SUMMARY
Multiple somatic rearrangements are often found in cancer genomes. However, the underlying processes of rearrangement and their contribution to cancer development are poorly characterised. Here, we employed a paired-end sequencing strategy to identify somatic rearrangements in breast cancer genomes. There are more rearrangements in some breast cancers than previously appreciated. Rearrangements are more frequent over gene footprints and most are intrachromosomal. Multiple architectures of rearrangement are present, but tandem duplications are common in some cancers, perhaps reflecting a specific defect in DNA maintenance. Short overlapping sequences at most rearrangement junctions suggest that these have been mediated by non-homologous end-joining DNA repair, although varying sequence patterns indicate that multiple processes of this type are operative. Several expressed in-frame fusion genes were identified but none were recurrent. The study provides a new perspective on cancer genomes, highlighting the diversity of somatic rearrangements and their potential contribution to cancer development.
INTRODUCTION
Cytogenetic studies over several decades have shown that somatic rearrangements, in particular chromosomal translocations, occur in many human cancer genomes1-3. The prevalence of rearrangements is, however, variable with some cancer genomes exhibiting few and others, including the genomes of many common adult epithelial cancers, showing many.
Somatic rearrangement is a common mechanism for the conversion of normal genes into cancer genes1-5. Indeed, of the ~400 genes that are currently known to be somatically mutated and implicated in cancer development, most are altered by genomic rearrangement (http://www.sanger.ac.uk/genetics/CGP/Census/). These rearrangements usually result in the formation of a fusion gene, derived from two disrupted normal genes, from which a fusion transcript and protein is generated. In some instances, however, rearrangements place an intact gene under the control of new regulatory elements or cause internal reorganisation of a gene. These events usually result in activation of the protein to contribute to oncogenesis, as in the paradigm of the BCR-ABL fusion gene in chronic myeloid leukaemia6.
Most of the currently known fusion genes are operative in leukaemias, lymphomas and sarcomas1,3 although similar cancer-causing rearrangements in RET, NTRK1, NTRK3, BRAF and TFE3 were reported in rare epithelial cancers many years ago (http://www.sanger.ac.uk/genetics/CGP/Census/). Activated fusion genes in common adult epithelial cancers such as those of the ETS transcription factor family in prostate cancer7 and the ALK gene in lung adenocarcinoma8 were discovered only recently and not through the conventional strategy of positional cloning of cytogenetically ascertained chromosomal breakpoints. Their late emergence primarily reflects the complexity of cytogenetically visible rearrangement patterns in the genomes of many adult epithelial cancers and the consequent difficulty in prior selection of driver rearrangements for further study among the many likely passenger changes9. Rearrangements also constitute a subset of mutational events that result in inactivation of recessive cancer genes (tumour suppressor genes) and underlie genomic amplifications that result in increased copy number of cancer genes10-12.
In recent years, the diversity of prevalence and pattern of point mutations and copy number changes in cancer genomes have been elucidated by systematic PCR-based resequencing studies and by application of high resolution copy number arrays respectively13-16. Understanding of the basic patterns of rearrangement in most cancers, however, remains rudimentary. We recently demonstrated that second generation sequencing of both ends of large numbers of DNA fragments generated from cancer genomes allows comprehensive characterisation of rearrangements17. Here we apply this approach to a series of breast cancers to explore patterns of rearrangement and their potential contribution to cancer development.
RESULTS
Landscapes of rearrangement
Twenty four breast cancers were investigated by sequencing both ends of ~65,000,000 randomly generated ~500bp DNA fragments from each cancer on Illumina GAII Genome Analysers (Supplementary Figure 1). The series included primary tumours and immortal cancer cell lines, examples of the commonest phenotypically defined subtypes and cases with high risk germline predisposition mutations in BRCA1, BRCA2 and TP53 (Table 1). Rearrangements were initially identified as discordant paired-end reads which did not map back to the reference human genome in the correct orientation with respect to each other and/or within ~500bp of each other. These were subsequently confirmed and evaluated for somatic or germline origin.
Table 1.
