Abstract
Murine norovirus (MNV), currently the only norovirus that efficiently replicates in cell culture, is often used as a model system to understand the molecular mechanisms of norovirus replication. MNV is a single stranded positive sense RNA virus of the Caliciviridae family. Replication of MNV involves the synthesis of both full length genomic and sub-genomic RNAs. The replication of these RNAs involves the synthesis of negative strand intermediates. To understand the molecular mechanism of RNA replication and the role of viral and host factors in virus replication, it is necessary to quantify accurately both positive and negative sense RNA molecules of the viral RNA during replication. Increasingly, strand specific reverse transcription-quantitative PCR (RT-qPCR) is becoming the method of choice for this kind of quantitation. Many strategies have been developed to avoid the false priming property of reverse transcriptase and to amplify specifically one strand in the presence of excess opposite strand. In this report, a SYBR based, real time RT-qPCR assay was developed to detect and quantify specifically the negative and the positive sense RNAs of MNV genomic RNA. This assay is based on using a tagged RT primer containing a non-viral sequence at the 5′ end of the viral strand specific sequence. This non-viral sequence is then used to amplify selectively the strand specific cDNA at the PCR stage. This assay can be used for a range of MNV strains including MNV-1 and 3, as these are now widely accepted for use in molecular studies. The specificity of this assay was determined by its ability to quantify one strand in the presence of up to 106 copies of competitor opposite sense RNA. Using this assay, the production of both strands of MNV-1 RNA was monitored during viral single step growth curve.
Keywords: Reverse transcription-quantitative PCR (RT-qPCR), strand-specific, norovirus
1. Introduction
Murine norovirus (MNV) is currently used as a model for studying human noroviruses (HuNV) due to its genetic similarity and the fact that unlike HuNV, MNV can be propagated in cell culture, has reverse genetics systems and a small genetically defined animal model readily available (Chaudhry, Skinner, and Goodfellow, 2007; Karst et al., 2003; Vashist et al., 2009; Ward et al., 2007; Wobus et al., 2004; Yunus et al., 2010). MNV is a positive sense single stranded RNA virus belonging to the Caliciviridae family. The RNA genome is approximately 7.4 kb long with a poly A tail at the 3′ end and is covalently attached to the viral protein VPg at the 5′ end (Chaudhry et al., 2006). To start the infectious cycle, after entering the host cell, viral genomic RNA acts as a template for VPg-dependent translation, producing the various viral proteins required for replication (Chaudhry et al., 2006; Daughenbaugh et al., 2003; Daughenbaugh, Wobus, and Hardy, 2006; Goodfellow et al., 2005; Hardy, 2005). The genome also acts as a template for genome replication via the synthesis of a negative strand RNA intermediate. The synthesis of negative strand RNA acts as a marker for the active replication of positive strand RNA viruses in host cells and tissues.
RT-qPCR is now widely accepted as the method of choice for the quantitation of viral RNA. Many RT-qPCR assays for MNV have been developed to estimate the viral load in infected tissues, cells or stool samples (Baert et al., 2008; Bailey et al., 2010; Karst et al., 2003; Stals et al., 2009). A significant advantage of RT-qPCR is its high sensitivity of detection and reproducibility. However, standard RT-qPCR assays do not provide strand specificity to the detection and hence do not determine the absolute quantity of viral RNA copies in a given sample because of the presence of both positive and negative strands of RNA. The lack of specificity has been attributed to the false priming property of reverse transcriptase that can occur due to false annealing of the primers or even in the absence of any primers (Bessaud et al., 2008; Kawakami et al., 2011; Lanford et al., 1994).
Strand specific detection of both positive and negative sense RNAs can be used as a marker for active viral replication and also provides a tool to understand the molecular mechanism of viral RNA replication. Several studies have recently reported the development of strand specific detection and quantitation of RNA from a number of viral systems (Bessaud et al., 2008; Horsington and Zhang, 2007; Kawakami et al., 2011; Komurian-Pradel et al., 2004; Plaskon, Adelman, and Myles, 2009; Purcell et al., 2006). In these studies, various strategies have been employed to grant specificity to the reverse transcription. These include: a) using a tagged RT primer that contains a non-viral sequence at the 5′ end of a viral strand specific sequence, b) use of reverse transcriptase enzymes with improved properties that minimise false priming, c) performing cDNA synthesis at a higher temperature and d) preventing the carry-over of RT primer and enzyme to the qPCR reaction. It is also important to remove DNA templates from in vitro transcripts used as standards using multiple rounds of DNase treatment and purification.
This study reports the development of a strand specific RT-qPCR assay for detection of both of positive and negative sense RNA molecules of the MNV genome. Furthermore this assay can be used to detect several strains of MNV including MNV-1 and MNV-3, currently widely used for many molecular studies.
