Abstract
Spermatogenesis is an essential precursor for successful sexual reproduction. Recently, there has been an expansion in our knowledge of the genes associated with particular stages of normal, physiological testicular development and pubertal activation. What has been lacking, however, is an understanding of those genes that are involved in specifically regulating sperm production, rather than in maturation and elaboration of the testis as an organ. By utilising the reversible (seasonal) fertility of the Syrian hamster as a model system, we sought to discover genes which are specifically involved in turning off sperm production and not in tissue specification and/or maturation. Using gene expression microarrays and in situ hybridisation in hamsters and genetically infertile mice, we have identified a variety of known and novel factors involved in reversible, transcriptional, translational and post-translational control of testicular function, as well those involved in cell division and macromolecular metabolism. The novel genes uncovered could be potential targets for therapies against fertility disorders.
Keywords: seasonal, hamster, microarray, fertility, transcriptome, infertility
Introduction
Attempts to elucidate the molecular basis of spermatogenesis range from analysis of the contribution of individual gene products to broadly based screens of differential gene expression in testes (Matzuk and Lamb 2002). Such screens, most recently using DNA microarrays, have characterised suites of gene products associated with particular stages of normal, physiological testicular development and pubertal activation (Ostermeier et al. 2002; Shima et al. 2004). Microarrays have also been used to look for differentially expressed genes in pathological conditions in humans and animal models of genetically determined or idiopathic aspermatogenesis (Beissbarth et al. 2003; Ellis et al. 2004; Fox et al. 2003; Maratou et al. 2004; Rockett et al. 2004). These genes may contribute to the underlying pathology, or their contrasting patterns of expression could highlight critical factors associated with fertility. Such studies have thus helped to establish a framework to understand how programmed patterns of gene expression contribute to early testicular development and the initiation of spermatogenesis. Moreover, they have revealed a series of cell-type specific genes implicated in various endocrine responses of the testes (Chauvin and Griswold 2004; Eacker et al. 2007; Sadate-Ngatchou et al. 2004a, b; Schultz et al. 2003; Shima et al. 2004; Small et al. 2005; Zhou et al. 2005).
In most mammals, however, fertility is predominantly a reversible, seasonal phenomenon, regulated principally by changes in day length detected by the circadian (24 hour) timing system (Goldman 1999; Hastings 1991; Lincoln and Short 1980; Malpaux et al. 2001). Through the lifetime of an individual male, spermatogenesis is alternately activated and repressed on an annual basis due to photoperiodically driven changes in pulsatile gonadotrophin secretion. These are in turn dependent on photoperiodically driven changes in the duration of nocturnal secretion of melatonin by the pineal gland: the key seasonal stimulus to the neuroendocrine axis (Bartness et al. 1993; Goldman 2001; Lincoln et al. 2006). The seasonally inactivated seminiferous tubules are greatly reduced in size and lose almost all spermatocytes and spermatids, whilst the spermatogonia and Sertoli cells remain in an inactive condition (Berndtson and Desjardins 1974; Hikim et al. 1988). Moreover, steroidogenesis by Leydig cells is suspended. These dramatic changes are, however, completely reversible in a physiological fashion. Prolonged exposure to short day lengths is accompanied by a spontaneous reactivation of the hypothalamo-pituitary-gondal axis and full restoration of fertility appropriate to the animal’s seasonal niche (Almeida and Lincoln 1984; Berndtson and Desjardins 1974; Goldman 2001). This “photorefractory” state allows the animal to anticipate changes in daylength, initiating spermatogenesis in advance of exposure to stimulatory (long) daylengths.
The specific patterns of differential gene expression in the testis that underlie this physiological suspension of spermatogenesis, and its re-activation after a seasonal delay, are not known. The importance of these genes is that they could hold the key to understanding physiologically-regulated fertility. Unlike many genes previously identified in screens of testicular development, they are not necessarily involved in the initial specification or establishment of a testis. More likely, they are a select set of genes necessary for switching spermatogenesis. In particular, their analysis may highlight critical factors involved in physiological down-regulation of spermatogonial and Sertoli cell function.
The aim of this study, therefore, was to employ cDNA microarrays to study differential gene expression in the testis of a well characterised seasonal mammal, the Syrian hamster, in the context of reversible, seasonal infertility. We anticipated two principal groups of genes. Those genes whose expression is down-regulated in the involuted testis (in which spermatogenesis is suspended) would, by definition, be associated with the activation of spermatogenesis. Secondly, genes whose expression is relatively enriched in the involuted testis, may be connected with inactivation of sperm production (i.e. they are inhibitory genes) or alternatively, their expression may be maintained non-specifically in somatic tissue, which constitutes the bulk of the involuted testis. Furthermore, we tested the dependence of these changes on melatonin by performing pinealectomies in hamsters prior to exposure to inhibitory short daylengths. This would not interfere with their circadian synchronisation, but it would prevent testicular involution, which is dependent on the blood melatonin profile produced by the pineal gland (Hastings et al. 1985a, b).
Materials & Methods
Animals
All animal procedures were licensed by the Home Office (U.K.) under the Scientific Procedures (Animals) Act, 1986. Adult male Syrian Hamsters (100-120g, Harlan Olac, U.K.) were housed individually under a daily schedule of 16 hours bright white light (>250 lux) and 8 hours dim red light (<20 lux) for 6 weeks with food and water available ad libitum. Animals were then subjected to pinealectomy (PX) or sham surgery (INT) as described previously (Maywood et al. 1996). Upon recovery from surgery after 2 weeks, the hamsters were transferred to a short photoperiod (SD) of 8 h bright light and 16 h dim red light. Weekly testicular palpation was used to monitor the progression of gonadal involution in INT males exposed to SD. After 12 weeks of SD, gonadal involution in INT hamsters was complete in all hamsters. The animals were transferred to continuous dim red light for 24 h and then groups (n=6) of INT and PX animals were sacrificed by cervical dislocation at CT8 and CT20, i.e. 8 or 20 hours after projected lights on, and their testes dissected and immediately frozen on dry ice. For microarray studies, RNA was extracted from the testes of n=6 animals at each time point (CT8, CT20) in each treatment group (PX, INT), and then pooled in equal quantity before hybridisation to microarrays. Similarly, a further set of testes from an additional n=4 hamsters for each of the above groups were sectioned by cryostat for in situ hybridisation for genes of interest. A further group of sham-operated animals was left in SD until 22 weeks, by which time the testes had spontaneously recrudesced as a result of the “photorefractory” response, i.e. loss of response to SD. These were also killed by cervical dislocation at CT8 (n=4) and CT20 (n=4), and tissue treated as before for in situ hybridisation. Finally, testes from n=4 hypogonadal (hpg) mice (Cattanach et al. 1977) and their wild-type littermates aged 5-6 months were obtained a colony held at the School of Biomedical Sciences, University of Nottingham, UK, originating from Jackson Labs (Bar Harbor, ME). These were fixed in 4% paraformaldehyde, and wax-embedded sections of testes were used for in situ hybridisation as described.