The 24 breast cancers analysed for somatic rearrangements
| Sample name |
Sample Type |
Age at diagnosis |
Grade | ER status |
PR status |
ERBB2 status |
Germline mutations |
|---|---|---|---|---|---|---|---|
| HCC1143 | Cell Line | 52 | 3 | − | − | − | − |
| HCC1187 | Cell Line | 41 | 3 | − | − | − | − |
| HCC1395 | Cell Line | 43 | 3 | − | − | − | BRCA1 |
| HCC1599 | Cell Line | 44 | 3 | − | − | − | BRCA2 |
| HCC1937 | Cell Line | 23 | 3 | − | − | − | BRCA1 |
| HCC1954 | Cell Line | 61 | 3 | − | − | + | − |
| HCC2157 | Cell Line | 48 | 2 | − | + | + | − |
| HCC2218 | Cell Line | 38 | 3 | − | − | + | TP53 |
| HCC38 | Cell Line | 50 | 3 | − | − | − | − |
| PD3664a a | Primary | 87 | 3 | − | − | − | − |
| PD3665a a | Primary | 47 | 3 | − | − | − | − |
| PD3666a b | Primary | 86 | 2 | + | + | − | − |
| PD3667a b | Primary | 43 | 2 | + | + | − | − |
| PD3668a c | Primary | 72 | 2 | + | + | − | − |
| PD3669a c | Primary | 66 | 2 | + | + | − | − |
| PD3670a d | Primary | 66 | 3 | − | − | + | − |
| PD3671a d | Primary | 48 | 3 | − | − | + | − |
| PD3672a e | Primary | 78 | 2 | − | − | − | − |
| PD3687a | Primary | 39 | 3 | − | − | − | BRCA1 |
| PD3688a | Primary | 47 | 3 | − | − | − | BRCA1 |
| PD3689a | Primary | 50 | 2 | + | + | − | BRCA2 |
| PD3690a | Primary | 40 | 2 | + | + | − | BRCA2 |
| PD3693a | Primary | 48 | 3 | − | − | + | − |
| PD3695a | Primary | 66 | 3 | − | − | − | − |
All the breast cancer samples screened were invasive ductal carcinomas with the exception of PD3672a which was an invasive lobular carcinoma. ER status refers to expression of estrogen receptor. PR status refers to expression of progesterone receptor. A subset of samples have also been classified according to expression profiles
basal-type
luminal A
luminal B
ERBB2
normal like.
2,166 confirmed somatic rearrangements were identified among the 24 cancers (Supplementary Tables 1 and 2). The presence of multiple read pairs spanning each rearrangement (Supplementary Table 1), easily detectable copy number changes associated with many and targeted FISH studies on a subset indicate that most rearrangements are in cells of the dominant cancer clone and not in minority cell populations. By investigating whether rearrangements were found for 200 changes in genomic copy number, we estimate that ~50% rearrangements have been detected.
Some cancers carried many rearrangements. For example the breast cancer cell line HCC38 has at least 238 (Figure 1), many more than could have been predicted from cytogenetic studies (http://www.path.cam.ac.uk/~pawefish/BreastCellLineDescriptions/HCC38.html). However, there is substantial variation in prevalence, with some primary breast cancers carrying only a single rearrangement (Figure 1, Supplementary Tables 1 and 2, Supplementary Figure 2). Overall, breast cancer cell lines showed more rearrangements (median 101, range 58-245) than primary cancers (median 38, range 1-231). This difference may be due to the acquisition of additional rearrangements during in vitro culture. However, it may also reflect less sensitive detection of rearrangements in primary cancers due to contaminating normal tissue or the relative propensity of some subclasses of breast cancer, for example metastases generating pleural effusions, to become established in culture. In some cancers rearrangements are evenly distributed through the genome. By contrast, in others they cluster in and connect genomic regions showing amplification. The rearrangement architecture in such amplicons is often highly complex10,11.
Figure 1.
Somatic rearrangements observed in six of the 24 breast cancer samples screened. (a) Genome wide circos plots of somatic rearrangements. An idiogram of a normal karyotype is shown in the outer ring. A copy number plot is represented by the blue line shown inner to the chromosome idiogram. Within the inner ring each green line denotes an intrachromosomal rearrangement and each purple line an interchromosomal rearrangement. (b) The prevalence of rearrangement architectures in individual cancers: Deletion (dark blue), tandem duplication (red), inverted orientation (green), interchromosomal rearrangements (light blue), rearrangements within amplified regions (orange). (c) Extent of overlapping microhomology at rearrangement breakpoints. The number of base pairs of microhomology is plotted on the horizontal axis.
Architectures of rearrangement
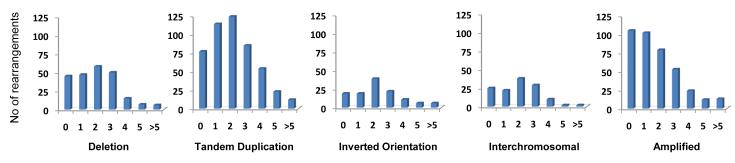
The orientations and relative chromosomal locations of the two genomic segments forming each rearrangement can be used as the basis of a rearrangement classification system. This may be further elaborated using information from copy number and other analyses that allow reconstruction of the genomic architecture associated with each rearrangement. For the purposes of this report, we have derived a simplified version of this system, classifying each rearrangement according to a) whether it is in an amplicon or not, b) if not in an amplicon whether it is interchromosomal or intrachromosomal and c) if intrachromosomal whether it results in a deletion, tandem duplication, or rearrangement of inverted orientation (Figure 1b).
There were 1,311 intrachromosomal and 239 interchromosomal rearrangements (excluding rearrangements within amplicons of which 397 were intrachromosomal compared to 219 interchromosomal) (Table 2, Supplementary Tables 1 and 2). Thus intrachromosomal rearrangements substantially outnumber interchromosomal rearrangements in breast cancer genomes. The breakpoints of 1574 out of 1708 intrachromosomal rearrangements were within 2Mb of each other. These patterns are presumably attributable to the greater likelihood of physical interaction between two positions on the same chromosome compared to positions on different chromosomes, perhaps due to individual chromosomes occupying domains in the nucleus, and between two locations that are in close proximity on the same chromosome. The predominance of intrachromosomal rearrangements has not previously been appreciated because of the limited resolution of cytogenetic studies and because FISH based approaches, such as spectral karyotyping, generally employ a single fluorescence colour per chromosome.
