Summary
Cellular life emerged ~3.7 billion years ago. With scant exception, terrestrial organisms have evolved under predictable daily cycles due to the Earth’s rotation. The advantage conferred upon organisms that anticipate such environmental cycles has driven the evolution of endogenous circadian rhythms that tune internal physiology to external conditions. The molecular phylogeny of mechanisms driving these rhythms has been difficult to dissect because identified clock genes and proteins are not conserved across the domains of life: Bacteria, Archaea and Eukaryota. Here we show that oxidation-reduction cycles of peroxiredoxin proteins constitute a universal marker for circadian rhythms in all domains of life, by characterising their oscillations in a variety of model organisms. Furthermore, we explore the interconnectivity between these metabolic cycles and transcription-translation feedback loops of the clockwork in each system. Our results suggest an intimate co-evolution of cellular time-keeping with redox homeostatic mechanisms following the Great Oxidation Event ~2.5 billion years ago.
Circadian rhythms are considered to be a feature of almost all living cells. When isolated from external stimuli, organisms exhibit self-sustained cycles in behaviour, physiology and metabolism, with a period of approx. 24 hours1. Circadian clocks afford competitive selective advantages that have been observed experimentally2,3, whilst disturbance of circadian timing in humans, as seen in rotational shift work and jet-lag, carries long-term health costs4. For all organisms in which the molecular timing mechanism has been investigated, a common model has arisen, namely a transcription-translation feedback loop (TTFL). TTFL components are not, however, shared between organisms. For example, the cyanobacterial clock is modelled around three proteins: KaiA, B and C. In the fungus Neurospora crassa, a loop involving FRQ and the white-collar (WC) complex is thought to drive cellular rhythms, whilst the plant TTFL involves elements including TOC1 and CCA11,5. Furthermore, although some multicellular organisms such as Drosophila and humans possess homologous components (e.g. the PERIOD proteins), their functions appear to differ between organisms6-8. Therefore, across phylogenetic kingdoms, there are apparently no common ‘clock’ components, suggesting that daily timekeeping evolved independently within different lineages. The converse, however, could equally be true, and the primary premise of this study was therefore to test the hypothesis that circadian clocks may instead have a common ancestry.
Conservation of PRX in circadian systems
Recent studies show that the oxidation state of highly-conserved peroxiredoxin (PRX) proteins exhibit circadian oscillations in cells from humans, mice and marine algae9,10, most likely reflecting an endogenous rhythm in the generation of reactive oxygen species (ROS)10. Since virtually all living organisms possess peroxiredoxins11, we hypothesised that this marker for circadian rhythms in metabolism may be functionally conserved across all three phylogenetic domains: Archaea, Bacteria and Eukaryota. Peroxiredoxins are peroxidases, whose activity is dependent upon oxidation of a key “peroxidatic” cysteine residue (Cp) in the active site, that is absolutely conserved, as are neighbouring proline and threonine/serine residues: conforming to a Pxxx(T/S)xxCP consensus12. Critically, the catalytic cysteine can become over- or hyper-oxidised (PRX-SO2/3), rendering the peroxiredoxin catalytically inactive, but able to participate in ROS signalling and chaperone activity13. Once overoxidised, PRX can be recycled by sulfiredoxin.
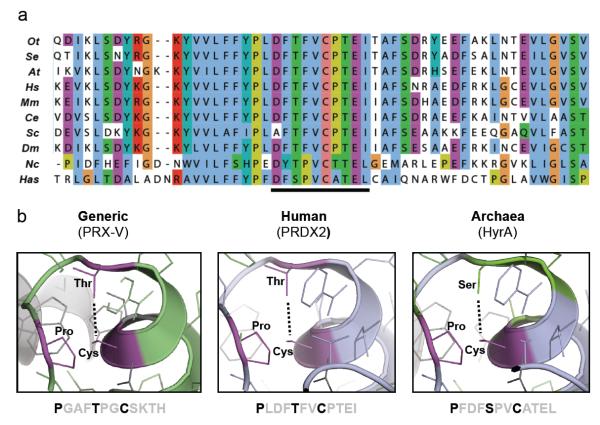
We previously characterised circadian cycles of peroxiredoxin oxidation using antiserum directed against the oxidised active site, which recognises both over- (PRX-SO2) and hyper-oxidised (PRX-SO3) forms9,10,14. To determine if this antiserum (raised against an oxidised DFTFVCPTEI peptide) could be used to assay over-/hyper-oxidation in diverse species, we performed multiple sequence alignments to compare peroxiredoxin protein sequences across a variety of circadian model organisms. This revealed a remarkable degree of conservation across all phylogenetic domains, especially within the active site (Fig. 1a, Supplementary Fig. S1 and Table S1-7). Indeed, even when we examined the structure of HyrA, the most distantly related peroxiredoxin orthologue in the archaeon Halobacterium salinarum NRC-1, we found that amino acid substitutions would not appear to perturb the geometry of the active site (Fig. 1b). Together, these findings suggested that the same antiserum could be used to probe oxidation rhythms in potentially any organism expressing a peroxiredoxin protein. This was confirmed by gene knockout and peroxide treatment in several representative organisms (Supplementary Fig. S3, S7-9).
Figure 1. The peroxiredoxin active site is highly conserved in all domains of life.
a, Multiple sequence alignment showing peroxiredoxin amino acid sequences. The highly conserved active site is underlined. Representatives shown from Eukaryota (Ot=Ostreococcustauri; At=Arabidopsis thaliana; Hs=Homo sapiens; Mm=Mus musculus; Ce=Caenorhabditis elegans; Sc=Saccharomyces cerevisiae; Dm=Drosophila melanogaster; Nc=Neurospora crassa), Bacteria (Se=Synechococcus elongatus PCC7942) and Archaea (Has=Halobacterium salinarum NRC-1). b, Critical residues in the active site of 2-Cys peroxiredoxins (in bold) are conserved in all organisms. Structures were derived from human PRX-V (PDB code: 1HD2)47 and human PRDX2 (PDB code: 1QMV)48, and modified with PyMOL to show the predicted structure for archaeal peroxiredoxin (HyrA, Genbank Accession: NP_280562.1).
Peroxiredoxin rhythms in eukaryotes
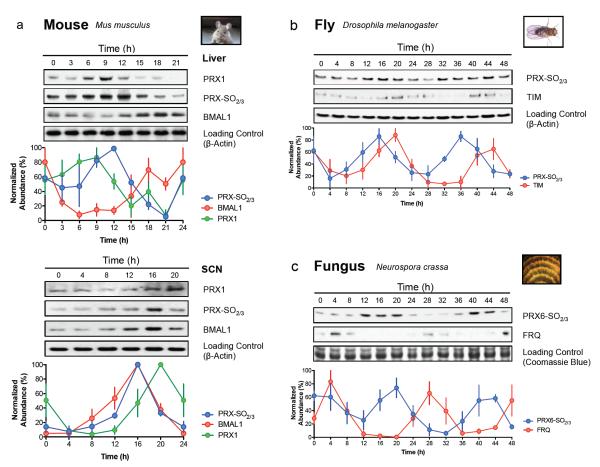
Using the PRX-SO2/3 antiserum, we first examined circadian time-courses from a range of eukaryotes under constant conditions (that is, in the absence of external timing cues). In mice, PRX-SO2/3 and total PRX1 exhibited a daily cycle in liver tissue and also in the central pacemaker, the suprachiasmatic nuclei (SCN) of the hypothalamus (Fig. 2a). Interestingly between the two tissues, peroxiredoxin oxidation rhythms were in distinct, and different, phase relationships with respect to total PRX1 and BMAL1 protein, suggesting a difference between brain and peripheral tissue (Fig. 2a), as observed previously for other clock components15.