cDNA microarrays
Approximately 50 mg of testicular tissue was homogenised in 1 ml of TRIZOL reagent (Invitrogen) at 4°C, and then RNA was isolated using phenol-chloroform extraction and alcohol precipitation. RNA was then further purified using RNeasy Mini columns (Qiagen). Total RNA from n=6 animals’ testes were pooled in equal quantity, and then this was transcribed into cDNA with incorporation of fluorescently-labelled (with Cy5 or Cy3 dye) dUTP by reverse transcription using SuperScript II (Invitrogen). Oligo-dT25, complementary to 3′ poly(A) tails of mRNA, was used as the primer for this reaction. The template RNA was degraded by alkaline hydrolysis and the resulting Cy5/Cy3-labelled single-stranded cDNA was cleaned using CentriSep columns (PE Biosystems) to remove enzymes and unincorporated primers/nucleotides before application to the spotted microarray for hybridization (Akhtar et al. 2002). After overnight hybridisation, arrays were subjected to a series of washes (in SSC) at room temperature, then dried by centrifugation before scanning. Analysis was then carried out using standard procedures; the basic analytical methods are described in (Turton et al. 2001). Briefly, feature sizes, background and feature fluorescence (either Cy3/Cy5 channels) were calculated using the automated default parameters of Imagene 4.1 software (Biodiscovery). The median fluorescence value for the pixels within the feature was calculated, and the raw data for each channel was then normalised by reference to the median fluorescence of the total feature set for that channel. A log2 (Cy5/Cy3) ratio was then calculated.
Microarray analysis
For the majority of our work, we used microarrays containing the well-known and characterised “NIA 15K” gene set to assay gene expression in PX and INT samples (Kargul et al. 2001). Samples from PX and INT animals were compared against each other at both CT8 and CT20. In both cases, two comparisons were made with “colour-swaps” (labelling the Cy3- and Cy5-labelled samples with the other dye and vice verse), giving a total of four arrays in total. The patterns of differential gene expression revealed at CT8 and CT20, and with the respective colour-swapped microarrays, were not significantly different and so data from the four microarrays were combined to generate a mean fold-difference change. We used a further independent microarray, with fewer probes (approx. 2100), to further assess differential gene expression in a similar fashion. We have used these arrays for circadian analyses previously (Akhtar et al. 2002). Genes were considered to be of interest if they showed at least a mean 2-fold change in expression between PX and INT samples. This was an arbitrary cut-off based on our previous experience with these microarrays and analysis of log2-scatterplots of the raw data using standard statistical algorithms within the Genespring software. Raw data are deposited in the NCBI Gene Expression Omnibus (GEO) Database under accession GSE10078.
In order to compare our expression patterns with published reports, the clones used on our own arrays, and the genes reported by other groups, were all identified by their most recent Unigene accession numbers. For all differentially expressed genes, a primary screen of previously reported tissue expression was made via the NCBI Gene Expression Omnibus (GEO), which for various genes provided details of expression in Sertoli and Leydig cells, spermatogonia, spermatocytes and spermatids (see Supplementary Table 1). In addition some previous studies had characterised expression within testicular somatic tissue. Functional analysis was carried out using Genetools eGOn (for Gene Ontology grouping) and DAVID (for functional group classification) (Beisvag et al. 2006; Dennis et al. 2003).
In situ hybridisation
In situ hybridisation was conducted on an independent series of tissue samples using probes for mouse circadian clock genes, previously validated for hamster tissue (Maywood et al. 1999), or by using riboprobes generated from IMAGE library clones corresponding to the microarray probes. For all novel probes, both sense and anti-sense transcripts were tested for hybridisation signal as described previously (Akhtar et al. 2002). None of the tested sense probes gave appreciable signal, whereas all the antisense probes used showed reproducible signal in testes and/or liver. A subset of these was further validated by testing for specific signals on mouse testes, giving identical results. The tissue distribution of hybridisation signal generated by the riboprobes was comparable between mouse and hamster testes (see Results section). NIH Image software was use to determine the intensity of hybridisation signal by use of 14C-microscales (Amersham, UK) that allowed optical density values from autoradiograms to be converted into quantitative nCi/g measurements (Akhtar et al. 2002). For comparability with the microarray study, which employed RNA extracts from the entire tissue, initial measures of hybridisation signal were integrated across the entire organ. In addition, for some genes with clearly heterogeneous tissue distribution, local intensity of gene expression was determined in regions of maximal expression. Relative intensity measures, derived from at least three hamsters per time point, were compared by ANOVA for the effects of circadian time and treatment (PX vs. INT) on gene expression. Post-hoc comparisons were made by Dunnett’s t-test.
Results
Photoperiodic regulation of testicular gene expression revealed by microarray
As anticipated, exposure of sham pinealectomised, intact (INT) Syrian hamsters to short photoperiods for 12 weeks led to testicular involution (testicular diameter, mean+/-SEM, INT week 1: 10.7+/-0.2 mm, week 12: 4.5+/-0.1 mm). This effect was blocked by pinealectomy (PX week 1: 9.8+/-0.2 mm, week 12: 11.1+/-0.2 mm). Histological analysis of the regressed testes of INT animals confirmed reduction of seminiferous tubules, with closure of the lumen and loss of spermatocytes, spermatids and spermatozoa (data not shown).