Table 2.
Summary of rearrangement patterns found in 24 breast cancers
| Rearrangement Class | Number in cell lines | Number in primaries | Total (%) |
|---|---|---|---|
| Deletion | 214 | 143 | 357 (16.5) |
| Mean per case (Range) | 23.8 (9-35) | 9.5 (0-41) | |
| Tandem Duplication | 370 | 369 | 739 (34) |
| Mean per case (Range) | 41.1 (4-138) | 24.6 (0-158) | |
| Inverted orientation | 113 | 102 | 215 (10) |
| Mean per case (Range) | 12.6 (4-24) | 6.8 (0-18) | |
| Inter-Chromosomal | 147 | 92 | 239 (11) |
| Mean per case (Range) | 16.3 (2-39) | 6.1 (0-27) | |
| Amplified | 308 | 308 | 616 (28.5) |
| Mean per case (Range) | 34.2 (0-208) | 20.5 (0-191) | |
| Total | 1152 | 1014 | 2166 (100) |
| Mean per case (Range) | 128 (58-245) | 67.6 (1-231) | |
| Base-pairs of microhomology at rearrangement junctions | |||
| Rearrangement Class | Mean (Range) | ||
| Deletion | 2.03 (0-14) | ||
| Tandem Duplication | 2.10 (0-9) | ||
| Inverted orientation | 2.50 (0-21) | ||
| Inter-chromosomal | 2.00 (0-9) | ||
| Amplified | 1.71 (0-9) | ||
| Total | 2.00 (0-21) | ||
| Base-pairs of non-templated sequence at rearrangement junctions | |||
| Rearrangement Class | Mean (Range) | ||
| Deletion | 3.27 (0-42) | ||
| Tandem Duplication | 3.46 (0-48) | ||
| Inverted orientation | 5.04 (0-45) | ||
| Inter-chromosomal | 3.63 (0-60) | ||
| Amplified | 3.83 (0-154) | ||
| Total | 3.71 (0-154) | ||
The most commonly observed architecture of rearrangement was tandem duplication (there were 739 tandem duplications, 357 deletions and 215 rearrangements with inverted orientation, Table 2, Supplementary Tables 1 and 2). The evidence that these are truly tandem insertions derives from the structure of the genomic rearrangement itself supported by cDNA and FISH studies (Tables 3 and 4, Supplementary Figures 3 and 4). The duplicated segments ranged in size from 3kb to greater than 1Mb (Supplementary Table 1). Despite being the commonest class of rearrangement in breast cancer, tandem duplications have previously been overlooked because they are intrachromosomal and involve small chromosomal segments beyond the resolution of cytogenetics or previous generations of copy number arrays.
Table 3.
Expressed in-frame fusion genes found in the 24 breast cancers
| Sample name |
5′ gene |
3′ gene |
Sample name |
5′ gene |
3′ gene |
|---|---|---|---|---|---|
| HCC1187b | PLXND1 | TMCC1 | HCC2157 | SMYD3 | ZNF695 |
| HCC1187 | RGS22 | SYCP1 | HCC38 | ACBD6 | RRP15 |
| HCC1395 | EFTUD2 | KIF18B | HCC38 | LDHC | SERGEF |
| HCC1395 | ERO1L | FERMT2 | HCC38 | MBOAT2 | PRKCE |
| HCC1395a | KCNQ5 | RIMS1 | HCC38 | SLC26A6 | PRKAR2A |
| HCC1395 | PLA2R1 | RBMS1 | HCC38 | SMF | PPARGC1B |
| HCC1599 | CYTH1 | PRPSAP1 | PD3664a | RAF1 | DAZL |
| HCC1937 | NFIA | EHF | PD3670a | AC141586.2 | CCNF |
| HCC1954 | STRADB | NOP58 | PD3670a | SEPT8 | AFF4 |
| HCC2157 | INTS4 | GAB2 | PD3688a | ETV6 | ITPR2 |
| HCC2157 | RASA2 | ACPL2 | PD3693aa | HN1 | USH1G |
Gene accession numbers and exons fused are outlined in Supplementary Table 6.
Gene fusion is amplified.
Predicted to be an out-of-frame gene fusion. However, RT-PCR across the exon-exon fusion boundary demonstrated both an out-of-frame and an in-frame gene fusion due to alternative splicing.
Table 4.
Expressed in frame rearranged genes found in the 24 breast cancers
| Sample name | Genes |
|---|---|
| HCC1187 | F8, FBXL20, GMDS, MED13L, RB1 |
| HCC1395 | CADPS2, DYNC2H1, KIAA0802, MBTPS1, TLE1 |
| HCC1937 | LRBA, RUNX1, SEMA3D, SSBP3 |
| HCC38 | EPHA3, EPS15, FRY, KCNMB2, REPS1, SLC4A4, TNRC15 |
| PD3664a |
C12orf35, C8orf70, GABRP2, HOMER2, INADL, KCNMA1, NFE2L3, ODZ1, PDE4B SVIL, VPS8 |
| PD3665a | DAPK1 |
| PD3668a | PLCB1, SYNJ1 |
| PD3671a | KIAA0146 |
| PD3687a | GP1, WRN |
| PD3693a | MACROD2 |
Gene accession numbers and exons fused are outlined in Supplementary Table 7.