2. Material and Methods
2.1 Cell culture and cell lines
The murine microglial BV2 cell line (Blasi et al., 1990) was kindly provided by Jennifer Pocock (University College London). BV2 cells were maintained in Dulbecco’s modified eagle’s medium (DMEM) (Gibco, New York, USA) supplemented with 10% fetal calve serum (FCS) (Biosera, East Sussex, UK), 2 mM L-glutamine, 0.075% sodium bicarbonate (both Gibco) and antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin).
2.2 Generation of RNA standards representing positive and negative strands of genomic RNA
The standards representing positive and negative strands of genomic RNA were synthesized using T7 RNA polymerase. The DNA representing both RNAs of MNV-1 was PCR amplified from an infectious clone pT7:MNV 3′Rz already described (Chaudhry et al., 2007) using specific primers with the forward primer containing a T7 promoter sequence at the 5′ end (Table 1a). The PCR products were gel purified and used for in vitro transcription reactions at 37°C for 4 hours. DNase I was then added to the reaction in presence of 10X DNase buffer and incubated at 37°C for 30 minutes. The RNA transcript was purified over poly-acrylamide gel; phenol chloroform extracted, precipitated using ethanol and dissolved in RNA storage solution (Applied Biosystems, California, USA). The RNA was then quantitated using spectrophotometry and the concentration of each RNA strand was adjusted to values equivalent to 1011 copies per μl, aliquoted and stored at −20°C for further use.
Table 1.
A) Primers used for production template for in vitro transcription of standard RNAs. The position of primers binding to MNV-1 strain is shown. The sequence of the T7 RNA polymerase promoter sequence in forward primer is underlined. B) Primers used for reverse transcription and qPCRs. The unique non-viral tag sequence in the RT primer sequence is underlined. The asterisk highlights that the genomes positions are with reference to the MNV-1 CW.1 genomic RNA sequence.
| A. Primers used for generation of standard RNAs: | |||
|---|---|---|---|
| Standard RNA | Name | Sequence 5′ -3′ | Position* |
| G positive | Gpos-F | GCGTAATACGACTCACTATAGGGCTTTTGGCCTCACCTCTG | 1085-1104 |
| Gpos-R | CCAAGATGAAATTGATGTGGCTGTAATCGGGCC | 1954-1986 | |
| G negative | Gneg-F | GCGTAATACGACTCACTATAGGGTGCCAAGATGAAATTGATG | 1970-1990 |
| Gneg-R | GCTTTTGGCCTCACCTCTG | 1086-1104 | |
| B. Primers used for RT-qPCR: | ||||
|---|---|---|---|---|
| RNA | Name | Sequence 5′ -3′ | Position* | |
| G positive | RT | TposGpos |
CGGGAAGGCGACTGGAGTGCCCAAACATCTTTCCCT TGTTC |
1760-1779 |
| qPCR - F | Tpos | CGGGAAGGCGACTGGAGTGCC | Non-viral | |
| qPCR -R | Gneg | TGGACAACGTGGTGAAGGAT | 1678-1697 | |
| G negative | RT | TnegGneg |
GGCCGTCATGGTGGCGAATAATGGACAACGTGGTG AAGGAT |
1678-1697 |
| qPCR - F | Tneg | GGCCGTCATGGTGGCGAATAA | Non-viral | |
| qPCR -R | Gpos | CAAACATCTTTCCCTTGTTC | 1760-1779 | |
2.3 Growth kinetic samples
BV-2 cells were seeded at 3 ×105 cells per well of a 24-well plate and subsequently infected with the MNV-1 virus at multiplicity of infection (MOI) of 5 TCID50 per cell. At given time points the cells were collected for RNA isolation and washed with PBS. Total RNA was isolated using Genelute Total RNA isolation kit (Sigma, Missouri, USA) as per the manufacturer’s protocol. The concentration of RNA isolated from each time point was determined using spectrophotometry and adjusted to 50 ng/μl for cDNA synthesis.
2.4 cDNA synthesis
A series of pre-determined copies of standard RNA or 100ng of total RNA from each time point of infection were used for reverse transcription using Superscript-III enzyme (Invitrogen, New York, USA). The reverse transcription primer (RT-primer) contained a non-viral tag sequence attached at the 5 end of the strand specific viral sequence (see table 1b). The RT reaction contained, 5X buffer, 20mM DTT, 0.5mM dNTPs, 100nM RT-primer and 2 units of SUPERSCRIPT-III enzyme in a volume of 20 μl. The reaction was carried out at 55°C for 30 minutes, stopped by heating at 90°C for 5 minutes and then diluted to 200 μl (1:10) with nuclease free water. A previously described method was used to generate cDNA for the purpose of qPCR using non-tagged primers (Bailey et al., 2010; McFadden et al., 2011).