To determine gene expression patterns in activated and inactivated testes, we conducted analyses using microarrays containing the NIA 15K gene set (Kargul et al. 2001). Consistently, across four independent microarray hybridisations, we found 94 transcripts significantly up-regulated in the active testes by at least a factor of 2, and 103 genes relatively down-regulated by a factor of >2 (see Supplementary Table 1). A small number of these differentially regulated genes were represented by multiple spots on the array. Comparison between these multiple spots of the mean change in expression across the four hybridisations revealed a high level of consistency in the data. We also conducted a second, complementary study using microarrays that we have previously used to assay circadian gene expression (Akhtar et al. 2002), with relatively fewer probes (approx. 2120) that did not overlap significantly with the NIA 15K probes, thus increasing the overall transcript coverage in our study. This additional microarray revealed only a few up/down-regulated transcripts between PX and INT groups. The only gene overlapping between the two studies was Y-box protein 2 (Ybx2). For further analysis, we therefore focused on genes from one or the other study.
Characteristic genes expressed in the active testes included A-kinase anchor protein 1 (Akap1) and Y-box protein 2 (Ybx2), the latter being an upstream regulator of protamine expression, implicated in male infertility (Hammoud et al. 2008). About 60% (n=56) of the transcripts enriched in the active testes were of unknown function, although 82% (n=77) of the total of these transcripts had been previously reported as being expressed in testes – determined by analysing existing data sets in the NCBI GEO microarray database (see Materials and Methods). Of the transcripts enriched in the inactive testis, 45% (n=46) were of unknown function, but again over 80% (n=86) of the total were previously identified as testicular products. Based on the available cell and tissue expression data from the NCBI GEO database, there was a clear difference in the likely cellular localisation of the regulated transcripts. Of the genes enriched in the active testis, 49% (38 of 77) were spermatid products. In contrast, only a handful (7 of 86) of putative spermatid products were represented in the inactive testis, and of these all but one are expressed in other testicular cell types (see Supplementary Table 1).
Conversely, a large proportion of the transcripts enriched in the inactive testis (49%) were likely Sertoli cell products, whereas these accounted for less than 30% of the transcripts relatively enriched in spermatogenically active tissue. A similar pattern was evident for spermatogonial genes, with a relatively higher incidence in the inactive (34%) compared with the active testis (17%). These global changes in gene expression are therefore consistent with the histological changes that occur across the seasonal cycle, with the loss and then reappearance of spermatocytes and spermatids. Consequently, genes associated with the presence of spermatocytes and spermatids were expressed at far higher levels in the PX as opposed to INT samples. Moreover, Sertoli cell genes expressed at higher levels in the PX animal are likely associated with activation/support of spermatogenesis (Supplementary Table 1). The enrichment of Sertoli and spermatogonial genes in the inactive testis may represent either a simple consequence of the relative enrichment of these cell types in the organ, or a real up-regulation implying that increased expression of these genes is associated with suppression of spermatogenesis.
In situ hybridisation of known genes validates microarray findings and reveals cellular localisation of differentially expressed genes in the testis
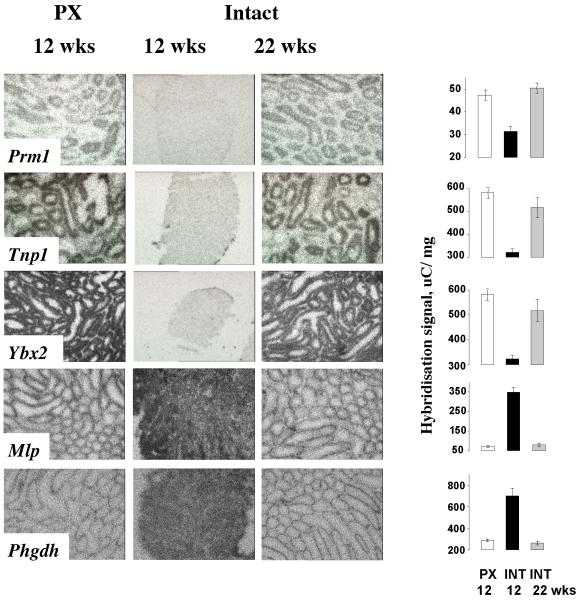
To validate the microarray data, and to determine likely cellular patterns of expression, we performed in situ hybridisation for selected genes on sections of testes from an independent set of PX and INT hamsters exposed to a short photoperiod. Also included were sections from testes of intact, but “photorefractory”, hamsters that had been exposed to short photoperiod for 22 weeks, leading to spontaneous re-activation of spermatogenesis. As anticipated, the known testicular genes Prm1, Tnp1 and Ybx2 were highly expressed within the tubules of PX hamsters. The expression levels of Prm1 and Tnp1 varied between individual tubules, being stage-specific, whereas signal intensity of Ybx2 was comparable in all tubules. In the testes of INT hamsters, expression of all three genes was at background level, whilst the reactivation of spermatogenesis following prolonged exposure to short photoperiods was accompanied by renewed expression of all three genes (Figure 1).
Figure 1. Location: 2nd page, Width: 2 columns, Height: 6”.
In situ hybridisation for genes identified by DNA microarray as up- or down-regulated in hamster testis during gonadal involution. Prm1, Tnp1 and Ybox2 are known markers for spermatogenesis and are clearly expressed within spermatogenically active tubules in PX animals and lost in intact animals after 12 weeks in short photoperiod. Mlp and Phgdh are not expressed in the inner tubule of active testis and become relatively enriched upon gonadal involution in intact animals after 12 weeks. The effects of short photoperiod are reversed in intact animals following spontaneous gonadal reactivation after 22 weeks in short photoperiod. Graphical data for signal intensity are presented as mean +SEM. Representative images from testes from animals sacrificed at CT20 are shown, but identical changes were seen in testes from CT8.
The expression of two genes identified by the microarray as relatively down-regulated in the active testes, MARCKS-like protein (Mlp), which encodes a secreted signalling factor, and 3-phosphoglycerate dehydrogenase (Phgdh), the product of which initiates serine biosynthesis, showed a very different tissue distribution to the up-regulated genes. Hybridisation signal was absent from the inner tubules and restricted to the outer tubule, and possibly interstitial cells (Figure 1). In mice, both of these genes are expressed in spermatogonia, and the expression of Mlp is reduced with the onset of spermatogenesis. With regression of the inner tubules in INT hamsters, this likely spermatogonial signal remained and so was relatively enriched. Upon reactivation of the inner tubules in refractory animals this relative enrichment was reversed and expression was again evident in the outer tubule (Figure 1). These in situ analyses therefore corroborate the microarray data but also highlight the potential impact of spatially restricted, cell-specific gene expression on the interpretation of such data derived from whole tissue homogenates. Specifically, does a relative enrichment in the INT testes represent an increase in signal per cell, or simply an enrichment of the total RNA sample by transcripts from these cells, which are relatively more abundant in the INT testis? To address this, we compared the intensity of hybridisation signal determined locally over the region of expression in individual tubules rather than across the entire testis. For both genes, the down-regulation remained, with INT:PX ratios of 1.5 (Mlp) and 3.5 (Phgdh), indicating that they were both expressed at higher levels in the relevant cells of the inactive testis.