There were major differences between individual breast cancers in the number of tandem duplications (Figure 1b, Supplementary Figure 5). Some exhibited more than one hundred while others showed few or none. The mechanistic basis for these differences is unknown, but may be due to a defective DNA maintenance process which confers a “mutator phenotype”. This would be similar, in principle, to the defects in DNA mismatch repair that are responsible for microsatellite instability in some cancers. This putative mutator phenotype is unlikely to be directly related to deficiencies in BRCA1 or BRCA2 mediated DNA repair as some cancers arising in individuals with germline mutations in these genes have few tandem duplications. Large numbers of tandem duplications were generally observed, however, in cancers that do not express estrogen and progesterone receptors.
Sequences at rearrangement junctions
DNA sequence across the rearrangement junction was obtained from 1,821 (3,642 breakpoints) of the 2,166 confirmed rearrangements (Supplementary Table 3). The sequences 100bp either side of each breakpoint were examined for the presence of motifs and sequence content. No striking signatures were observed, although there was a slight deficit of C:G base pairs compared to the genome as a whole (p<0.001) and modest enrichment of some motifs (Supplementary Tables 4 and 5). However, no single motif was commonly found in any class of rearrangement.
The sequences either side of each rearrangement junction were then compared to each other. In most instances the two contributing DNA segments showed a short stretch of identical sequence, known as an overlapping microhomology, immediately adjacent to the rearrangement junction which was present only once in the rearranged DNA (Figure 1c, Table 2, Supplementary Table 3 and Supplementary Figure 6). Approximately 15% rearrangements showed non-templated sequence at the rearrangement junction (Table 2, Supplementary Table 3 and Supplementary Figure 7). In many, this is only a few base pairs long, although the longest segment of this type was 154bp. A further 3.8% of rearrangements included one or more small fragments of DNA (<500bp) from elsewhere in the genome interposed between the rearrangement breakpoints identified by the paired end sequencing. We have previously termed these small DNA fragments “genomic shards”10,17.
Overlapping microhomologies and non-templated sequences at rearrangement junctions are often considered to be signatures of a non-homologous end-joining (NHEJ) DNA double strand break repair process18-21. The segments of overlapping microhomology are believed to mediate alignment of the two DNA fragments that are joined. It has, however, recently been proposed that complex germline rearrangements with genomic shards and overlapping microhomology might be due to replicative mechanisms21. The small proportion of complex rearrangements with genomic shards may indicate that this mechanism is relatively infrequently operative in breast cancer.
It has previously been suggested that there exist multiple NHEJ repair processes which may be characterised by different lengths of overlapping microhomology at rearrangement junctions21,22. To investigate this possibility we examined the distribution of microhomology lengths in each breast cancer (Figure 1c, Supplementary Figure 6). In some breast cancers, rearrangements with zero base pairs of microhomology were most frequent, whilst in others rearrangements with two or more base pairs were the commonest class. Rearrangements with zero base pairs of microhomology were most common in amplicons, in contrast to all other classes of rearrangement in which the modal class of microhomology was 2bp (Figure 2). These differences are unlikely to be due to chance (p<0.001) and suggest that there are at least two classes of NHEJ repair which are operative to different extents in different breast cancers21.
Figure 2.
ETV6-ITPR2, an expressed, in frame fusion gene generated by a 15Mb inversion in the primary breast cancer PD3668a. (a) Across-rearrangement PCR to confirm the presence of the somatic rearrangement. (b) RT-PCR of RNA between ETV6 exon 2 and ITPR exon 35 to confirm the presence of a chimeric expressed transcript; (c) Schematic diagram of the protein domains fused in the predicted ETV6/ITPR2 fusion protein. (d) Sequence from RT-PCR product shown in (b) confirming ETV6 exon 2 fused to ITPR2 exon 35.
Because the analysis of paired-end sequences requires alignment to the reference human genome and because sequences within repetitive elements are more likely to misalign it is conceivable that we have missed classes of rearrangement mediated by repeats. To investigate this possibility further we constructed libraries from the breast cancer cell line HCC1187 in which the sequenced ends were 3kb rather than 500bp apart. The 3kb paired ends will flank the majority of common repeats and thus allow detection of rearrangements mediated by them. Although additional rearrangements were detected, a distinct class of repeat mediated rearrangement was not found (data not shown).
Rearrangements of protein coding genes
Fifty per cent of rearrangements fell within the footprint of a protein coding gene compared to 40% expected by chance (p<10−7). The reasons for this striking enrichment of rearrangements in genic regions are not clear. Since rearrangements that confer selective advantage on a cancer clone are a priori more likely to be located in genes it is conceivable that some of this effect is due to selection and that a subset of rearrangements is implicated in cancer development. However, it may be more plausible that there are structural properties of genic regions that increase the likelihood of a DNA double strand break occurring, perhaps dependent on active transcription or chromatin configuration.
