelièvre A, Desbiens X, Pelczar H, Defossez PA, de Launoit Y. Differential expression patterns of the PEA3 group transcription factors through murine embryonic development. Oncogene. 1997;15:937–952. doi: 10.1038/sj.onc.1201261. [DOI] [PubMed] [Google Scholar]
- 16.Arber S, Ladle DR, Lin JH, Frank E, Jessell TM. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000;101:485–498. doi: 10.1016/s0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- 17.Laing MA, Coonrod S, Hinton BT, Downie JW, Tozer R, Rudnicki MA, et al. Male sexual dysfunction in mice bearing targeted mutant alleles of the PEA3 Ets gene. Mol Cell Biol. 2000;20:9337–9345. doi: 10.1128/mcb.20.24.9337-9345.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, et al. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature. 2005;436:1030–1034. doi: 10.1038/nature03894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xin JH, Cowie A, Lachance P, Hassell JA. Molecular cloning and characterization of PEA3, a new member of the Ets oncogene family that is differentially expressed in mouse embryonic cells. Genes Dev. 1992;6:481–496. doi: 10.1101/gad.6.3.481. [DOI] [PubMed] [Google Scholar]
- 20.Schlesser HN, Simon L, Hofmann MC, Murphy KM, Murphy T, Hess RA, et al. Effects of ETV5 (ets variant gene 5) on testis and body growth, time course of spermatogonial stem cell loss, and fertility in mice. Biol Reprod. 2008;78:483–489. doi: 10.1095/biolreprod.107.062935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eo J, Shin H, Kwon S, Song H, Murphy KM, Lim JH. Complex ovarian defects lead to infertility in Etv5-/- female mice. Mol Hum Reprod. 2011;17:568–576. doi: 10.1093/molehr/gar021. [DOI] [PubMed] [Google Scholar]
- 22.Morrow CM, Tyagi G, Simon L, Carnes K, Murphy KM, Cooke PS, et al. Claudin 5 expression in mouse seminiferous epithelium is dependent upon the transcription factor ets variant 5 and contributes to blood-testis barrier function. Biol Reprod. 2009;81:871–879. doi: 10.1095/biolreprod.109.077040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon L, Ekman GC, Garcia T, Carnes K, Zhang Z, Murphy T, et al. ETV5 regulates sertoli cell chemokines involved in mouse stem/progenitor spermatogonia maintenance. Stem Cells. 2010;28:1882–1892. doi: 10.1002/stem.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyagi G, Carnes K, Morrow C, Kostereva NV, Ekman GC, Meling DD, et al. Loss of Etv5 decreases proliferation and RET levels in neonatal mouse testicular germ cells and causes an abnormal first wave of spermatogenesis. Biol Reprod. 2009;81:258–266. doi: 10.1095/biolreprod.108.075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 26.Kanatsu-Shinohara M, Ogonuki N, Iwano T, Lee J, Kazuki Y, Inoue K, et al. Genetic and epigenetic properties of mouse male germline stem cells during long-term culture. Development. 2005;132:4155–4163. doi: 10.1242/dev.02004. [DOI] [PubMed] [Google Scholar]
- 27.Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 2004;101:16489–16494. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, Goodyear SM, Tobias JW, Avarbock MR, Brinster RL. Spermatogonial stem cell self-renewal requires ETV5-mediated downstream activation of Brachyury in mice. Biol Reprod. 2011;85:1114–1123. doi: 10.1095/biolreprod.111.091793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii K, Kanatsu-Shinohara M, Toyokuni S, Shinohara T. FGF2 mediates mouse spermatogonial stem cell self-renewal via upregulation of Etv5 and Bcl6b through MAP2K1 activation. Development. 2012;139:1734–1743. doi: 10.1242/dev.076539. [DOI] [PubMed] [Google Scholar]
- 30.Niu Z, Goodyear SM, Rao S, Wu X, Tobias JW, Avarbock MR, et al. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 2011;108:12740–12745. doi: 10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costantini F. GDNF/Ret signaling and renal branching morphogenesis: From mesenchymal signals to epithelial cell behaviors. Organogenesis. 2010;6:252–262. doi: 10.4161/org.6.4.12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu BC, Cebrian C, Chi X, Kuure S, Kuo R, Bates CM, et al. Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat Genet. 2009;41:1295–1302. doi: 10.1038/ng.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawamura K, Ye Y, Kawamura N, Jing L, Groenen P, Gelpke MS, et al. Completion of Meiosis I of preovulatory oocytes and facilitation of preimplantation embryo development by glial cell line-derived neurotrophic factor. Dev Biol. 2008;315:189–202. doi: 10.1016/j.ydbio.2007.12.029. [DOI] [PubMed] [Google Scholar]