2.5 Real Time qPCR
Real time qPCRs were performed on a ViiA 7 real time PCR machine and analyzed using software version ViiA™v1.1 (Applied Biosystems, Californial, USA). qPCR reactions were prepared using the MESA Blue qPCRMasterMix Plus for Syber Assay (Eurogentech, Seraing, Belgium). Briefly, 2 μl of diluted cDNA was mixed with 2X Mesa blue master mix and the respective primers (see table 1b) for each strand at a final concentration of 125 nM, prior to enzyme activation by incubation at 95°C for 10 min. Reactions were then subjected to 50 cycles of 94°C, 15 sec; 58°C, 20 sec; 72°C, 20 sec. The standard curve was generated using in vitro transcribed RNA representing each strand, serially diluted from 107 to 25 copies in the presence of 10ng yeast total RNA as carrier. The viral genome copy numbers were determined by interpolation of the standard curve for the respective strand of RNA. The specificity of this assay was determined by including cDNA representing 106 copies of opposite strand in all the samples for the standard curve. Each sample and standard curve was run in triplicate to ensure reproducibility.
2.6 Primer Design
A sequence alignment of 38 strains of MNV was performed using AlignX (Invitrogen, new York, USA). This alignment was used to design primers for qPCR (Gpos and Gneg) using the software Visual OMP (DNA Software Inc). The primers used in this study are presented in table 1b. The list of MNV strains containing exact sequence match with these primer pairs include DQ223042, EU004654, EU004655, EU004656, EU004657, EU004460, EU004462, EU004658, EU004659, EU004661, EU004662, AY228235. This primer pair can also be used for many other strains of MNV that have 1 mismatched base in the primer sets. Two different non-viral tag sequences were added at the 5′ end of either primers to make RT primers for respective strands.
3. Results
3.1 Generation of standard RNAs representing MNV positive and negative sense genomic RNA
Standard RNA representing 902 bases of the positive and 905 bases of the negative strand of genomic RNA were synthesized by in vitro transcription as described in materials and methods. The RNA region was selected to include the qPCR primer binding sites and to include any regions predicted to fold into RNA structures in the region surrounding primer-binding sites (Figure 1). Including the complexity of RNA folding in the surrounding sequence was chosen to determine primer-binding to standard RNA in a near native state of viral RNA i.e. where primer accessibility may be reduced to local RNA structure. The MNV-1 infectious clone pT7:MNV 3′Rz, already described (Chaudhry et al., 2007), was used as template for PCR amplification of the region of interest. The primers used for PCR amplification are shown in table 1a. In each case, the forward primers contained a T7 RNA polymerase promoter. The purified PCR product was subjected to in vitro transcription using T7 RNA polymerase. The RNA preparation was digested twice with DNase I to ensure complete DNA removal and was extracted three times using phenol and chloroform. The RNA was then precipitated using ethanol, dissolved in RNA storage solution and quantified using spectrophotometry. The RNA was diluted to 1011 copies/μl, aliquoted and stored for later use. The integrity of RNA was checked by denaturing PAGE and single band was detected (data not shown).
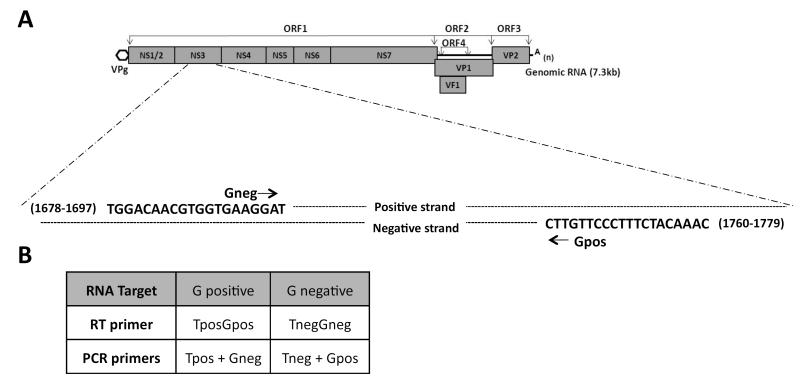
Figure 1. Schematic illustration of the positions of the primers used in this study.
The MNV genome is approximately 7.3 kbases, which encodes four open reading frames that are processed into two structural and seven non-structural proteins as depicted in the genome cartoon. The approximate positions of the PCR primers (Gpos and Gneg) used in this study are shown on the positive and negative strands of RNA. In order to generate the RT primer, a specific tag sequence (table 1b) was added to each respective primer at the 5′ end.