Twenty-nine rearrangements were predicted to generate in-frame gene fusions. Using exon-exon RT-PCR, rearranged transcripts from 19/22 in-frame fusion genes in non-amplified regions and from 2/6 (1 not determined) in amplified regions were found (Table 3). Thus most in-frame rearranged genes from non-amplified regions have the requisite 5′ and 3′ DNA sequences for transcript formation and stability. Conversely most from amplified regions do not and these rearrangements probably represent fragments of one or both genes reflecting the high density of rearrangements often present in these regions10. Sixty-six in-frame internally rearranged genes were also identified. 39/58 assessed showed rearranged transcripts (Table 4). In some cancers multiple in-frame rearranged and expressed genes are present (Tables 3 and 4, Supplementary Tables 6 and 7).
Several in-frame fusion genes are potentially of biological interest as candidates for new cancer genes. Notably, two were members of the ETS family of transcription factors. ETV6 is rearranged to form cancer genes with multiple different fusion partners in leukaemias23, congenital fibrosarcoma24 and myelodysplastic syndrome. It also forms a rearranged cancer gene with NTRK3 in the rare subclass of secretory breast cancer25. Here, ETV6 was fused to ITPR2 (Figure 3) through an inversion involving intron 2, a site previously reported in other cancers23, and was rearranged in a further breast cancer without clearly forming an in-frame fusion gene. ITPR2 encodes Inositol 1,4,5-triphosphate receptor Type 2 which is involved in signal transduction and regulation of cellular calcium fluxes. The second rearrangement fused EHF, which has not been previously implicated in cancer development, to NFIA a transcription factor involved in adenovirus replication (Supplementary Figure 3).
Figure 3.
Extent of overlapping microhomology at different architectural classes of rearrangement junctions. The number of base pairs of microhomology is plotted on the horizontal axis.
Fusion genes implicated in cancer development are likely to be recurrent. However, none of the novel fusion genes we identified was present in more than one out of the 24 cancers screened. Three expressed, in-frame fusion genes were examined by FISH (ETV6-ITPR2, NFIA-EHF and SLC26A6-PRKAR2A) and 20 by RT-PCR across the rearranged exon-exon junction in 288 additional breast cancer cases. No further examples were found indicating that they are either passenger events or that they contribute infrequently to breast cancer development.
Rearrangements were found in several known cancer genes including BRAF, PAX3, PAX5, NSD1, PBX1, MSI2 and ETV6 (see above). Each of these is a partner in a fusion gene in other classes of human cancer and was rearranged in two of the 24 samples analysed, although in many cases an in-frame fusion gene was not obviously generated (Supplementary Table 8). Rearrangements found in RB, APC, FBXW7 and other recessive cancer genes may have resulted in gene inactivation to contribute to cancer development.
Several other genes were rearranged in multiple cancers (Supplementary Table 9). Some are in amplified regions surrounding ERBB2 (for example ACCN1 which is rearranged in four out of the 24 breast cancers) or other known targets of genomic amplification in breast cancer. It is likely that these are recurrently rearranged because of the high density of rearrangements associated with these regions of recurrent genomic amplification. Others, however, are not in regions of genomic amplification. For example, SHANK2 was rearranged in five of the 24 breast cancers, while IGF1R, GRHL2, EFNA5, and MACROD2 were each rearranged in four. These recurrently rearranged genes generally have large genomic footprints and may simply represent bigger targets for randomly positioned rearrangements (Supplementary Table 9). For some, however, an elevated local rate of DNA double strand breakage (“fragility”) may also contribute to the clustering of rearrangements.
DISCUSSION
This study has generated the most comprehensive insight thus far into patterns of somatic rearrangement in cancer genomes. Most rearrangements in breast cancer are intrachromosomal. Tandem duplications appear to be the most common subclass and are known to form activated cancer genes in other cancer types26,27. The high prevalence of tandem duplications in a subset of cancers suggests the presence of a defect in DNA maintenance which generates this particular class of rearrangement. The underlying abnormality responsible for this phenotype is unknown. It may reside in the licensing mechanisms responsible for defining, priming and monitoring origins of DNA replication28.
Breast cancers are highly heterogeneous and are subclassified on the basis of estrogen receptor, progesterone receptor and ERBB2 expression and by mRNA expression profiles29,30. Subclasses defined in these ways show correlations with patterns of genomic alteration31,32. Breast cancers with many tandem duplications are usually estrogen and progesterone receptor negative and classified by expression profile as basal-like. In contrast, cancers with few rearrangements or with rearrangements within amplicons (other than those involving ERBB2) are usually estrogen receptor positive and classified as Luminal A and Luminal B types respectively.
Many novel in-frame fusion genes or internally rearranged genes were identified, most of which were expressed. None, however, were found to be recurrent. Approximately 2% rearrangements would be expected to generate an in-frame fusion gene by chance, compared to 1.6% observed. It is therefore likely that most are passenger events. Nevertheless, as previously suggested for somatic point mutations13,14 it may be that multiple, infrequently rearranged cancer genes are operative in breast cancer as they are in leukaemia2. Furthermore, detailed analysis of rearrangement breakpoints will be necessary to investigate the possibility of fusions between promoters/regulatory elements and intact genes that result in deregulation of expression. Much larger series will be required to investigate comprehensively the possibility of recurrent cancer-causing rearrangements in breast cancer.