In situ hybridisation of differentially expressed novel testes genes
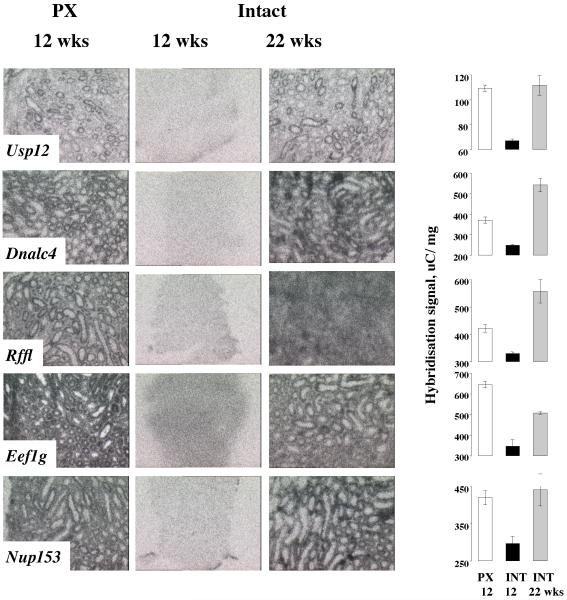
To determine the spatial expression of novel genes within the testis, and to further validate our microarray analyses, we conducted in situ hybridisation for ten randomly selected genes of unknown function at the time of analysis. Five were preferentially expressed in the spermatogenically active testes, and five were down-regulated in the active testis (Figures 2 & 3). The genes up-regulated in the spermatogenically active testis were strongly expressed in the inner and outer tubules, and the hybridisation signal dropped to background levels in the regressed testes. The effect was clearly reversible because strong tubular signal was restored in the testes of fertile, photorefractory hamsters (Figure 2). In all cases, expression was developmentally regulated, the intensity of signal varying between tubules as a function of spermatogenic stage. In the mouse, the Rffl gene is expressed in spermatids and so is massively up-regulated after the onset of spermatogenesis, as is Dynein light chain 4 (Dnalc4), which also appeared to be in spermatids in the hamster (confirmed by emulsion autoradiography; data not shown). Eukaryotic translation elongation factor 1 gamma (Eef1g) is expressed in mouse spermatogonia, spermatocytes and Sertoli cells, whilst Nucleoporin 153 (Nup153) expression is reported in Sertoli cells. Both genes were tightly regulated by photoperiod in the hamster, being down-regulated in the involuted testis (Figure 2). Finally, expression of Ubiquitin specific protease 12 (Usp12) was clearly stage-specific in hamster tubules, presumably in spermatids, and was absent from the regressed testis but restored in fertile, photorefractory animals (Figure 2).
Figure 2. Location: 3rd page, Width: 2 columns, Height: 7”.
In situ hybridisation for novel genes identified by DNA microarray as up-regulated in spermatogenically active hamster testis. All genes are expressed within active tubules of PX animals and expression in lost in intact animals with gonadal involution. This effect of photoperiod is reversible, with reactivated expression in intact hamsters after 22 weeks in short photoperiod. Graphical data for signal intensity are presented as mean +SEM. Representative images from testes from animals sacrificed at CT20 are shown.
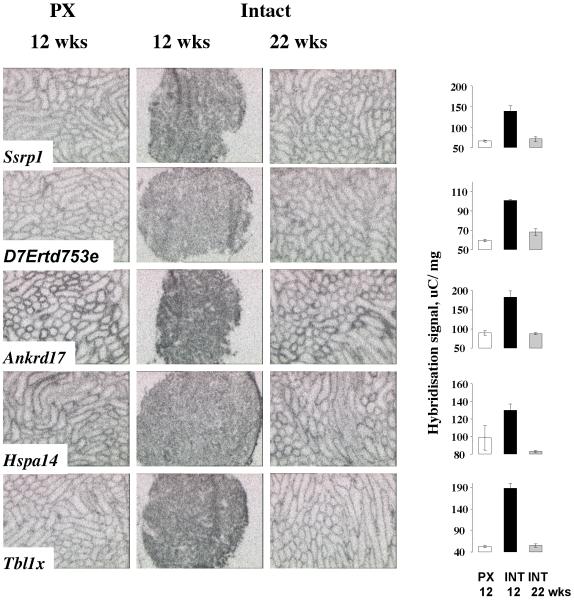
Figure 3. Location: 4th page, Width: 2 columns, Height: 7”.
In situ hybridisation for novel genes identified by DNA microarray as up-regulated in inactive hamster testis. This effect of photoperiod is reversible, with reactivated expression in intact hamsters after 22 weeks in short photoperiod. Graphical data for signal intensity are presented as mean +SEM. Representative images from testes from animals sacrificed at CT20 are shown.
The expression of genes identified by the microarrays as relatively down-regulated in the active hamster testis, i.e. enriched in the inactive testis, was restricted to the outer tubules and interstitial regions of the active testis (Figure 3). With the loss of spermatogenic function in the involuted testes, the overall signal intensity for these genes increased, whilst reduced and localised expression was again evident in the re-activated testes of refractory animals. A particular feature of this set of transcripts is the number reported in the mouse to be expressed in Sertoli cells. These include Ankyrin repeat domain 17 (Ankrd17), which was clearly stage-specific in the hamster, varying in intensity between tubules (Figure 3). Levels were decreased in the photoperiodically activated testes when measured both across the testis (ratio PX:INT 0.49) and locally on tubules (ratio PX:INT 0.58).