3.2 A conventional real-time RT-qPCR assay is unable to quantify specifically one strand of RNA in the presence of an opposite sense strand
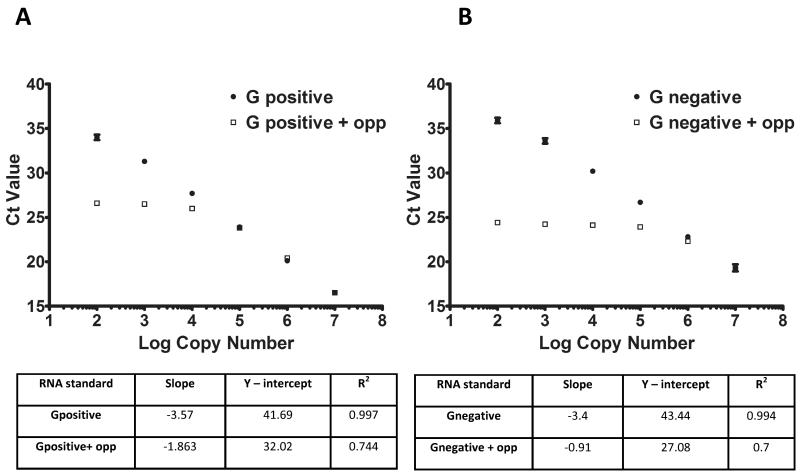
Due to the polarity of positive strand RNA virus replication at any given time during virus replication, the positive strand infectious viral RNA is present at higher levels than the negative strand. Due to this bias, the quantitation of positive strand RNA using a standard RT-qPCR does not typically pose any problem. A 2 step RT-qPCR method has been used previously to quantify the positive strand of viral genomic RNA (Bailey et al., 2010; McFadden et al., 2011). In this previous method, cDNA was generated using reverse transcription (RT) with an antisense primer that was designed to bind positive strand RNA specifically (Gpos, Figure 1). The cDNA was later amplified by qPCR using same primer and a negative strand specific primer (Gneg, Fig 1), as forward and reverse primers respectively (Table 1b). This method was able to reproducibly detect as few as 100 copies of the positive strand genomic RNA within a complex RNA mixture, consisting of RNA from the opposite strand and yeast total RNA. To examine if this assay was capable of detecting only positive or negative sense RNA genomes of MNV, the specificity and sensitivity of this assay was examined in the presence of an excess of the opposite polarity RNA. A known amount of either the positive or negative sense standard RNA was serially diluted 10 fold in the presence or absence of 106 copies of the opposite strand. Strand specific primers (Gpos for positive strand and Gneg for negative strand) were used for the RT of each RNA dilution to obtain strand specific cDNA synthesis of either strand. After a 10-fold dilution, the cDNAs were then amplified by qPCR using the primers Gpos/Gneg. The standard curve was plotted with RNA copy number on the X axis and Ct values on the Y axis (Figure 2). Here presented results indicate that in the absence of an excess of opposite polarity RNA, the standard curve has a linear range with a detection limit of at least 100 copies. However, in the presence of an excess of the opposite sense RNA, the standard curve does not remain linear and the parameters of an efficient qPCR (slope and R2) deviate from the expected values (Figure 2). This suggests that, due to false priming, the presence of high concentrations of the opposite strand hinders the accurate quantitation of either strand.
Figure 2. Standard RT-qPCR does not display strand specificity.
The specificity of normal RT-qPCR was tested by generating standard curves of either positive (Panel A) or negative strand (Panel B) in presence and absence of a fixed amount of in vitro transcribed opposite polarity RNA. The experiment was performed in triplicate and mean Ct value with standard deviation were plotted against RNA absolute copy number. The correlation coefficients of the qPCRs are presented in a table below each curve.
3.3 Strand Specificity cannot be obtained by a superior RT enzyme and high temperature
It is well documented in the literature that false priming is a common problem associated with quantitation of strand specific RNA (Kawakami et al., 2011; Moison, Arimondo, and Guieysse-Peugeot, 2011; Tuiskunen et al., 2010). It has been demonstrated that RT enzymes can cause false priming, which affects detection of low copy number of RNA in presence of an excess of the opposite sense RNAs (Moison et al., 2011). Previous reports suggest that nature of the RT enzyme and incubation temperature can be used to grant some specificity. To test if using an enzyme with increased activity at higher incubation temperatures improved the specificity of the previously established method, these conditions were tested on 108 copies of positive and negative strand standard RNAs. Both standards were amplified using Gpos as well as Gneg as RT primers using Superscript-III instead of Superscript-II, as used in figure 2. The enzyme Superscript-III is known to function at increased incubation temperatures, minimising false priming and potentially increasing the specificity of the assay (Moison et al., 2011), therefore the RT temperature was raised to 55°C and the reaction time reduced to 30 minutes. After RT, the cDNAs were used for qPCR using the primers Gpos and Gneg to determine the Ct value (Table 2). The Ct values obtained from either the positive or the negative sense control RNAs were similar, irrespective of which strand was being detected, confirming that the modified RT conditions did not improve specificity.
Table 2.
| RNA | RT primer | PCR primers | Ct Valuea) | St. Dev. |
|---|---|---|---|---|
| G positive | Gpos | Gneg + Gpos | 14.9 | 0.1 |
| Gneg | Gneg + Gpos | 16.1 | 0.1 | |
| G negative | Gpos | Gneg + Gpos | 18.0 | 0.5 |
| Gneg | Gneg + Gpos | 17.0 | 0.1 |
CT values obtained by RT-qPCR using untagged RT- primer. 108 copies of the positive or negative strand control RNA were subjected to RT using an untagged primer specific for both strand and amplified by qPCR using PCR primers. The respective Ct value with standard deviation of three individual replicates is displayed. The experiment was repeated at least three times.