Exhaustive sequencing of substantial numbers of cancer genomes to yield complete catalogues of all classes of somatic mutations will gather pace over the next few years. The current study offers insight into the complexity of rearrangement patterns that will be encountered in solid tumour genomes, demonstrates the potential for generation of active rearranged genes that may be implicated in cancer development and illustrates the types of information that will emerge on mutational processes that have been operative during the development of individual cancers.
Methods
Library construction and paired-end sequencing
Genomic libraries from nine breast cancer cell lines and fifteen primary breast cancers were generated using 5μg of total genomic DNA17. Briefly, 5μg of genomic DNA was randomly fragmented to between 200 and 700bp by focused acoustic shearing (Covaris Inc.). These fragments were electrophoresed on a 2% agarose gel wherefrom the 450-550bp fraction was excised and extracted using the Qiagen gel extraction kit (with gel dissolution in chaotropic buffer at room temperature to ensure recovery of AT rich sequences). The size fractionated DNA was end repaired using T4 DNA polymerase, Klenow polymerase and T4 polynucleotide kinase. The resulting blunt ended fragments were A-tailed using a 3′-5′ exonuclease-deficient Klenow fragment and ligated to Illumina paired-end adaptor oligonucleotides in a ‘TA’ ligation at room temperature for 15 minutes. The ligation mixture was electrophoresed on a 2% agarose gel and size-selected by removing a 2 mm horizontal slice of gel at ~600bp using a sterile scalpel blade. DNA was extracted from the agarose as above. 10ng of the resulting DNA was PCR amplified for 18 cycles using 2 units of Phusion polymerase. PCR cleanup was performed using AMPure beads (Agencourt BioSciences Corporation) following the manufacturer’s protocol. We prepared Genome Analyzer paired-end flow cells on the supplied Illumina cluster station and generated 37bp paired-end sequence reads on the Illumina Genome Analyser platform following the manufacturer’s protocol. Images from the Genome Analyzer were processed using the manufacturer’s software to generate FASTQ sequence files. These were aligned to the human genome (NCBI build 36.2) using the MAQ algorithm v0.4.333.
Reads removed from structural variant analysis
Reads which failed to align in the expected orientation or distance apart were further evaluated using the SSAHA algorithm34 to remove mapping errors in repetitive regions of the genome. In addition, during the PCR enrichment step, multiple PCR products derived from the same genomic template can occasionally be sequenced. To remove these, reads where both ends mapped to identical genomic locations (plus or minus a single nucleotide), were considered PCR duplicates, and only the read pair with the highest mapping quality retained. Further, erroneous mapping of reads originating from DNA present in sequence gaps in NCBI build 36.2 were removed by excluding the highly repetitive regions within 1Mb of a centromeric or telomeric sequence gap. Additional read pairs, where both ends mapped to within less than 500bp of one another, but in the incorrect orientation were excluded from analysis, unless support for a putative rearrangement was indicated by additional read pairs. The majority of these singleton read pairs are likely to be artifacts resulting from either intramolecular rearrangements generated during library amplification or mispriming of the sequencing oligonucleotide within the bridge amplified cluster. Finally, read pairs where both ends mapped to within 500bp of a previously identified germline structural variant were removed from further analysis, as these were likely to represent the same germline allele.
Generation of genome wide copy number plots
Full methods for generation of high resolution, genome wide copy number information can be found in reference17. Briefly, the human reference genome was divided into bins of ~15kb of mappable sequence and high quality, correctly mapping read pairs, with a MAQ alternative mapping quality ≥35, were assigned to their correct bin and plotted. A binary circular segmentation algorithm originally developed for genomic hybridisation microarray data35 was applied to these raw plots to identify change-points in copy number by iterative binary segmentation.
PCR confirmation of putative rearrangements
The following criteria were used to determine which incorrectly mapping reads pairs were evaluated by confirmatory PCR: (i) Reads mapping ≥10kb apart spanned by ≥2 read independent read pairs (where at least one read pair had an alternative mapping quality ≥35); (ii) Reads mapping ≥10kb apart spanned by 1 read pair (with an alternative mapping quality ≥35), with both ends mapping to within 100kb of a change-point in copy number identified by the segmentation algorithm; iii) Reads mapping ≥600bp apart spanned by ≥2 read independent read pairs (where at least one read pair had an alternative mapping quality ≥35) with both ends mapping to within 100kb of a change-point in copy number identified by the segmentation algorithm; iv) Selected read pairs mapping between 600bp and 10kb apart spanned by ≥2 read independent read pairs (where at least one read pair had an alternative mapping quality ≥35).
Primers were designed to span the possible breakpoint by locating them in the 1 kb outside region the paired-end reads, for a maximum product size of 1kb. PCR reactions were performed on tumor and normal genomic DNA for each set of primers at least twice, using the following thermocycling parameters: 95°C x 15min (95°C x 30s, 60°C x 30s, 72°C x 30s) for 30 cycles, 72°C x 10min. Products giving a band were sequenced by conventional Sanger capillary methods and compared to the reference sequence to identify breakpoints. Somatically acquired rearrangements were defined as those generating a reproducible band in the tumor DNA with no band in the normal DNA following PCR amplification, together with unambiguously mapping sequence data suggesting a rearrangement.