Figure 2. Peroxiredoxin oxidation cycles are conserved in eukaryotic models of the circadian clock.
Representative immunoblots probed for oxidised/hyperoxidised 2-Cys peroxiredoxin (PRX-SO2/3 or PRX6-SO2/3) are shown for a, mouse (Mus musculus), b, fly (Drosophila melanogaster), and c, fungus (Neurospora crassa). For each model system, the organism was sampled under free-running conditions. Loading controls show either β-actin immunoblots or Coomassie Blue stained gels loaded with identical samples used for immunoblotting. Immunoblot quantification by densitometry is shown below each panel (mean ± SEM) for n=3 biological replicates. See Supplementary Fig. S2 for plant rhythms, and Supplementary Table S9 for cycle period estimates (by harmonic regression) and detailed statistics (by ANOVA).
To extend these findings beyond vertebrates, we examined peroxiredoxin rhythms in the fruit fly, Drosophila melanogaster. We pooled whole heads from insects maintained in constant darkness (DD) over two circadian cycles after they had been stably entrained to 12 hours light, 12 hours dark cycles (LD 12:12). Again, circadian oscillations in PRX-SO2/3 immunoreactivity were observed, as well as in the clock protein TIMELESS (TIM) (Fig. 2b). Similarly, seedlings from the plant Arabidopsis thaliana exhibited robust PRX-SO2/3 oscillations in free-running conditions of constant light (LL), which were also seen in the filamentous fungus Neurospora crassa, another well-characterised clock model system (Fig. 2c, Supplementary Fig. S2). Therefore, just as in tissue from ‘complex’ vertebrates, this range of ‘simpler’ eukaryotic systems displayed robust PRX oxidation cycles, with peaks tending to occur around anticipated dawn.
Peroxiredoxin rhythms in prokaryotes
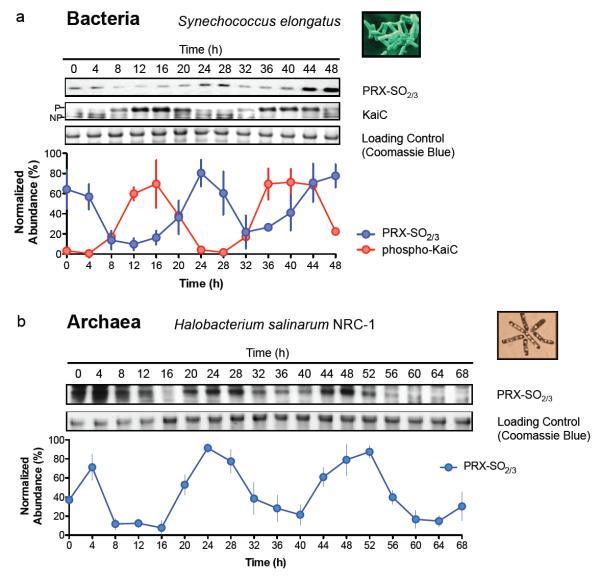
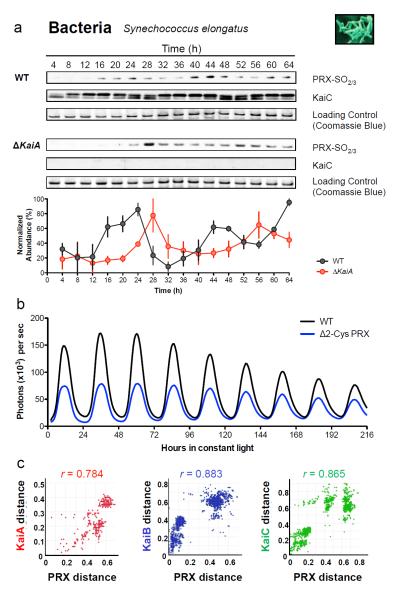
Having observed ~24 hour PRX oxidation rhythms in organisms with nucleated cells, we next sought to examine representative prokaryotes from each major domain – Bacteria and Archaea. For bacteria, we utilised the best characterised prokaryotic clock model system, Synechococcus elongatus PCC794216. The major proteins involved in the cyanobacterial clockwork are encoded by the KaiABC cluster from which, remarkably, circadian oscillations of phosphorylation can be reconstituted in vitro17. However, since all three Kai proteins are only expressed together in a small number of bacterial species, and in no known archaea18, this system cannot represent a general prokaryotic clock mechanism. We postulated that, regardless of timekeeping mechanism, the PRX oxidation cycles we observe reflect an absolutely conserved rhythmic cellular output. We tested this by assaying PRX-SO2/3 under free-running conditions (constant light, LL) for 48 hours, and observed cycles of cyanobacterial PRX oxidation, peaking later than phospho-KaiC (Fig. 3a).
Figure 3. Peroxiredoxin oxidation cycles are conserved in prokaryotic models of the circadian clock.
Representative immunoblots probed for oxidised/hyperoxidised 2-Cys peroxiredoxin (PRX-SO2/3) are shown for a, bacteria (Synechococcus elongatus PCC7942), and b, archaea (Halobacterium salinarum NRC-1). Prior to sampling under free-running conditions (constant light, LL), cyanobacteria were synchronised with a 12 hr dark pulse whereas archaea were stably entrained to 12 hr light: 12 hr dark cycles. Loading controls show Coomassie Blue stained gels loaded with identical samples used for immunoblotting. Immunoblot quantification by densitometry is shown below each panel (mean ± SEM) for n=3 biological replicates. See Supplementary Table S9 for cycle period estimates and detailed statistics. P=phosphorylated KaiC; NP=non-phosphorylated KaiC.
Finally, we extended our studies to the third phylogenetic domain, Archaea, assaying rhythms of peroxiredoxin oxidation in Halobacterium salinarum NRC-1. Whilst no clock mechanisms have been identified for any archaeon, diurnal transcriptional rhythms were recently observed in H. salinarum19, and it thus represents an ideal platform to test whether PRX oxidation constitutes a universal marker for cellular rhythms. After entraining the archaea for three cycles in LD12:12, we placed them into LL at constant temperature. We observed robust, high amplitude circadian oscillations of PRX-SO2/3 for three cycles (Fig. 3b). Together, our findings in evolutionarily diverse prokaryotes provide compelling evidence that rhythmic peroxiredoxin oxidation is a conserved circadian marker across phylogenetic domains.
Relations between PRX cycles and TTFLs
In all circadian model systems, the proposed clock mechanism revolves around a transcription-translation feedback loop (TTFL)5,20,21. Therefore, how do the metabolic rhythms observable through peroxiredoxin oxidation relate to, and interact with, the known transcriptional clockwork in different organisms? Again, we employed several model organisms, with available ‘clock’ mutants, to answer this question.