3.4 Use of a tagged RT primers increase the specificity of the detection of in vitro transcribed RNAs
Another common strategy used to increase the strand specificity is the addition of a non-viral tag sequence to the 5′ end of each strand specific RT primer, such that the tag sequence can then be used as one of the primers in PCR amplification. Since the tag sequence is not present in the viral RNA, the PCR will only amplify the cDNA synthesised by the specific tagged RT primer. Thus, even if cDNA synthesis occurs due to false priming, it would not be able to participate as a template in the PCR and hence would not be detected. This strategy has been shown to render specificity to the RT-PCR for a number of RNA viruses, including foot and mouth disease virus and dengue virus (Escaffre, Queguiner, and Eterradossi, 2010; Horsington and Zhang, 2007; Purcell et al., 2006; Tuiskunen et al., 2010). One issue with this approach however is possible carry-over of the tagged RT primer into the PCR reaction i.e. if the RT primer is carried forward for PCR, it may amplify the non-specific cDNA produced due to false priming. To avoid carry-over of the RT primer, only the minimum amount required was included in the RT reaction. Further, the cDNA reaction was diluted to reduce the net amount of the RT primer carried through to the PCR reaction, as well as all other constituents of the RT reaction that might interfere with efficiency of the PCR.
Using this approach, two tagged RT primers were designed; TposGpos and TnegGneg, for the generation of cDNA from positive and negative strand viral RNA respectively. To test if employing tagged RT primers was sufficient to achieve strand specificity, the assay was again used to quantify 108 copies of in vitro transcribed RNAs. The primers TposGpos or TnegGneg were used as RT primers for their respective RNAs using the Superscript-III enzyme as described in the material and methods. qPCR was then performed using primers pairs that hybridised to the non-viral tag sequences and a corresponding primer (virus specific) on the opposite strand; Tpos/Gneg or Tneg/Gpos for the detection of positive and negative sense RNAs respectively (Figure 1B and Table 3). Using this approach a substantially improved specificity was observed, with an increase in the differential Ct values when detecting one strand with the opposite polarity RT qPCR assay (compare with section 3.3 and table 2); a difference in Ct of 18, equivalent to an approximately 5.5 log10 difference in copy number, was obtained when detecting 108 copies of in vitro transcribed positive strand RNA. Similarly, the amplification of the negative strand standard RNA using TnegGneg as the RT primer and qPCR using Tneg/Gpos, resulted in a Ct differential of 18.4. These data confirmed that the combination of using tagged primers during the synthesis of cDNA and other changes to the experimental setup was able to grant specificity to the detection of positive and negative sense viral RNA.
Table 3.
| RNA | RT primer | PCR primers | Ct Valuea) | St.Dev. |
|---|---|---|---|---|
| G positive | TposGpos | Tpos + Gneg | 13.6 | 0.1 |
| Tneg + Gpos | 31.5 | 0.2 | ||
| G negative | TnegGneg | Tpos + Gneg | 34.0 | 1.2 |
| Tneg + Gpos | 15.6 | 0.0 |
108 copies of the positive or negative strand were subjected to RT using a tagged primer specific for each strand and quantified by qPCR using PCR primers as described in the text. The respective Ct value with standard deviation of three individual replicates is shown. The experiment was repeated at least three times.
3.5 Standard curves generated using in vitro transcribed RNAs have high strand specificity and sensitivity
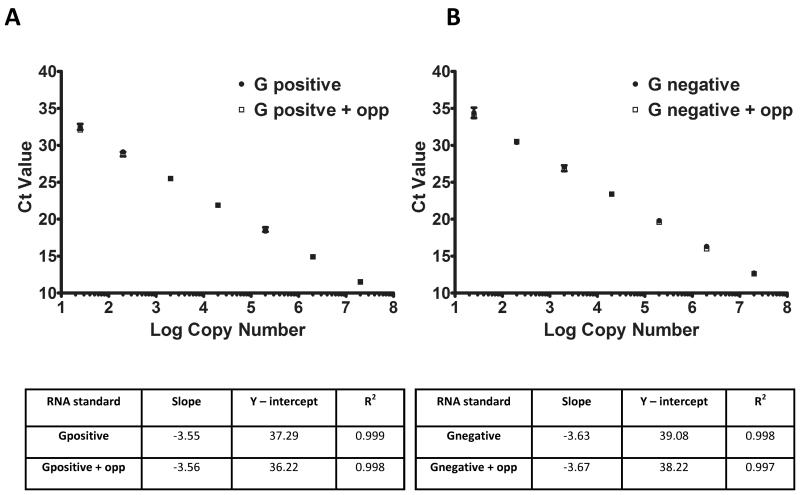
To determine the sensitivity of the strand specific assays, serial dilutions of in vitro transcribed RNAs were used for cDNA synthesis using the tagged RT primers in the absence or presence of an excess of opposite polarity RNA (105 copies). cDNA reactions were diluted tenfold to minimise any inhibitory effect of the components of the cDNA synthesis reaction and to reduce the carry-over of the unused RT primer. Two μl of the diluted cDNA reactions containing, in addition to the serial dilution of the standard RNA, 106 copies of the opposite strand, was used in the qPCR reaction to generate standard curves for both positive and negative strand of the genome (Figure 3a and b). This revised strategy was able to quantify RNA in a strand specific manner even in the presence of an excess of the opposite polarity RNA. The standard curve displayed a linear response down to 25 copies of either strand in presence or absence of 106 copies of opposite strand. The parameters of qPCR are presented in figure 3. The detection limit of the assay was set to 25 copies, the minimum amount of standard RNA used in this assay. To further confirm the robustness of this method, known quantities of either strand of standard RNA in the presence of excess amount of opposite strand were tested. The data suggests that this method is able to detect ~12.5 copies of RNA with acceptable standard deviation (Table 4).