RT-PCR and cloning
Total RNA (100ng) from the tumor and matched constitutional DNA/lymphoblastoid cell lines was reverse transcribed into single stranded cDNAs using Reverse Transcriptase II (Invitrogen) and Oligo (dT)12-18 (Invitrogen) in 20μl reaction at 25 °C for 10 min, 42°C for 50min, 72°C for 15min. The cDNA was then diluted with 30μl of distilled water before subsequent PCR amplification. Resulting bands were sequenced to confirm the specificity of the reaction and the presence of the aberrant transcript. To detect fusion transcripts, we used forward primers in the putative 5′ partner gene and reverse primers from the 3′ partner. To detect rearranged transcripts, we used forward primers and reverse primers corresponding to the predicted exons fused. When multiple bands, possibly suggestive of splice variants were detected, all bands were excised from the gel and capillary sequenced separately.
DNA probes and Fluorescence in situ hybridisation (FISH)
FISH based on the split apart or fusion probe strategy was used to validate NFIA-EHF gene aberrations. For the NFIA-EHF fusion probe, BAC clones (http://www.ensembl.org/, Ensembl release 54) RP11-32I17, chromosome 1: 61,191,261-61,339,873; RP11-364M11, chromosome 1: 61,064,196-61,228,554 (red) and RP11-64P01, chromosome 1: 34,699,699-34,860,527; RP11-277N08, chromosome 1: 34,722,104-34,965,946 (green) were used. For the EHF split-apart probe, BAC clones RP11-64P01, chromosome 11:34,699,699-34,860,527; RP11-277N08, chromosome 11: 34,772,104-34,965,946 (green) and RP11-567H10, chromosome 11: 34,123,423-34,294,895; RP11-278N12, chromosome 11: 34,086,610-34,248,310 and RP11-686L07, chromosome 11: 33,936,895-34,109,642 (red) were used.
Dual colour FISH was used to detect the SLC26A6-PRKAR2A tandem duplication using BAC clones RP11-527M10, Chromosome 3: 45,948,424-46,115,480 (green); RP11-148G20, Chromosome 3: 48,575,991-48,781,362 (red).
BAC clones were purchased from BACPAC resource (Children’s Hospital Oakland;http://bacpac.chori.org/), and DNA from all BAC clones was purified, labelled and individually verified for specificity by FISH and direct sequencing as described previously36. BAC DNA was labelled with either biotin or digoxygenin-l I-dUTP (Roche) using the Bioprime kit (Invitrogen) and FISH was performed as described previously36. Biotinylated probes were detected with Cy5–Streptavidin (Invitrogen, Zymed Laboratories) and digoxigenin-labeled BACs, with anti-digoxigenin-fluorescein, Roche (green). Nuclei and chromosomes were counterstained with DAPI. Images were captured with a Zeiss Axioplan 2 microscope equipped with a CCD camera (Applied Imaging Diagnostic Instruments) and Cytovision software, version 2.81 (Applied Imaging). Only morphologically intact and non-overlapping nuclei were analyzed.
Breakpoint analysis
All 1832 breakpoints defined to the basepair level were used in the analysis of breakpoint sequence context, excluding shards and overlapping regions. Analysis was performed on all breakpoints together, and also on subsets divided into deletions, tandem duplications, amplicons, other intrachromosomal events, and all interchromosomal events. 10bp and 100bp on either side of the breakpoint sites were extracted for analysis. As a control, for each real breakpoint, 100 sequences of the same length were extracted from the regions extending from 10,000 to 20,000bp away from either side of the break. These matched control sequences were used as a comparison in the analysis to account for any regional differences such as large variations in GC or repeat content. The length of nucleotide tracts (polynucleotide, polypurine/polypurimidine, and alternating polypyrimidine/polypurine) were compared in the breakpoint and control regions using a one-tailed Mann-Whitney U test and the average GC content and presence of known motifs associated with DNA breaks37 were compared using a Fisher exact test.
Enrichment of breakpoints in genes
To determine whether breakpoints were enriched in genic regions, we compared the number of breakpoints falling within genes to an empirically-derived expected proportion. We classified breakpoints as genic or intergenic based on if their coordinates fell within a gene as annotated by Ensembl (http://www.ensembl.org/, Ensembl release 54). To account for the fact that some areas of the genome will be difficult to sequence align to with short reads, we derived the expected proportion of breakpoints that should fall within a gene from the actual proportion of read pairs that aligned to genic regions. Treating each breakpoint of a rearrangement independently, we then compared the number of breakpoints falling within a gene to this expected proportion using a Chi-squared test to obtain a p-value for the overrepresentation of breakpoints in genes.
Inter-individual heterogeneity in the patterns of microhomology
Comparison of microhomology and non-templated sequence distributions across individual samples was performed using Scholz and Stephens’ k-sample generalisation of the Anderson-Darling goodness-of-fit test, with 10,000 data permutations to generate the statistic’s null distribution38.