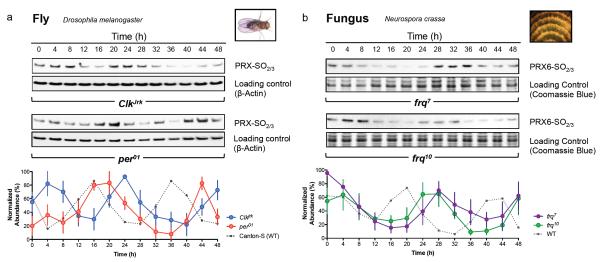
The Drosophila transcriptional clockwork is structurally similar to the mammalian TTFL, although there are some important differences22. In flies, CLOCK and CYCLE (orthologous to mammalian BMAL1) comprise the positive limb, driving oscillatory expression of PERIOD (PER) and TIMELESS (TIM). PER and TIM negatively regulate their own expression, closing the loop5,6. This circuit can be disrupted by many mutations23. Two mutants, per01 and ClkJrk, are behaviourally arrhythmic, and show non-cycling expression of circadian components, including PER and TIM24,25. In order to examine peroxiredoxin oxidation patterns in these mutants, they were entrained as described above for wild type (Canton-S) flies. We observed two circadian cycles of PRX-SO2/3 oscillation, with an altered circadian phase relative to wild type. This indicates the presence of an underlying capacity for circadian timing in both mutant strains, which was clearly perturbed by the absence of functional transcriptional feedback circuitry (Fig. 4a).
Figure 4. Peroxiredoxin oxidation cycles in circadian clock mutants.
Representative immunoblots probed for oxidised/hyperoxidised peroxiredoxin (PRX-SO2/3 or PRX6-SO2/3) are shown for a, fly (Drosophila melanogaster) and b, fungus (Neurospora crassa). For each model system, organisms were sampled under free-running conditions (constant darkness, DD). Loading controls show either β-actin immunoblots or Coomassie Blue stained gels loaded with identical samples used for immunoblotting. Immunoblot quantification by densitometry is shown below each panel (mean ± SEM) for n=3 biological replicates. See Supplementary Table S9 for cycle period estimates (by harmonic regression) and detailed statistics (by ANOVA), as well as Supplementary Fig. S10 for TIM and FRQ immunoblots for fly and fungus respectively.
To establish the wider relevance of these findings, we also examined similar mutants in the fungus Neurospora crassa. The frequency (frq) locus encodes a critical element in the TTFL of Neurospora, in addition to the PAS-containing white collar (WC) transcription factors26. In the long-period frq7 mutant27, PRX oxidation rhythms exhibited a similarly lengthened period, with an altered phase relative to rhythms in FRQ protein abundance (Fig. 4b). Deletion of the frq locus characterises the frq10 strain, and measurable markers of clock output such as its spore-forming (‘conidiation’) rhythm, are profoundly perturbed in these fungi, although apparently stochastic oscillations can re-emerge under various growth conditions27-29. Circadian rhythms of PRX oxidation were, however, clearly seen in frq10 mutants sampled in constant darkness (Fig. 4b), with a delayed phase relative to wild type (bd) fungi. This illustrates that PRX rhythms represent an alternative readout for an oscillator that persists in the absence of a FRQ-dependent clock.
We next examined the phenotypes of mutant circadian transcriptional regulators in photosynthetic eukaryotes and prokaryotes. The transcriptional clockwork of the plant Arabidopsis thaliana and the alga Ostreococcus tauri are very similar and rely on circadian oscillation of TOC1. Accordingly, overexpressing TOC1 in either species disrupts transcriptional rhythms30,31. In such strains, under LL, we observed persistent oscillations of PRX oxidation, albeit with altered amplitude and phase relative to controls (Supplementary Fig. S2,3). Finally, in cyanobacteria we assayed PRX oxidation in the arrhythmic KaiA deletion strain, AMC702 32. Strikingly, an approx. 24 hour rhythm of PRX oxidation persisted despite a functional Kai-based oscillator being absent, again in an altered phase relative to wild type (Fig. 5a). Taken together, these observations imply that metabolic rhythms remain closely aligned to transcriptional feedback mechanisms when those mechanisms are present. Critically, however, metabolic rhythms persist even when cycling clock gene transcription is abolished (summarised in Supplementary Table S9).
Figure 5. Relationships between peroxiredoxins and the cyanobacterial Kai-based oscillator.
a, Representative immunoblots for oxidised/hyperoxidised 2-Cys peroxiredoxin (PRX-SO2/3) are shown for wild-type (WT; strain AMC149) and KaiA deletion mutant (ΔKaiA; strain AMC702) cyanobacteria. Immunoblot quantification by densitometry is shown below each panel (mean ± SEM), n=3 biological replicates. See Supplementary Table S9 for detailed analysis. b, Bioluminescence traces for cyanobacteria cultures of wild-type (WT) or 2-Cys peroxiredoxin knockout strains (Δ2-Cys PRX), as reported by psbAIp::luxAB49. c, Co-evolution of cyanobacteria oscillator components and peroxiredoxin proteins. Interspecies plots correlate inter-protein distances between Kai proteins and 2-Cys peroxiredoxin41,50. r=correlation coefficient (P< 1×10−6). See Supplementary Table S8 and Supplementary Fig. S4-6 for details.
Having determined the TTFL influence on peroxiredoxin oxidation rhythms, we reciprocally tested whether rhythmic PRX oxidation is required for timekeeping, using TTFL components as markers of the clockwork. We assayed reporter bioluminescence and delayed fluorescence in mutant S. elongatus and A. thaliana lines, respectively, that were deficient in 2-CysPRX (Synechococcus Δ2-CysPRX, Accession AAP49028; Arabidopsis double mutant: Δ2-CysPRXA/Δ2-CysPRXB, Accessions NM_111995, NM_120712)33. In these mutants, circadian rhythms persisted with wild type period, albeit significantly perturbed in either phase or amplitude, relative to controls (Fig. 5b, Supplementary Fig. S9). This suggests that peroxiredoxins are not required for oscillator function in systems that possess an alternative timing mechanism, namely a transcription-translation feedback loop (TTFL). On the other hand, our findings in TTFL mutants reveal that cellular components that are required for rhythmic outputs are not essential to rhythms in redox metabolism. Given the information that we have about the above model organisms, employed commonly to study clock biology, we suggest that both PRX and TTFL components of the circadian system are important, but potentially individually dispensable for circadian rhythms at the cellular level. Moreover, the phenotypes of these mutants suggest that cellular ROS balance is important for robust clock function, as was described recently in Neurospora34.