Structure specific recognition protein (Ssrp1) and Transducin-like 1 X-linked (Tbl1x) are also Sertoli cell products in the mouse, the former also reported from spermatogonia (Sadate-Ngatchou et al. 2004a, b; Shima et al. 2004; Small et al. 2005) – see Supplementary Table 1 for summary. Local measurements indicated expression of Ssrp1 was not especially sensitive to photoperiod in the hamster (local ratio 0.84) whereas Tbl1x clearly was (local ratio 0.55). These changes are consistent with the up-regulation of this gene observed in the inactive tubules of the hamster testes. Heat shock protein 70-14 (Hspa14) also declines after testicular maturation in the mouse, and it is also relatively enriched in interstitial cells, which is presumably the origin of expression in the hamster testis (Sadate-Ngatchou et al. 2004b; Small et al. 2005). The local expression of this gene, however, did not appear to be photoperiodically regulated (local ratio 0.92). Finally, the function of D7Ertd753e is not known but expression was restricted to the very outer tubule and/ or the interstitial tissue of the testis and so was relatively increased on the microarray and in situ analyses, but not appreciably so when analysed locally (local ratio 0.96).
Regulated expression of photoperiodically controlled genes in testes of hypogonadal mice
The foregoing in situ hybridisation studies provided independent confirmation of photoperiodically driven changes in gene expression from a representative set of transcripts in the hamster testis. As a further validation, we determined whether the changes observed in the context of this physiologically regulated, reversible, spermatogenesis arrest could be observed in a congenital aspermatogenic condition where testicular development is arrested at a neonatal stage, due to the lack of hypothalamic-pituitary stimulation (Cattanach et al. 1977; Ebling et al. 2006). We therefore performed in situ hybridization to determine the impact of the hypogonadal (hpg) mutation in the mouse on the expression of a subset of these seasonally regulated genes. As anticipated, the robust expression of Prm1, Tnp1 and Per1 in wild-type mouse testis was absent in hpg tissue (Figure 4). Moreover, the expression of Rffl, Dnalc4, Eef1g and Nup153 were all suppressed in the testes of mutant mice (Figure 4). Conversely, the expression levels of the genes relatively enriched in the photoperiodically suppressed testis, D7Ertd753e, Ankrd17, Ssrp, Tbl1x and Hspa14 were also increased in the hpg testis. On the basis of this cross-validation by in situ hybridisation, using independent samples of hamster and mouse testes, and with additional biological data from photoperiodically refractory hamsters and the hpg mouse, we propose that the changes in gene expression revealed by DNA microarray of the photoperiodically regulated testes of hamster represent a series of potential targets for the reversible control of male fertility.
Figure 4. Location: 5th page, Width: 2 columns, Height: 6½”.
In situ hybridisation in testes from wild-type and hypogonadal mice for photoperiodically genes identified by DNA microarray as up- or down-regulated in spermatogenically active hamster testis. Genes associated with spermatogenesis are expressed the active tubules of wild-type mice and expression is lost in mutant mice.
Discussion
The set of differentially expressed genes associated with seasonal fertility and infertility in the Syrian hamster are a component of a transcriptomic set associated with reversible activation of spermatogenesis. They therefore complement previous reports in the mouse of gene sets specifically activated with the onset of spermatogenesis and testicular specification (Chauvin and Griswold 2004; Eacker et al. 2007; Sadate-Ngatchou et al. 2004a, b; Schultz et al. 2003; Shima et al. 2004; Small et al. 2005; Zhou et al. 2005). By utilising the hamster as a model system, it is conversely possible to discover genes that are associated with reversibly turning off testicular function, which is a unique property of seasonal reproduction.
We used microarrays that did not specifically select for testes-enriched transcripts. An advantage of this approach was that unexpected and important transcriptional pathways, which have hitherto not been associated with activation/inhibition of testicular activity, could be identified. Overall, we found that ~2% of transcripts were differentially regulated. This is a relatively low number compared to studies looking at, for example, differential expression of genes during testicular maturation, in which one study found ~20% of genes assayed were regulated (Shima et al. 2004). The difference in “hit rate” no doubt reflects the fact that, in our study, genes specifically associated with switching spermatogenesis on/off were detected, whereas in other studies, there was obviously a significant contribution of genes important in tissue specification and maturation. Another potential reason for this difference could have been because of the interspecific use of microarrays, originally designed for murine analysis, in assaying hamster transcription (Bar-Or et al. 2007). However, we do not think that this was a significant factor for a number of reasons. First, expression levels and overall hybridisation signals across the microarrays were similar to that seen in similar murine analyses. Secondly, validation experiments using in situ hybridisation, with riboprobes directed against mouse mRNA, reproducibly targeted hamster tissues with the same tissue distribution as that seen in wild-type mouse testis. Therefore, we are confident that the relatively low number of regulated transcripts revealed is not the result of technical factors and/or species variability.
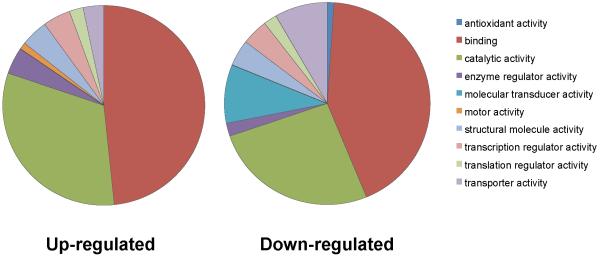
Functionally, genes differentially expressed in the active testes of pinealectomised animals fell into a number of distinct categories, according to their Gene Ontology groupings (Figure 5). Most functional categories contained a similar proportion of genes that were up- or down-regulated in the active testis, e.g. genes involved in binding DNA/protein and those regulating catalytic activity and structural processes. In contrast, motor activity genes (regulating flagellar assembly/function) were up-regulated in active testes, whereas genes mediating antioxidant activity, molecular transduction and transport were down-regulated (Figure 5). Thus, using simple Gene Ontology (GO) characteristics, there are clear pathways that are favoured and suppressed in the spermatogenically active testis (Beisvag et al. 2006).
Figure 5. Location: 6th page, Width: 2 columns, Height: 3”.
Functional categories of genes up- and down-regulated in testes of pinealectomised hamsters. A subset of Gene Ontology (GO) categories is shown.