Figure 3. Strand specific RT-qPCR of MNV genomic RNA.
The standard curves were generated by quantifying in vitro transcribed RNA using the strand specific RT-qPCR as described in the text. cDNAs synthesised by reverse transcription of either positive (Panel A) or negative strand (Panel B) in presence or absence of a fixed amount of opposite strand using tagged RT primer were quantified using qPCR. The experiment was performed in triplicate and mean Ct value with standard deviation were plotted against RNA absolute copy number. The correlation coefficients of the qPCRs are presented in table below each curve.
Table 4.
| RNA | Absolute Quantity |
Quantity extrapolated from St. curve (Mean)a) |
St. Dev. (%) |
|---|---|---|---|
| G Positive | 2.0 E+08 | 1.9 E+08 | 1.4 |
| 100 | 110.7 | 8.1 | |
| 50 | 57.1 | 11.6 | |
| 12 | 14.1 | 7.6 | |
| G Negative | 2.0 E+08 | 1.5 E+08 | 1.9 |
| 100 | 122.6 | 16.7 | |
| 50 | 50.4 | 15 | |
| 12 | 18.2 | 14.3 |
Known amounts of either strand of RNA were re-quantified standard curve in figure 3. The standard deviation in % is also presented to determine the accuracy of the quantitation. The values presented are representative of experiment repeated at least twice.
3.6 Absolute quantitation of viral positive and negative strands of MNV genomic RNA during virus replication in cell culture
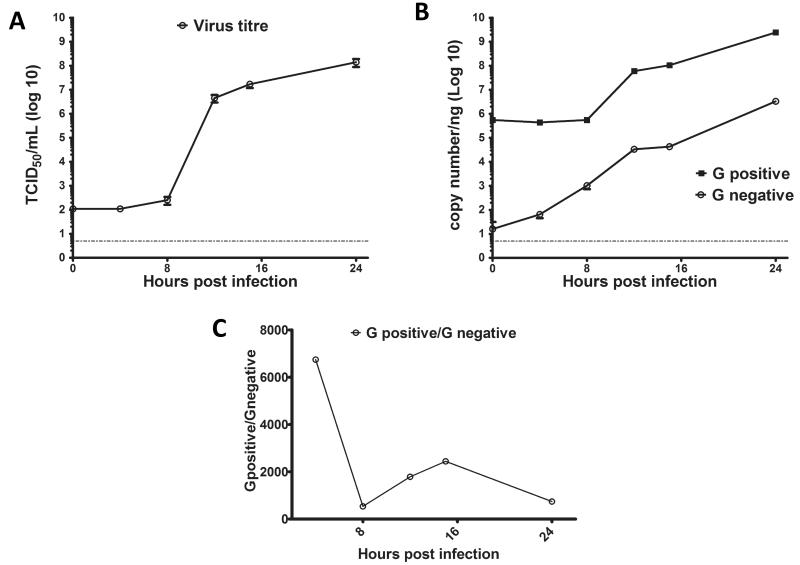
To begin to understand the dynamics of norovirus RNA synthesis during virus replication, the assay developed above was used to characterise the kinetics of RNA synthesis during MNV replication in cell culture. The MNV-permissible microglial cell line BV-2 (Cox, Cao, and Lu, 2009) was infected with MNV-1 at MOI of 5 and cells were harvested at various time-points post infection for virus titre and RNA extraction. A viral growth curve was generated from the samples collected at various time points by plotting the viral titre against time (Figure 4a). Total cellular RNA was extracted from the infected cells at various times post infection and subjected to the strand specific RT-qPCR as described above. The copy number of each RNA strand in samples was determined by extrapolation from standard curves (Figure 4b). Prior to 8 hours post infection, the positive strand RNA quantity remained constant while that of the negative strand RNA increased. After 8 hours, the positive strand RNA as well as negative strand RNA synthesis continued to increase. The ratio of positive to negative strand RNA was found to vary over the viral life cycle (Figure 4c); the ratio of positive to negative strand decreases initially, before subsequently increasing after 15 hours, followed by a subsequent reduction again in the later stages of the viral life cycle.