Implications for clock evolution
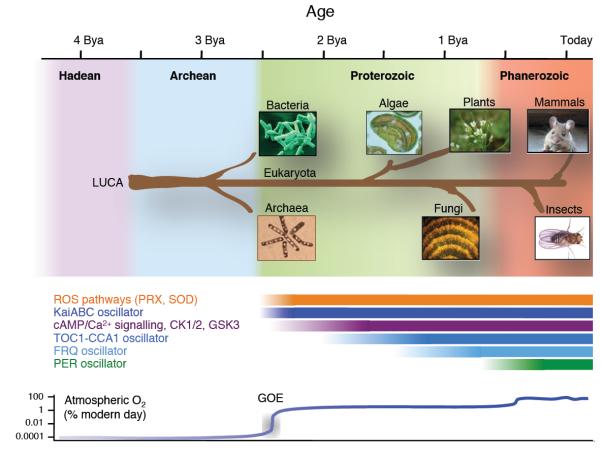
We have observed ~24 hour cycles of peroxiredoxin oxidation-reduction in all domains of life and consequently, the possibility that cellular rhythms share a common molecular origin seems increasingly plausible. Since the cellular role of peroxiredoxins principally involves removal of toxic metabolic by-products (ROS), we hypothesised that the ability to survive cycles of oxidative stress may have contributed a selective advantage from the beginnings of aerobic life.
Approximately 2.5 billion years ago, photosynthetic bacteria acquired the capacity for photo-dissociation of water, leading to the geologically rapid accumulation of molecular oxygen during the Great Oxidation Event (GOE), when anaerobic life underwent a catastrophic decline35. Evidently, organisms that survived the transition to an aerobic environment were those that respired and/or evolved oxygen. Since electron transport chains involving oxygen inevitably produce toxic superoxide anions as by-products36, during the GOE, successful organisms had to acquire ROS removal systems or were relegated to anaerobic niches35. Superoxide dismutase, which converts superoxide to hydrogen peroxide, is ubiquitous and, like PRX (the major cellular H2O2 ‘sink’), is estimated to have arisen around the time of the GOE37, the same era during which the most ancient known clock mechanism (the Kai oscillator) evolved. Importantly, we note that (1) during the GOE, rhythms of O2 production/consumption and ROS generation would have been driven by the solar cycle, as they are today38-40; (2) metabolic/oxidation rhythms appear to be present in every organism with a circadian clock, all of which are aerobes, and these rhythms persist in the absence of transcriptional cycles; (3) circadian timekeeping confers a selective advantage when it facilitates anticipation of environmental change (noxious or otherwise).
We believe that the most reasonable interpretation is that cellular metabolism in the most competitive early aerobes adapted to confer anticipation of, and resonate with, environmental cycles in energy supply and oxidative stress. We presume that the echoes of this ancient evolutionary adaptation are revealed by the conserved PRX oxidation cycles we now observe in disparate organisms, implying that over the last 2.5 billion years, ROS and metabolic pathways must have co-evolved with the cellular clockwork, and are likely interlinked. On top of this, additional cellular mechanisms appear to have been incorporated, when advantageous, as they arose over time. For example, in eukaryotes, the timekeeping contribution made by certain post-translational mechanisms (such as casein kinase) is conserved across multiple disparate organisms, whereas clock gene transcription factors are widely divergent, having evolved and been introduced more recently (see Fig. 6).
Figure 6. Phylogenetic origins of circadian oscillatory systems.
A timeline is shown at the top of the schematic (billion years ago, Bya), with the geological era illustrated with a coloured background. A schematic phylogenetic tree shows the origins of each organism studied, stemming from the last universal common ancestor (LUCA). The putative epoch over which each oscillator system has existed is illustrated by the labelled bars. ROS, reactive oxygen species; PRX, peroxiredoxin; SOD, superoxide dismutase; CK1/2, casein kinase 1 or 2; GSK3, glycogen synthase kinase 3.
If there was significant pressure for the co-evolution of metabolic/ROS pathways with cellular timekeeping systems, then evidence for this should exist in the phylogenetic trees of their component mechanisms. To substantiate this we used the Mirrortree algorithm 41 to assess the degree of co-evolution between the 2-Cys peroxiredoxin family, representing metabolism/ROS pathways, with the most ancient characterised clock mechanism: the three cyanobacterial Kai proteins. Since the Kai proteins are found exclusively in prokaryotes, we focused on these for our analysis18. All three components of the cyanobacterial oscillator appear to have co-evolved with 2-Cys peroxiredoxins, as revealed by the strong correlation between the distances of respective proteins within each phylogenetic tree (KaiA, r=0.784; KaiB, r=0.883; KaiC, r=0.865; p< 1 × 10−6 for all) (Fig. 5c and Supplementary Fig. S4). Most significantly, when evolution of KaiC (the most ancient member) was compared with other absolutely conserved protein families, the three highest correlations observed were for the other two clock components (KaiA and KaiB) and PRX (Supplementary Fig. S5 and Table S8). This implies that similarities in the evolutionary profiles of these cellular mechanisms go beyond those that would be expected simply based on time since a common ancestry because even highly conserved proteins had significantly inferior correlations to PRX (Supplementary Fig. S6 and Table S8)42. Given that Kai proteins are not found in eukaryotic systems, but metabolic rhythms such as those observed in peroxiredoxin oxidation are, our results imply that metabolic rhythms are at least as ancient as, and pre-date most, phylum-specific timekeeping mechanisms, but are intrinsically integrated with them in modern organisms.
Concluding remarks
Placed within the context of what we know about transcription factor components of the eukaryotic clockwork, a striking convergence upon oxygen and redox metabolism is apparent. It has long been recognised that oxygen-sensing PAS-domain proteins are involved in the clockwork of many eukaryotes, but the rationale behind this has remained elusive23,43-45. In light of our current findings, we speculate that sensing and responding to oxidative cycles in cellular environments could have driven the evolution of circadian rhythms, and maintained the intrinsic link between clocks and metabolism (Fig. 6). A direct prediction therefore, is that organisms that lack ROS detoxification systems will not have circadian rhythms. At least one such class of organism exists on Earth, an example being the hyperthermophilic archaea Methanopyri (NCBI Taxonomy ID: 183988). Given its distinct anoxic environmental niche and methanogenic metabolism46, there may never have been a selective evolutionary pressure to develop circadian timekeeping as we know it.
Materials and Methods
Organisms and Strains
Arabidopsis thaliana
Surface-sterilised Arabidopsis thaliana seeds (Ws) were plated on solid medium (1.2% agar plus 0.5 × Murashige & Skoog medium (Duchefa Biochemie, Leiden, the Netherlands), pH 5.8), vernalised at 4 °C for 4 days, and grown for 7 days under 12h :12h light-dark cycles under cool-white fluorescent tubes (70-100 μEm−2s−1) at 20 °C before transfer to constant light conditions at ZT0. Plantlets were sampled every 4h for 3 days by snap-freezing 15 seedlings per sample in liquid N2. Tissue was crushed in a Tissue Lyser (Eppendorf) using a 1/8 inch chrome ball (Spheric-Trafalgar Ltd., London, UK) and tissue was thawed in extraction buffer (8M Urea, 300 mMNaCl, 100 mMTris pH 7.5, 10 mM EDTA, 4% Poly(vinylpolypyrrolidone), 1% NP40, and 2x Complete protease inhibitors (Roche)) and incubated on ice for 15 minutes, vortexing every 2 minutes before centrifugation at 16,000 g for 10 minutes. Supernatants were loaded onto gels after adding SDS loading buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromphenol blue and0.125 M Tris HCl, pH approx. 6.8) and heating to 100°C for 5mins. Equal protein loading was confirmed by gel electrophoresis and Coomassie staining of gels loaded with equal volumes of lysate from each time-point in each replicate set. Delayed fluorescence was performed as reported previously51.