We also employed a more sophisticated classification strategy (DAVID) to decipher clusters of genes that are co-regulated and functionally linked (Dennis et al. 2003). This revealed some complementary findings to those from the GO analysis. In the active testis, protein catabolism but RNA/DNA elaboration appears to be favoured (Table 1). This is revealed by activation of a number of genes involved in protein degradation, by both the proteasome (Usp12, Usp37) and independently of it (Nrd1, Thop1). Concurrently, there is deactivation of protein synthesis, as illustrated by significantly decreased expression of ribosomal proteins (Rpl10, Rpl36a, Rps26), an elongation factor (Gfm1) and a translation termination factor (Etf1). Furthermore, four molecular chaperones (Cct8, Chaperonin, Hspa14, Hspa8), essential for proper protein folding, were down-regulated in the spermatogenically active tissue (Table 1). The active testis also up-regulates multiple genes associated with transcriptional regulation/chromatin structure, including Hoxb1, Pias2, Tfam and Ybx2, as well as those involved in cell division (e.g. Cdc34). In synergy, there is increased expression of factors involved in macromolecule metabolism (including DNA/RNA) in coordination with the presumed upturn in demand for nucleotides etc. for these processes (Table 1).
Table 1.
Functionally classified groups of genes up- and down-regulated in spermatogenically active testes as determined by DAVID (Dennis et al. 2003).
| UP-regulated in active testes | |
|---|---|
|
Protein metabolism and degradation
| |
| Nrdl | NARDILYSIN, N-ARGININE DIBASIC CONVERTASE, NRD CONVERTASE 1 |
| Thopl | THIMET OLIGOPEPTIDASE 1 |
| Usp12 | UBIQUITIN SPECIFIC PEPTIDASE 12 |
| Usp37 | RIKEN CDNA C330008N13 GENE |
|
Transcriptional regulation
| |
| Clpb | CLPB CASEINOLYTIC PEPTIDASE B HOMOLOG (E. COLI) |
| Csda | COLD SHOCK DOMAIN PROTEIN A |
| Hoxbl | HOMEO BOX B1 |
| Nargl | NMDA RECEPTOR-REGULATED GENE 1 |
| Pias2 | PROTEIN INHIBITOR OF ACTIVATED STAT 2 |
| Tfam | TRANSCRIPTION FACTOR A, MITOCHONDRIAL |
| Ybx2 | Y BOX PROTEIN 2 |
|
Post-translational modification
| |
| Prkcd | PROTEIN KINASE C, DELTA |
| Stk17b | SERINE/THREONINE KINASE 17B (APOPTOSIS-INDUCING) |
| Uhmk1 | U2AF HOMOLOGY MOTIF (UHM) KINASE 1 |
| Wnk1 | WNK LYSINE DEFICIENT PROTEIN KINASE 1 |
|
Macromolecule metabolism
| |
| Bat3 | HLA-B-ASSOCIATED TRANSCRIPT 3 |
| Birc6 | BACULOVIRAL IAP REPEAT-CONTAINING 6 |
| Cdc34 | CELL DIVISION CYCLE 34 HOMOLOG (S. CEREVISIAE) |
| Herpud1 | HOMOCYSTEINE-INDUCIBLE, ENDOPLASMIC RETICULUM STRESS-INDUCIBLE, UBIQUITIN-LIKE DOMAIN MEMBER 1 |
| Rffl | RING FINGER AND FYVE LIKE DOMAIN CONTAINING PROTEIN |
| Rnf139 | RING FINGER PROTEIN 139 |
| Ube2j1 | UBIQUITIN-CONJUGATING ENZYME E2, J1 |
| DOWN-regulated in active testes | |
|---|---|
|
Protein synthesis
| |
| Etf1 | EUKARYOTIC TRANSLATION TERMINATION FACTOR 1 |
| Fbxw17 | F-BOX AND WD-40 DOMAIN PROTEIN 17 |
| Gfm1 | G ELONGATION FACTOR 1 |
| Rpl10 | RIBOSOMAL PROTEIN 10 |
| Rpl36a | RIBOSOMAL PROTEIN L36A |
| Rps26 | RIBOSOMAL PROTEIN S26 |
|
Molecular chaperones
| |
| Cct8 | CHAPERONIN SUBUNIT 8 (THETA) |
| Hspa14 | HEAT SHOCK PROTEIN 14 |
| Hspa8 | HEAT SHOCK PROTEIN 8 |
| Hspd1 | HEAT SHOCK PROTEIN 1 (CHAPERONIN) |
What do these gene expression changes mean for the active testis? Spermatogenesis involves a number of stages including mitosis, meiosis and cellular remodelling to finally create spermatids, the carriers of male hereditary information. Sertoli cells support and regulate this process and we would thus expect them to have a different molecular milieu, suited to their particular role. This is indicated in this study, with for example, factors associated with DNA packaging and chromatin remodelling (e.g. Protamine1/2, Pias2, Ybx2), expressed in late-stage cells (spermocytes and spermatids) but not in spermatogonia or Sertoli cells. Conversely, factors important for cell division (e.g. Cdc34) are up-regulated in early-stage cells (spermatogonia) but not in spermatids (see Supplementary Table 1). Protein degradation is favoured overall in the active testis, which might be anticipated, as the major function of the testis is to make and package DNA into spermatids. However, it is likely that specific proteins are degraded at different stages of the spermatogenic cycle, and accordingly expression of different protein degradation factors is up-regulated in specific compartments. Sertoli cells express Nrd1 at increased levels, whereas spermatids elaborate more Thop1 transcript. A number of ribosomal genes, important for protein synthesis in general, are down-regulated in cell types (see Table 1 and Supplementary Table 1). Overall, the gene expression changes we observed complement the biological function of the testis, and its subcomponents, in active spermatogenesis.
More than two hundred mouse genes are currently known to result in some form of infertility when mutated (Matzuk and Lamb 2002). Many of these have been discovered by phenotypic analysis of transgenic and knockout mice. It is highly likely that many more such fertility loci remain to be discovered, not only by further targeted approaches but also by analysis of spontaneous mutants (Cha et al. 2004; Clark et al. 2004; Harada et al. 2003; Lalouette et al. 1996; Meng et al. 2002; Rule et al. 1999) and by focussed chemical mutagenesis screens (Lessard et al. 2004). The uncharacterised ESTs exhibiting differential expression identified in this study may thus be candidates for the genes underlying uncharacterised fertility loci associated with defects in spermatogenesis.