Supplementary Material

ACKNOWLEDGEMENTS
The authors are grateful to Maryou Lambros, Felipe Geyer and Radost Vatcheva for their assistance in the FISH experiments. We would like to acknowledge the support of the Kay Kendall Leukaemia Fund under Grant KKL282, Human Frontiers Award reference LT000561/2009-L, the Dana-Farber/Harvard SPORE in breast cancer under NCI grant reference CA089393, Breakthrough Breast Cancer, the Research Council of Norway Grants no 155218 and 175240, and the Wellcome Trust under grant reference 077012/Z/05/Z.
Footnotes
Supplementary Information is included with this manuscript.
Author Contributions M.R.S., P.A.F., P.J.C. and P.J.S. designed the experiment. S.E., D.J.M, P.J.S., M-L.L, I.V. L.J.M., J.B, M.A.Q., H.S., C.C., R.N., A.M.S., A.L., J.W.M.M., and C.L. carried out laboratory analyses. J.A.F., J.S.R-F., L.vV., A.L.R. D.P.S. and A.L.B-D. provided clinical samples. P.J.S., D.J.M., I.V., M-L.L., E.D.P., J.T.S., L.A.S., C.L., C.D.G., M.J., J.W.T., K.W.L., P.J.C., P.A.F., J.S.R-F., J.W.M.M., A.M.S., J.A.F., M.R.S., H.E.G.R., A.L.R., A.L.B-D., L.vV., A.L., P.J.C., P.A.F. performed data analysis, informatics and statistics. M.R.S., wrote the manuscript with comments from P.J.S., P.A.F., P.J.C., A.L.B-D., J.S.R-F., J.A.F., A.L.R., D.P.S., L.vV.
REFERENCES
- 1.Kaye FJ. Mutation-associated fusion cancer genes in solid tumors. Mol Cancer Ther. 2009;8:1399–408. doi: 10.1158/1535-7163.MCT-09-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat Genet. 2004;36:331–4. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- 3.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–45. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 4.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawyers CL. Chronic myeloid leukemia. N Engl J Med. 1999;340:1330–40. doi: 10.1056/NEJM199904293401706. [DOI] [PubMed] [Google Scholar]
- 7.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 8.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 9.Hoglund M, Gisselsson D, Sall T, Mitelman F. Coping with complexity. multivariate analysis of tumor karyotypes. Cancer Genet Cytogenet. 2002;135:103–9. doi: 10.1016/s0165-4608(01)00645-8. [DOI] [PubMed] [Google Scholar]
- 10.Bignell GR, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volik S, et al. End-sequence profiling: sequence-based analysis of aberrant genomes. Proc Natl Acad Sci U S A. 2003;100:7696–701. doi: 10.1073/pnas.1232418100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruan Y, et al. Fusion transcripts and transcribed retrotransposed loci discovered through comprehensive transcriptome analysis using Paired-End diTags (PETs) Genome Res. 2007;17:828–38. doi: 10.1101/gr.6018607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 15.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weir BA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–8. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–9. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–24. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 19.van Gent DC, van der Burg M. Non-homologous end-joining, a sticky affair. Oncogene. 2007;26:7731–40. doi: 10.1038/sj.onc.1210871. [DOI] [PubMed] [Google Scholar]
- 20.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 2005;4:639–48. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–64. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–82. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 23.Bohlander SK. ETV6: a versatile player in leukemogenesis. Semin Cancer Biol. 2005;15:162–74. doi: 10.1016/j.semcancer.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 24.Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet. 1998;18:184–7. doi: 10.1038/ng0298-184. [DOI] [PubMed] [Google Scholar]
- 25.Lannon CL, Sorensen PH. ETV6-NTRK3: a chimeric protein tyrosine kinase with transformation activity in multiple cell lineages. Semin Cancer Biol. 2005;15:215–23. doi: 10.1016/j.semcancer.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Jones DT, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–7. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basecke J, Whelan JT, Griesinger F, Bertrand FE. The MLL partial tandem duplication in acute myeloid leukaemia. Br J Haematol. 2006;135:438–49. doi: 10.1111/j.1365-2141.2006.06301.x. [DOI] [PubMed] [Google Scholar]
- 28.Blow JJ, Gillespie PJ. Replication licensing and cancer--a fatal entanglement? Nat Rev Cancer. 2008;8:799–806. doi: 10.1038/nrc2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 30.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chin K, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 32.Bergamaschi A, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–40. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. (2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venkatraman E, Olshen A. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 36.Lambros M, et al. Unlocking pathology archives for molecular genetic studies: a reliable method to generate probes for chromogenic and fluorescent in situ hybridization. Lab Invest. 2006;86:398–408. doi: 10.1038/labinvest.3700390. [DOI] [PubMed] [Google Scholar]
- 37.Abeysinghe SS, et al. Translocation and gross deletion breakpoints in human inherited disease and cancer I: Nucleotide composition and recombination-associated motifs. Hum Mutat. 2003;22:229–244. doi: 10.1002/humu.10254. [DOI] [PubMed] [Google Scholar]
- 38.Scholz FW, Stephens MA. K-sample Anderson-Darling Tests. Am J Stat Assoc. 1987;82:918–924. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.