Drosophila melanogaster
Canton-S (CS) wild-type flies and congenic per01and ClkJrk mutants were raised on standard medium at 25°C in 12h:12h light-dark (LD 12:12) cycles and their behavioural phenotypes validated using DAM5 activity monitors (TriKinetics, Waltham, MA)52. Adult male flies were entrained for an additional three days in light boxes at 25°C prior to transfer into constant darkness (DD) at 25°C. Flies were harvested over two circadian cycles every 4 h, their heads dissociated on dry ice in the light, and these frozen at -80°C until use. Protein was extracted from n=20 fly heads per time-point in CHAPS/Urea buffer (8 M urea, 4% w/w CHAPS, 5 mM magnesium acetate, 10 mM Tris [pH 8.0]) with Complete protease inhibitors (Roche)53. Samples were ground in a 1.5ml microfuge tube with a miniature pestle, and then protein assays (Bio-rad RC DC kit) performed to correct for any variability in protein concentration. Protein-adjusted supernatants were loaded onto gels after adding appropriate volumes of 4X LDS loading buffer (Invitrogen) to bring the final concentration to 2X, and then heating to 70°C for 10 mins. Equal protein loading was confirmed by gel electrophoresis and Coomassie staining of gels loaded with equal volumes of lysate from each time-point in each replicate set.
Halobacterium salinarum NRC-1
Wild type Halobacterium salinarum NRC-1 was cultured from a single colony in Complete Medium (CM) at 37°C with shaking at 125 rpm (Innova Waterbath, New Brunswick Scientific, Edison, NJ)54. We performed experiments using n=3 different starting colonies (biological replicates). Cells were incubated under entrainment conditions for five days (12:12 LD cycle; daylight was simulated with full spectrum light at 150 μEm−2s−1) prior to sampling. Post-entrainment, samples (2 ml) were collected every 4 h for three days in continuous light, at constant cell density. The cell density was maintained by replacing a fixed volume of the culture with equivalent fresh CM (30-80 mls) every 4 h 55. Absorbance at 600nm (OD600) was recorded at each time-point, prior to dilution, to determine how much medium to add, and to also calculate cell numbers for lysate preparation. As previously described, comparative analysis with a similarly processed control culture discounted any unaccounted perturbations that could have been introduced by this periodic dilution54. Cell pellets were harvested by centrifugation at 1600 rcf for 2 min, decanted, flash-frozen in liquid N2 and stored at −80° until protein extraction. Cell pellets were lysed with 2X denaturing LDS sample buffer (Invitrogen) without reducing agent. Total protein was normalised by the addition of 2X LDS sample buffer on the basis of cell numbers in each sample (determined from OD600 measurements). Lysates were then passed through a 27g x ½ inch needle by syringing (to reduce viscosity of the samples), prior to heating to 70°C for 10 mins to fully denature proteins before loading on gels. Equal protein loading was confirmed by gel electrophoresis and Coomassie staining of gels loaded with equal volumes of lysate from each time-point in each replicate set.
Mus musculus
All animal experimentation was licensed by the Home Office under the Animals (Scientific Procedures) Act 1986, with Local Ethical Review by the Medical Research Council and the University of Cambridge.
For free-run liver experiments
Liver tissue was harvested from n=4 adult male C57Bl/6 mice once every 3 h on the second cycle after transfer from 12L:12DR to DR:DR (L, light [220 μW cm−2] and DR, dim red light [< 5 μW cm−2]) and immediately frozen and then stored at -80°C prior to use. Prior to sampling, animals were stably entrained to a 12 h L: 12 h DR cycle for 3-4 weeks. Lysates were prepared in denaturing 2X LDS sample buffer (Invitrogen) with 1:10 β-mercaptoethanol to a final protein concentration 2 μg/ul, and heated to 70°C for 10 mins prior to loading on gels. 10 μg protein per lane was loaded for immunoblotting.
For SCN organotypic slice culture experiments
Wild-type, ~ 10 day old pups on a PER2:LUC, C57BL/6 genetic background56 were used for experiments. Brains were removed from pups and sectioned at 300 μm with a McIlwain ‘Tissue Chopper’. Slices were sorted and trimmed to contain principally SCN tissue and placed onto a Millipore membrane insert (PICMORG) for culture at 37°C in 5% CO2 as described previously57. Slices were transferred to 1.1 ml HEPES buffered medium with 100 μM beetle luciferin (Promega)58 in a glass-bottomed Petri dish sealed with a coverslip and vacuum grease. Total bioluminescence was recorded with Hamamatsu photomultiplier tube assemblies housed in a light-tight 37 °C incubator and recordings were expressed as counts per second integrated over 6 min sample bins59. SCN slices were harvested “around the clock” every 4 h according to the phase of the PER2:LUC bioluminescence cycle. CT0 was operationally defined as the nadir in bioluminescence signal, and CT12 was taken to be at the peak. Individual SCN slices (n=3 per time-point) were immersed in 50 μl 2X LDS sample buffer (Invitrogen), and heated to 70°C for 10 mins prior to loading on gels. 10 μl of lysate per lane was loaded for immunoblotting. Equal protein loading was confirmed by gel electrophoresis and Coomassie staining of gels loaded with equal volumes of lysate from each time-point in each replicate set.
Neurospora crassa
We used the following strains: ‘wild type’ (bd, matA FGSC#1858) ,frq10(bd;frq10, matA FGSC#7490), frq7(bd;frq7, matA FGSC#4898) and PRX-KO (NCU06031, matA FGSC#20012). All strains were maintained on Vogel’s minimal media with 1.5% sucrose as a carbon source. Strain manipulation and growth media followed standard procedures60. For circadian experiments, cultures were initiated by inoculating 106 conidia in 25 ml of 1X Vogel’s medium containing 2% glucose, 0.5% arginine, 10 ngml−1 biotin and 0.2% Tween 80. Plates were incubated under constant light for 36 h at 30°C. Disks (1.2 cm diameter) were cut from the cohesive mycelial pad and three disks were placed in each of a series of 50 ml tubes (or 100 ml flasks) containing 30 ml (or 50 ml) of 1X Vogel’s medium with 0.03% glucose, 0.05% arginine and 10 ngml−1 biotin61. These were incubated at 25 °C under constant light for at least 2 h before staggered transfers to constant darkness (25°C) with shaking at 150 rpm. Mycelia were then harvested at 4 h intervals, dried on paper, frozen in liquid nitrogen, and stored at -80°C. Harvests were consolidated over 8 h to control for development/age of the tissue. All manipulations in the dark were performed under safe red light. For immunoblot analysis, tissue was ground in liquid nitrogen with a mortar and pestle and suspended in ice-cold extraction buffer (50 mM HEPES [pH 7.4], 137 mMNaCl, 10% glycerol, 5 mM EDTA containing 50 μgml−1 phenylmethyl-sulphonylfluoride (PMSF), 3 μgml−1 leupeptin and 3 μgml−1 pepstatin A62 and a phosphatase inhibitors cocktail [PhosSTOP, Roche]) at a ratio of 0.5 ml of buffer per 0.5 ml of mycelia powder. For immunoblotting, an antiserum directed against human PRX6 (1-Cys) was employed63 (at 1:5000 dilution) since no typical 2-Cys PRXs are annotated in the N. crassa genome to date, and thus antibody specificity could not otherwise be assured. Equal protein loading was confirmed by gel electrophoresis and Coomassie staining of gels loaded with equal volumes of lysate from each time-point in each replicate set.