Supplementary Material
Supplementary Table 1. All unambiguously identified genes/transcripts up- or down-regulated in spermatogenically active testes. Putative molecular function (based on Gene Ontology groups), and cell-specific expression patterns are shown, as determined from previous microarray expression studies (Chauvin and Griswold 2004; Sadate-Ngatchou et al. 2004a, b; Shima et al. 2004; Small et al. 2005; Zhou et al. 2005).
Acknowledgements
ABR, MHH and ESM are supported by the MRC. ABR is supported by the Wellcome Trust (WT083643MA) and the MRC Centre for Obesity and Related metabolic Diseases (MRC-CORD). SC-E was supported by an MRC training grant. CPK gratefully acknowledges a Royal Society Wolfson Research Merit Award. CPK and MHH wish to thank the BBSRC for a project grant used to fund this work (S09882).
References
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- Almeida OF, Lincoln GA. Reproductive photorefractoriness in rams and accompanying changes in the patterns of melatonin and prolactin secretion. Biology of reproduction. 1984;30:143–158. doi: 10.1095/biolreprod30.1.143. [DOI] [PubMed] [Google Scholar]
- Bar-Or C, Czosnek H, Koltai H. Cross-species microarray hybridizations: a developing tool for studying species diversity. Trends Genet. 2007;23:200–207. doi: 10.1016/j.tig.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Bartness TJ, Powers JB, Hastings MH, Bittman EL, Goldman BD. The timed infusion paradigm for melatonin delivery: what has it taught us about the melatonin signal, its reception, and the photoperiodic control of seasonal responses? J Pineal Res. 1993;15:161–190. doi: 10.1111/j.1600-079x.1993.tb00903.x. [DOI] [PubMed] [Google Scholar]
- Beissbarth T, Borisevich I, Horlein A, Kenzelmann M, Hergenhahn M, Klewe-Nebenius A, Klaren R, Korn B, Schmid W, Vingron M, Schutz G. Analysis of CREM-dependent gene expression during mouse spermatogenesis. Mol Cell Endocrinol. 2003;212:29–39. doi: 10.1016/j.mce.2003.09.023. [DOI] [PubMed] [Google Scholar]
- Beisvag V, Junge FK, Bergum H, Jolsum L, Lydersen S, Gunther CC, Ramampiaro H, Langaas M, Sandvik AK, Laegreid A. GeneTools--application for functional annotation and statistical hypothesis testing. BMC Bioinformatics. 2006;7:470. doi: 10.1186/1471-2105-7-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndtson WE, Desjardins C. Circulating LH and FSH levels and testicular function in hamsters during light deprivation and subsequent photoperiodic stimulation. Endocrinology. 1974;95:195–205. doi: 10.1210/endo-95-1-195. [DOI] [PubMed] [Google Scholar]
- Cattanach BM, Iddon CA, Charlton HM, Chiappa SA, Fink G. Gonadotrophin-releasing hormone deficiency in a mutant mouse with hypogonadism. Nature. 1977;269:338–340. doi: 10.1038/269338a0. [DOI] [PubMed] [Google Scholar]
- Cha KB, Karolyi IJ, Hunt A, Wenglikowski AM, Wilkinson JE, Dolan DF, Dootz G, Finnegan AA, Seasholtz AF, Hankenson KD, Siracusa LD, Camper SA. Skeletal dysplasia and male infertility locus on mouse chromosome 9. Genomics. 2004;83:951–960. doi: 10.1016/j.ygeno.2003.12.020. [DOI] [PubMed] [Google Scholar]
- Chauvin TR, Griswold MD. Androgen-regulated genes in the murine epididymis. Biology of reproduction. 2004;71:560–569. doi: 10.1095/biolreprod.103.026302. [DOI] [PubMed] [Google Scholar]
- Clark AT, Firozi K, Justice MJ. Mutations in a novel locus on mouse chromosome 11 resulting in male infertility associated with defects in microtubule assembly and sperm tail function. Biology of reproduction. 2004;70:1317–1324. doi: 10.1095/biolreprod.103.020628. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Eacker SM, Shima JE, Connolly CM, Sharma M, Holdcraft RW, Griswold MD, Braun RE. Transcriptional profiling of androgen receptor (AR) mutants suggests instructive and permissive roles of AR signaling in germ cell development. Molecular endocrinology (Baltimore, Md. 2007;21:895–907. doi: 10.1210/me.2006-0113. [DOI] [PubMed] [Google Scholar]
- Ebling FJ, Nwagwu MO, Baines H, Myers M, Kerr JB. The hypogonadal (hpg) mouse as a model to investigate the estrogenic regulation of spermatogenesis. Human fertility (Cambridge, England) 2006;9:127–135. doi: 10.1080/14647270500509103. [DOI] [PubMed] [Google Scholar]
- Ellis PJ, Furlong RA, Wilson A, Morris S, Carter D, Oliver G, Print C, Burgoyne PS, Loveland KL, Affara NA. Modulation of the mouse testis transcriptome during postnatal development and in selected models of male infertility. Mol Hum Reprod. 2004;10:271–281. doi: 10.1093/molehr/gah043. [DOI] [PubMed] [Google Scholar]
- Fox MS, Ares VX, Turek PJ, Haqq C, Reijo Pera RA. Feasibility of global gene expression analysis in testicular biopsies from infertile men. Mol Reprod Dev. 2003;66:403–421. doi: 10.1002/mrd.10364. [DOI] [PubMed] [Google Scholar]
- Goldman BD. The circadian timing system and reproduction in mammals. Steroids. 1999;64:679–685. doi: 10.1016/s0039-128x(99)00052-5. [DOI] [PubMed] [Google Scholar]
- Goldman BD. Mammalian photoperiodic system: formal properties and neuroendocrine mechanisms of photoperiodic time measurement. Journal of biological rhythms. 2001;16:283–301. doi: 10.1177/074873001129001980. [DOI] [PubMed] [Google Scholar]
- Hammoud S, Emery BR, Dunn D, Weiss RB, Carrell DT. Sequence alterations in the YBX2 gene are associated with male factor infertility. Fertil Steril. 2008 doi: 10.1016/j.fertnstert.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Harada M, Kishimoto K, Furuhashi T, Naito K, Nakashima Y, Kawaguchi Y, Hiraoka I. Infertility observed in reproductive toxicity study of N-acetyl-L-cysteine in rats. Biology of reproduction. 2003;69:242–247. doi: 10.1095/biolreprod.102.013862. [DOI] [PubMed] [Google Scholar]