Figure 4. Quantitation of MNV positive and negative sense genomic RNA during replication in cell culture.
BV-2 cells were infected at MOI of 5 TCID50 per cell and samples for TCID50 and RNA isolation were harvested in triplicate at stated time points post infection. The virus titre was plotted over time to generate one step growth curve of MNV (A). The production of positive and negative sense genomic RNA during one step growth curve were estimated by extrapolation of standard curves generated in figure 3 and plotted against time-points post infection (B). The ratio of positive and negative strands of genomic RNA during the course of infection(C).
4. Discussion
The development of an assay to distinguish specifically the negative strand viral RNA produced during virus replication is essential for the study of various aspects of the norovirus life cycle and provides an additional tool to indicate that active replication is occurring. In this study a strand specific RT-qPCR assay for the accurate quantitation of both positive and negative sense RNA genomes of the MNV was developed enabling the synthesis of negative sense genomic RNA to be accurately monitored during an MNV infection in vitro for the first time.
The synthesis of non-specific cDNA during an RT reaction, hampering strand specific detection, has previously been reported (Beiter et al., 2007; Gunji et al., 1994; Lanford et al., 1994; Peyrefitte et al., 2003). These cDNAs are typically generated by false priming events that might arise due to the presence of short nucleic acids in the samples prepared from infected cells/tissues, secondary structures in RNA, or in fact from the RT enzyme itself (Haddad et al., 2007; Moison et al., 2011; Piche and Schernthaner, 2003; Plaskon et al., 2009; Timofeeva and Skrypina, 2001; Tuiskunen et al., 2010). It is shown in this report that the absolute quantitation of a standard RNA in the presence of high concentrations of the opposite strand is not possible when unmodified primers are used for the RT (Figure 2a and b).
The quantitation of positive sense viral RNA during infection of any positive strand RNA virus is usually not affected by false priming as the ratio of positive to negative polarity RNA is generally high at any given point during infection. However, for the same reason, the quantitation of negative strand RNA is inhibited. Previous reports have used a variety of methods to overcome this issue including the use of a two-step RNase protection assay where excess positive sense RNA is first removed by RNAses digestion leaving a 1:1 ratio of negative to positive strand. This improves the availability of the negative strand for hybridisation with the probe in the second step, with minimal interference form the positive strand (Novak and Kirkegaard, 1991). Recent work has however relied on RT qPCR as a more high-throughput method for the quantitation of viral RNA. In the current report, the use of tagged RT primers, along with other modifications, was found to be essential to achieve strand specificity. These additional modifications included using an RT enzyme with improved properties that minimise false priming by functioning at a higher extension temperature. Using these modifications, it was possible to quantify very low levels of positive and negative strands of standard RNA, as well as viral RNA, during authentic virus replication in host cells. Previous reports have described using a strategy of column purifying the cDNA reaction to remove any inhibitory components prior to qPCR (Bessaud et al., 2008; Escaffre et al., 2010). However, in the assay described here, this additional modification did not improve the qPCR reaction beyond that obtained by the dilution of the cDNA reactions (data not shown). Furthermore, this additional step could contribute to increased variation in the assay by virtue of minor differences in the column yield.
In the assay described above, an excess of at least 5 log10 of the opposite strands was maintained to determine specificity in the standard curves. The RT-qPCR assays for both the positive and negative sense strands of the MNV genome were evaluated for its specificity, sensitivity and reproducibility. The standard curves generated both in the presence as well as the absence of the opposite strands was evaluated for the mean linear correlation coefficients (y intercept, expected Ct value at quantity equal to 1; slope, indicates PCR efficiency and R2 value, closeness of fit between the regression line and individual data points). The correlation coefficients were calculated from the regression line in standard curves both in the presence as well as the absence of an excess amount of opposite strand (Figures 2 and 3). The use of tagged RT-primers lead to robust and reproducible values for the correlation coefficients even in the presence of an excess of the opposite strand, between 107 to 25 copies per sample (Figure 3). Known amounts of the standard RNA of both RNAs were used to evaluate the accuracy of the standard curve. The amounts extrapolated from the standard curve matched with the input known amounts (Table 4).
It is worth noting that T7 RNA polymerase produces low amount of opposite sense RNA during in vitro transcription (Konarska and Sharp, 1989; Melton et al., 1984; Schenborn and Mierendorf, 1985). The degree to which this occurs is dependent on the reaction conditions, incubation time and the sequence/structure adopted by the RNA transcript. Whilst any appreciable levels of opposite strand was not detected in the preparations of standard RNA in this study, further improvements could be made by shifting from a SYBR Green based assay to TaqMan DNA hydrolysis probe based assay. SYBR Green dye non-specifically detects dsDNA produced during higher cycle number of qPCR decreasing its sensitivity. TaqMan probe based assay has been shown to improve the sensitivity by at least one log (Plaskon et al., 2009).