Ostreococcus tauri
Ostreococcus tauri cells were cultured as previously described64 and entrained in a 12:12 light-dark cycle of blue light (17.5 μEm−2s−1, Lee lighting filter 724) at a constant temperature of 20 °C. Cultures of the arrhythmic TSL8 line (TOC1-overexpressing) and its parent line CCA1-LUC65 were transferred into constant light at ZT0 and sampled every 4h for three days. 5 ml of cells were chilled on ice and pelleted at 4,500 g at 4 °C for 10 minutes. The resulting pellet was re-suspended in 50 μl sea water64, and cells were lysed by adding 50 μl of 2X extraction buffer (Sigma-Aldrich, LUC-1 kit) and then 100 μl SDS loading buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromphenol blue and 0.125 M TrisHCl, pH approx. 6.8) whilst vortexing vigorously. Samples were heated to 100°C for 5 mins to denature proteins before loading on gels. Equal protein loading was confirmed by gel electrophoresis and Coomassie staining of gels loaded with equal volumes of lysate from each time-point in each replicate set. For H2O2 treatment, cells were incubated for 30 minutes at the stated concentrations diluted in artificial sea water under normal culture conditions prior to lysis (described above).
Synechococcus elongatus
The cyanobacterial strains used were Synechococcus elongatus PCC7942 wild type (AMC149) and ΔKaiA mutant (AMC702). Cells were grown with aeration in constant light (LL) of 100 μE s−1 m−2 at 30 °C until the optical density at 730 nm (OD730) of culture reached 0.3. The OD730 was maintained between 0.27 and 0.45 by dilution with fresh BG-11 medium66,67. The culture was exposed to 12 h of constant darkness (DD) to synchronize the circadian clock, and then returned to LL. At 4 h intervals under LL, cells were harvested, immediately frozen, and stored at −80 °C. Samples were prepared for immunoblotting as described previously68. Equal protein loading was confirmed by gel electrophoresis and Coomassie staining of gels loaded with equal volumes of lysate from each time-point in each replicate set. A single knockout for the 2-Cys PRX gene69 was generated by inserting an expression cassette for the kanamycin resistance gene into the EcoRI site near the N-terminal coding region of the gene. For bioluminescence recordings, psbAIp::luxAB was used as a reporter, using previously described protocols70. The period of all the PRX knockout strain showed no significant difference to that of WT. For H2O2 treatment, cells were incubated for 30 minutes at 1 mM H2O2 vs. vehicle prior to cell collection.
Gel Electrophoresis and Western Blotting
We used NuPAGE Novex 4–12% Bis-Tris gradient gels (Life Technologies), and ran them using the manufacturer’s protocol with a non-reducing MES SDS buffer system, allowing characterization of proteins between 10–260 kDa64,71. Protein transfer to nitrocellulose for blotting was performed using the iBlot system (Life Technologies), with a standard (P3, 7 min) protocol. Nitrocellulose was then washed briefly, and then blocked for 30 min in 0.5% w/w BSA/non-fat dried milk (Marvel) in Tris buffered saline/0.05% Tween-20 (TBST). After three brief washes in TBST, membranes were incubated in antibody diluted in blocking buffer (0.5% milk/BSA) overnight at 4 °C. The following day, membranes were washed for 5 min three times (in TBST) and then incubated with 1:10,000 HRP-conjugated secondary antibody (Sigma-Aldrich) for 30 min. Four more 10-min washes were then performed before performing chemiluminescence detection using Immobilon reagent (Millipore), or ECL Plus reagent (GE Healthcare). To check protein loading was even in the gels, they were stained with Coomassie SimplyBlue (Life Technologies). Antisera against peroxiredoxins were obtained from Abcam (Cambridge, UK) and used at the following dilutions (PRX-SO2/3, ab16830 1:10,000 and PRX1, ab59538 1:2,000) in blocking buffer. Rabbit anti-Bmal1 antiserum was used at 1:2,000 in 0.5% BSA (Santa Cruz Antibodies, sc-48790). Rabbit anti-PER or anti-TIM were used at 1:10,000 dilution in blocking buffer. Anti-PER and anti-TIM were kindly provided by Ralf Stanewsky (Queen Mary College, London, UK) and Francois Rouyer (Gif-sur-Yvette, University of Paris, France) respectively. A mouse monoclonal β-actin antibody (Santa Cruz Antibodies, sc-47778) was used at 1:5,000 in 0.5% milk/BSA. Mouse anti-FRQ antibody was used at a dilution of 1:40 in blocking buffer. Anti-KaiC antiserum was used as described previously 68.
Phylogenetic analyses
We used the Mirrortree online server, using input FASTA sequences for human PRDX2 (GenBank: CAG46588.1) and compared this serially to Synechococcus elongatus PCC 7942 proteins:KaiA (GenBank: AAM82684.1), KaiB (GenBank: AAM82685.1) and KaiC (GenBank: AAM82686.1) 72. Similar analyses were performed for KaiC comparisons with other conserved bacterial proteins. Interspecies plots were generated, which contain a simplified representation of the correlation between the inter-protein distances in phylogenetic trees for each protein being compared 73.
The Mirrortree web server implements the Mirrortree algorithm for calculating the tree similarity between two protein families, which has been demonstrated to be a good predictor of the interaction or functional relationships between them72,73. Phylogenetic trees are obtained from these alignments with the neighbor-joining (NJ) algorithm implemented in ClustalW73 using bootstrap (100 repetitions) and excluding gaps for the calculation. The distance matrices are obtained by summing the branch lengths that separate each pair of proteins in the tree. Instead of calculating the complete matrices, only the proteins of organisms present in both trees are used. The similarity of trees between the two families is calculated as the correlation between their distance matrices according to the equation:
| where: | n is the number of elements of the matrices, that is, n = (N2−N)/2 |
| N is the number of common organisms | |
| Riare the elements of the first distance matrix | |
| Si is the corresponding value for the second matrix | |
| and are the mean values of Ri and Sirespectively. |
Correlation coefficients obtained for analyses of KaiC distance vs other conserved bacterial proteins (including KaiB, C and PRX2) were compared by converting them to a normally distributed metric using Fisher’s r-to-z transformation:
| where: | r is the Mirrortree correlation coefficient |
| r′ is the Fisher-transformed correlation coefficient |
Transformed coefficients (r′) were then compared to each other to generate the test statistic, z:
| where: | r1′ is the first Fisher-transformed correlation coefficient |
| r2′ is the second Fisher-transformed correlation coefficient | |
| N1is number of common organisms in the first correlation | |
| N2is number of common organisms in the second correlation |
We computed z with r1′ representing the KaiC-PRX2 correlation coefficient, obtaining negative values for higher correlations, and positive values for lower ones (see Supplementary Table S8 and Supplementary Fig. S6); p values were then computed using standard tables (http://faculty.vassar.edu/lowry/rdiff.html).