- Hastings MH. Neuroendocrine rhythms. Pharmacol Ther. 1991;50:35–71. doi: 10.1016/0163-7258(91)90072-t. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Herbert J, Martensz ND, Roberts AC. Annual reproductive rhythms in mammals: mechanisms of light synchronization. Ann N Y Acad Sci. 1985a;453:182–204. doi: 10.1111/j.1749-6632.1985.tb11810.x. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Herbert J, Martensz ND, Roberts AC. Melatonin and the brain in photoperiodic mammals. Ciba Found Symp. 1985b;117:57–77. doi: 10.1002/9780470720981.ch5. [DOI] [PubMed] [Google Scholar]
- Hikim AP, Bartke AJ, Russell LD. The seasonal breeding hamster as a model to study structure-function relationships in the testis. Tissue Cell. 1988;20:63–78. doi: 10.1016/0040-8166(88)90008-0. [DOI] [PubMed] [Google Scholar]
- Kargul GJ, Dudekula DB, Qian Y, Lim MK, Jaradat SA, Tanaka TS, Carter MG, Ko MS. Verification and initial annotation of the NIA mouse 15K cDNA clone set. Nat Genet. 2001;28:17–18. doi: 10.1038/ng0501-17. [DOI] [PubMed] [Google Scholar]
- Lalouette A, Lablack A, Guenet JL, Montagutelli X, Segretain D. Male sterility caused by sperm cell-specific structural abnormalities in ebouriffe, a new mutation of the house mouse. Biology of reproduction. 1996;55:355–363. doi: 10.1095/biolreprod55.2.355. [DOI] [PubMed] [Google Scholar]
- Lessard C, Pendola JK, Hartford SA, Schimenti JC, Handel MA, Eppig JJ. New mouse genetic models for human contraceptive development. Cytogenet Genome Res. 2004;105:222–227. doi: 10.1159/000078192. [DOI] [PubMed] [Google Scholar]
- Lincoln GA, Clarke IJ, Hut RA, Hazlerigg DG. Characterizing a mammalian circannual pacemaker. Science (New York, NY. 2006;314:1941–1944. doi: 10.1126/science.1132009. [DOI] [PubMed] [Google Scholar]
- Lincoln GA, Short RV. Seasonal breeding: nature’s contraceptive. Recent Prog Horm Res. 1980;36:1–52. doi: 10.1016/b978-0-12-571136-4.50007-3. [DOI] [PubMed] [Google Scholar]
- Malpaux B, Migaud M, Tricoire H, Chemineau P. Biology of mammalian photoperiodism and the critical role of the pineal gland and melatonin. Journal of biological rhythms. 2001;16:336–347. doi: 10.1177/074873001129002051. [DOI] [PubMed] [Google Scholar]
- Maratou K, Forster T, Costa Y, Taggart M, Speed RM, Ireland J, Teague P, Roy D, Cooke HJ. Expression profiling of the developing testis in wild-type and Dazl knockout mice. Mol Reprod Dev. 2004;67:26–54. doi: 10.1002/mrd.20010. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2002;4(Suppl):s41–49. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Bittman EL, Hastings MH. Lesions of the melatonin- and androgen-responsive tissue of the dorsomedial nucleus of the hypothalamus block the gonadal response of male Syrian hamsters to programmed infusions of melatonin. Biology of reproduction. 1996;54:470–477. doi: 10.1095/biolreprod54.2.470. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Mrosovsky N, Field MD, Hastings MH. Rapid down-regulation of mammalian period genes during behavioral resetting of the circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:15211–15216. doi: 10.1073/pnas.96.26.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Akutsu H, Schoene K, Reifsteck C, Fox EP, Olson S, Sariola H, Yanagimachi R, Baetscher M. Transgene insertion induced dominant male sterility and rescue of male fertility using round spermatid injection. Biology of reproduction. 2002;66:726–734. doi: 10.1095/biolreprod66.3.726. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Dix DJ, Miller D, Khatri P, Krawetz SA. Spermatozoal RNA profiles of normal fertile men. Lancet. 2002;360:772–777. doi: 10.1016/S0140-6736(02)09899-9. [DOI] [PubMed] [Google Scholar]
- Rockett JC, Patrizio P, Schmid JE, Hecht NB, Dix DJ. Gene expression patterns associated with infertility in humans and rodent models. Mutat Res. 2004;549:225–240. doi: 10.1016/j.mrfmmm.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Rule MC, Mutcherson RJ, 2nd, Foss AD, Nguyen TK, Myrie KA, King TR. Mouse male sterility and histoincompatibility (mshi) maps between the D10Mit51/168/212 cluster and D10Mit213. Mamm Genome. 1999;10:447–450. doi: 10.1007/s003359901021. [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Pouchnik DJ, Griswold MD. Follicle-stimulating hormone induced changes in gene expression of murine testis. Molecular endocrinology (Baltimore, Md. 2004a;18:2805–2816. doi: 10.1210/me.2003-0203. [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Pouchnik DJ, Griswold MD. Identification of testosterone-regulated genes in testes of hypogonadal mice using oligonucleotide microarray. Molecular endocrinology (Baltimore, Md. 2004b;18:422–433. doi: 10.1210/me.2003-0188. [DOI] [PubMed] [Google Scholar]
- Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- Small CL, Shima JE, Uzumcu M, Skinner MK, Griswold MD. Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biology of reproduction. 2005;72:492–501. doi: 10.1095/biolreprod.104.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turton NJ, Judah DJ, Riley J, Davies R, Lipson D, Styles JA, Smith AG, Gant TW. Gene expression and amplification in breast carcinoma cells with intrinsic and acquired doxorubicin resistance. Oncogene. 2001;20:1300–1306. doi: 10.1038/sj.onc.1204235. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Shima JE, Nie R, Friel PJ, Griswold MD. Androgen-regulated transcripts in the neonatal mouse testis as determined through microarray analysis. Biology of reproduction. 2005;72:1010–1019. doi: 10.1095/biolreprod.104.035915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. All unambiguously identified genes/transcripts up- or down-regulated in spermatogenically active testes. Putative molecular function (based on Gene Ontology groups), and cell-specific expression patterns are shown, as determined from previous microarray expression studies (Chauvin and Griswold 2004; Sadate-Ngatchou et al. 2004a, b; Shima et al. 2004; Small et al. 2005; Zhou et al. 2005).