Image and statistical analysis
Coomassie-stained gel images were obtained using a Licor Odyssey system, and immunoblot films were scanned using a back-illuminated flat-bed scanner. Densitometric quantification of images was performed using NIH ImageJ software. Signal was normalised against the respective loading control for each replicate at each time-point for grouped data. Sine curve fitting was performed using Circwave software74, using an harmonic regression method, and ANOVA was also performed as an independent measure of temporal variation. Statistical comparisons between Mirrortree correlation coefficients were performed as detailed above.
Supplementary Material
Acknowledgements
This work was primarily supported by the Wellcome Trust (083643/Z/07/Z and 093734/Z/10/Z), the European Research Council (ERC Starting Grant No. 281348, MetaCLOCK), and EMBO Young Investigators Programme, as well as the MRC Centre for Obesity and Related metabolic Disorders (MRC CORD), and the NIHR Cambridge Biomedical Research Centre. CPK and MHH acknowledge EC grant EUCLOCK (No. 018741) and BBSRC grant BB/C006941/1. SynthSys is funded by BBSRC and EPSRC award BB/D019621 to AJM and others. NSB was supported by ENIGMA, U. S. Department of Energy, under contract No. DE-AC02-05CH11231, and by a grant from NIH (P50GM076547). CHJ was supported by NIH (R01GM088595, R01GM067152 and R21HL102492). MM was supported by the NWO (Dutch Science Foundation VICI award and Open Programma), the University of Groningen (Rosalind Franklin Fellowship Program). We thank M. Jain, G. O’Neill and J. Chambers for helpful discussion about the manuscript, and S. G. Rhee, F. Rouyer and R. Stanewsky for the kind gifts of antisera.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions A.B.R. and J.S.O. conceived and designed the experiments, and wrote the manuscript. R.S.E, E.W.G., G.v.O., M.O., X.Q., Y.X., Y.Z., M.P., U.K.V., K.A.F. and E.S.M. performed experiments. M.H.H., N.S.B., C.H.J., M.M., A.J.M., and C.P.K. provided reagents.
Author information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 2.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks; an experimental assessment in cyanobacteria. Curr Biol. 2004;14:1481–6. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 3.Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–3. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 4.Barger LK, Lockley SW, Rajaratnam SM, Landrigan CP. Neurobehavioral, health, and safety consequences associated with shift work in safety-sensitive professions. Curr Neurol Neurosci Rep. 2009;9:155–64. doi: 10.1007/s11910-009-0024-7. [DOI] [PubMed] [Google Scholar]
- 5.Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–48. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 6.Allada R, Emery P, Takahashi JS, Rosbash M. Stopping time: the genetics of fly and mouse circadian clocks. Annu Rev Neurosci. 2001;24:1091–119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- 7.Rosbash M. The implications of multiple circadian clock origins. PLoS Biol. 2009;7:e62. doi: 10.1371/journal.pbio.1000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178:1147–55. doi: 10.1534/genetics.107.088658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Neill JS, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–8. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall A, Karplus PA, Poole LB. Typical 2-Cys peroxiredoxins--structures, mechanisms and functions. FEBS J. 2009;276:2469–77. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–3. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 13.Barranco-Medina S, Lazaro JJ, Dietz KJ. The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett. 2009;583:1809–16. doi: 10.1016/j.febslet.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 14.Woo HA, et al. Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid. Immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. J Biol Chem. 2003;278:47361–4. doi: 10.1074/jbc.C300428200. [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. Embo J. 1997;16:6762–71. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson CH, Mori T, Xu Y. A cyanobacterial circadian clockwork. Curr Biol. 2008;18:R816–R825. doi: 10.1016/j.cub.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–5. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 18.Dvornyk V, Vinogradova O, Nevo E. Origin and evolution of circadian clock genes in prokaryotes. Proc Natl Acad Sci U S A. 2003;100:2495–500. doi: 10.1073/pnas.0130099100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitehead K, Pan M, Masumura K, Bonneau R, Baliga NS. Diurnally entrained anticipatory behavior in archaea. PLoS One. 2009;4:e5485. doi: 10.1371/journal.pone.0005485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakin-Thomas PL. Transcriptional feedback oscillators: maybe, maybe not. J Biol Rhythms. 2006;21:83–92. doi: 10.1177/0748730405286102. [DOI] [PubMed] [Google Scholar]
- 21.Reddy AB, O’Neill JS. Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 2009 doi: 10.1016/j.tcb.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–15. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 23.Young MW. The molecular control of circadian behavioral rhythms and their entrainment in Drosophila. Annu Rev Biochem. 1998;67:135–52. doi: 10.1146/annurev.biochem.67.1.135. [DOI] [PubMed] [Google Scholar]
- 24.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 25.Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–22. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Dunlap JC, Loros JJ. How fungi keep time: circadian system in Neurospora and other fungi. Curr Opin Microbiol. 2006;9:579–87. doi: 10.1016/j.mib.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Aronson BD, Johnson KA, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 28.Granshaw T, Tsukamoto M, Brody S. Circadian rhythms in Neurospora crassa: farnesol or geraniol allow expression of rhythmicity in the otherwise arrhythmic strains frq10, wc-1, and wc-2. J Biol Rhythms. 2003;18:287–96. doi: 10.1177/0748730403255934. [DOI] [PubMed] [Google Scholar]
- 29.Lakin-Thomas PL, Brody S. Circadian rhythms in microorganisms: new complexities. Annu Rev Microbiol. 2004;58:489–519. doi: 10.1146/annurev.micro.58.030603.123744. [DOI] [PubMed] [Google Scholar]
- 30.Corellou F, et al. Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus. Plant Cell. 2009;21:3436–49. doi: 10.1105/tpc.109.068825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mas P, Alabadi D, Yanovsky MJ, Oyama T, Kay SA. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell. 2003;15:223–36. doi: 10.1105/tpc.006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ditty JL, Canales SR, Anderson BE, Williams SB, Golden SS. Stability of the Synechococcus elongatus PCC 7942 circadian clock under directed anti-phase expression of the kai genes. Microbiology. 2005;151:2605–13. doi: 10.1099/mic.0.28030-0. [DOI] [PubMed] [Google Scholar]
- 33.Pulido P, et al. Functional analysis of the pathways for 2-Cys peroxiredoxin reduction in Arabidopsis thaliana chloroplasts. J Exp Bot. 2010;61:4043–54. doi: 10.1093/jxb/erq218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida Y, Iigusa H, Wang N, Hasunuma K. Cross-Talk between the Cellular Redox State and the Circadian System in Neurospora. PLoS One. 2011;6:e28227. doi: 10.1371/journal.pone.0028227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M, et al. A universal molecular clock of protein folds and its power in tracing the early history of aerobic metabolism and planet oxygenation. Mol Biol Evol. 2011;28:567–82. doi: 10.1093/molbev/msq232. [DOI] [PubMed] [Google Scholar]
- 36.Nathan C, Ding A. SnapShot: Reactive Oxygen Intermediates (ROI) Cell. 2010;140:951–951. e2. doi: 10.1016/j.cell.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33:337–49. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 38.Mulholland PJ, Houser JN, Maloney KO. Stream diurnal dissolved oxygen profiles as indicators of in-stream metabolism and disturbance effects: Fort Benning as a case study. Ecological Indicators. 2005;5:243–252. [Google Scholar]
- 39.Venkiteswaran JJ, Wassenaar LI, Schiff SL. Dynamics of dissolved oxygen isotopic ratios: a transient model to quantify primary production, community respiration, and air-water exchange in aquatic ecosystems. Oecologia. 2007;153:385–398. doi: 10.1007/s00442-007-0744-9. [DOI] [PubMed] [Google Scholar]
- 40.Bamforth SS. Diurnal Changes in Shallow Aquatic Habitats. Limnology and Oceanography. 1962;7:348–353. [Google Scholar]
- 41.Ochoa D, Pazos F. Studying the co-evolution of protein families with the Mirrortree web server. Bioinformatics. 2010;26:1370–1. doi: 10.1093/bioinformatics/btq137. [DOI] [PubMed] [Google Scholar]
- 42.Peixoto AA, Campesan S, Costa R, Kyriacou CP. Molecular evolution of a repetitive region within the per gene of Drosophila. Mol Biol Evol. 1993;10:127–39. doi: 10.1093/oxfordjournals.molbev.a039993. [DOI] [PubMed] [Google Scholar]
- 43.McIntosh BE, Hogenesch JB, Bradfield CA. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu Rev Physiol. 2010;72:625–45. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- 44.Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–31. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 45.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–4. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 46.Shima S, Thauer RK, Ermler U. Hyperthermophilic and salt-dependent formyltransferase from Methanopyrus kandleri. Biochem Soc Trans. 2004;32:269–72. doi: 10.1042/bst0320269. [DOI] [PubMed] [Google Scholar]
- 47.Declercq JP, et al. Crystal structure of human peroxiredoxin 5, a novel type of mammalian peroxiredoxin at 1.5 A resolution. J Mol Biol. 2001;311:751–9. doi: 10.1006/jmbi.2001.4853. [DOI] [PubMed] [Google Scholar]
- 48.Schroder E, et al. Crystal structure of decameric 2-Cys peroxiredoxin from human erythrocytes at 1.7 A resolution. Structure. 2000;8:605–15. doi: 10.1016/s0969-2126(00)00147-7. [DOI] [PubMed] [Google Scholar]
- 49.Xu Y, Mori T, Johnson CH. Cyanobacterial circadian clockwork: roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. Embo J. 2003;22:2117–26. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pazos F, Valencia A. Similarity of phylogenetic trees as indicator of protein-protein interaction. Protein Eng. 2001;14:609–14. doi: 10.1093/protein/14.9.609. [DOI] [PubMed] [Google Scholar]
References
- 51.Gould PD, et al. Delayed fluorescence as a universal tool for the measurement of circadian rhythms in higher plants. Plant J. 2009;58:893–901. doi: 10.1111/j.1365-313X.2009.03819.x. [DOI] [PubMed] [Google Scholar]
- 52.Rosato E, Kyriacou CP. Analysis of locomotor activity rhythms in Drosophila. Nat Protoc. 2006;1:559–68. doi: 10.1038/nprot.2006.79. [DOI] [PubMed] [Google Scholar]
- 53.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–15. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 54.Whitehead K, Pan M, Masumura K, Bonneau R, Baliga NS. Diurnally entrained anticipatory behavior in archaea. PLoS One. 2009;4:e5485. doi: 10.1371/journal.pone.0005485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mori T, Binder B, Johnson CH. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci U S A. 1996;93:10183–8. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoo SH, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–46. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.House SB, Thomas A, Kusano K, Gainer H. Stationary organotypic cultures of oxytocin and vasopressin magnocellular neurons from rat and mouse hypothalamus. Journal of Neuroendocrinology. 1998;10:849–861. doi: 10.1046/j.1365-2826.1998.00272.x. [DOI] [PubMed] [Google Scholar]
- 58.Hastings MH, Reddy AB, McMahon DG, Maywood ES. Analysis of circadian mechanisms in the suprachiasmatic nucleus by transgenesis and biolistic transfection. Methods Enzymol. 2005;393:579–92. doi: 10.1016/S0076-6879(05)93030-9. [DOI] [PubMed] [Google Scholar]
- 59.Maywood ES, et al. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 60.Davis RH. Neurospora: Contributions of a Model Organism. Oxford University Press; New York: 2000. [Google Scholar]
- 61.Merrow M, Brunner M, Roenneberg T. Assignment of circadian function for the Neurospora clock gene frequency. Nature. 1999;399:584–6. doi: 10.1038/21190. [DOI] [PubMed] [Google Scholar]
- 62.Olmedo M, et al. A role in the regulation of transcription by light for RCO-1 and RCM-1, the Neurospora homologs of the yeast Tup1-Ssn6 repressor. Fungal Genet Biol. 2010;47:939–52. doi: 10.1016/j.fgb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 63.Woo HA, Rhee SG. Immunoblot Detection of Proteins That Contain Cysteine Sulfinic or Sulfonic Acids with Antibodies Specific for the Hyperoxidized Cysteine-Containing Sequence. Mary Ann Liebert, Inc.; 2010. p. 275. [DOI] [PubMed] [Google Scholar]
- 64.O’Neill JS, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–8. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corellou F, et al. Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus. Plant Cell. 2009;21:3436–49. doi: 10.1105/tpc.109.068825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ishiura M, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 67.Qin X, et al. Intermolecular associations determine the dynamics of the circadian KaiABC oscillator. Proc Natl Acad Sci U S A. 2010;107:14805–10. doi: 10.1073/pnas.1002119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu Y, et al. Intramolecular regulation of phosphorylation status of the circadian clock protein KaiC. PLoS One. 2009;4:e7509. doi: 10.1371/journal.pone.0007509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stork T, Laxa M, Dietz MS, Dietz KJ. Functional characterisation of the peroxiredoxin gene family members of Synechococcus elongatus PCC 7942. Arch Microbiol. 2009;191:141–51. doi: 10.1007/s00203-008-0438-7. [DOI] [PubMed] [Google Scholar]
- 70.Xu Y, Mori T, Johnson CH. Cyanobacterial circadian clockwork: roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. Embo J. 2003;22:2117–26. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ochoa D, Pazos F. Studying the co-evolution of protein families with the Mirrortree web server. Bioinformatics. 2010;26:1370–1. doi: 10.1093/bioinformatics/btq137. [DOI] [PubMed] [Google Scholar]
- 73.Pazos F, Valencia A. Similarity of phylogenetic trees as indicator of protein-protein interaction. Protein Eng. 2001;14:609–14. doi: 10.1093/protein/14.9.609. [DOI] [PubMed] [Google Scholar]
- 74.Oster H, Damerow S, Hut RA, Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms. 2006;21:350–61. doi: 10.1177/0748730406293053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.