The assay developed above was used to study the kinetics of the production of both strands of MNV genomic RNA during high MOI infection in cell culture (Figure 4b). To understand the kinetics of positive versus negative strand genomic RNA replication, the ratio was examined during the course of virus infection. Note that only data after 4 hours was used to determine the relative ratios as at 0 hours, the ratio of positive to negative strand does not indicate the ratio during replication but the ratio of RNAs present in the virions adsorbed on cells. Prior to 8 hours post infection, there was no detectable increase in production of infectious virus. Between 8 and 15 hours there was an exponential increase of infectious virus, prior to entering stationary phase by 24 hours, where extensive cytopathic effect and cell death was observed. The levels of viral positive sense genomic RNA remained largely unaltered during first 8 hours, whilst the negative sense RNA increased. The observed increase in viral negative strand RNA clearly reflects the initial rounds of viral RNA synthesis that occurs after the ‘pioneer round’ of translation of the incoming viral RNA. From 8 hours to 24 hours, both strands of genomic RNA production increases during the multiple cycles of positive and negative strand RNA synthesis. The ratio of positive to negative strand genomic RNA decreased during the early stages of infection. This ratio increased along with the production of viral negative strand RNA until 15 hours post infection, after which time although there is still production of both strands, the ratio begins to decline. The observed decline in the ratio of viral positive to negative sense RNA later in the viral life cycle is most likely due to the encapsidation and release of viral positive strand RNA. Data obtained at this later time point (24 hours post infection) is also further compromised due to the increased apoptosis and cell death that occurs as a result of virus replication.
In summary, an RT-qPCR assay was developed that can be used to quantify MNV positive and negative sense genomic RNA accurately and robustly. This assay will be a useful addition to the variety of methods currently available to monitor active replication of MNV during infection. In addition it may enable the finer details of viral and host factors that contribute to negative strand RNA synthesis to be understood.
Acknowledgements
This work was supported by grants from the Wellcome Trust and the Imperial College NIHR Biomedical Research Centre. IG is a Wellcome Senior Fellow.
References
- Baert L, Wobus CE, Van Coillie E, Thackray LB, Debevere J, Uyttendaele M. Detection of murine norovirus 1 by using plaque assay, transfection assay, and real-time reverse transcription-PCR before and after heat exposure. Appl. Environ. Microbiol. 2008;74:543–6. doi: 10.1128/AEM.01039-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D, Karakasiliotis I, Vashist S, Chung LM, Rees J, McFadden N, Benson A, Yarovinsky F, Simmonds P, Goodfellow I. Functional analysis of RNA structures present at the 3′ extremity of the murine norovirus genome: the variable polypyrimidine tract plays a role in viral virulence. J. Virol. 2010;84:2859–70. doi: 10.1128/JVI.02053-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiter T, Reich E, Weigert C, Niess AM, Simon P. Sense or antisense? False priming reverse transcription controls are required for determining sequence orientation by reverse transcription-PCR. Anal. Biochem. 2007;369:258–61. doi: 10.1016/j.ab.2007.06.044. [DOI] [PubMed] [Google Scholar]
- Bessaud M, Autret A, Jegouic S, Balanant J, Joffret ML, Delpeyroux F. Development of a Taqman RT-PCR assay for the detection and quantification of negatively stranded RNA of human enteroviruses: evidence for false-priming and improvement by tagged RT-PCR. J. Virol. Methods. 2008;153:182–9. doi: 10.1016/j.jviromet.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Blasi E, Barluzzi R, Bocchini V, Mazzolla R, Bistoni F. Immortalization of murine microglial cells by a v-raf/v-myc carrying retrovirus. J. Neuroimmunol. 1990;27:229–37. doi: 10.1016/0165-5728(90)90073-v. [DOI] [PubMed] [Google Scholar]
- Chaudhry Y, Nayak A, Bordeleau ME, Tanaka J, Pelletier J, Belsham GJ, Roberts LO, Goodfellow IG. Caliciviruses differ in their functional requirements for eIF4F components. J. Biol. Chem. 2006;281:25315–25. doi: 10.1074/jbc.M602230200. [DOI] [PubMed] [Google Scholar]
- Chaudhry Y, Skinner MA, Goodfellow IG. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J. Gen. Virol. 2007;88:2091–100. doi: 10.1099/vir.0.82940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox C, Cao S, Lu Y. Enhanced detection and study of murine norovirus-1 using a more efficient microglial cell line. Virol J. 2009;6:196. doi: 10.1186/1743-422X-6-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughenbaugh KF, Fraser CS, Hershey JW, Hardy ME. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J. 2003;22:2852–9. doi: 10.1093/emboj/cdg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughenbaugh KF, Wobus CE, Hardy ME. VPg of murine norovirus binds translation initiation factors in infected cells. Virol J. 2006;3:33. doi: 10.1186/1743-422X-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escaffre O, Queguiner M, Eterradossi N. Development and validation of four Real-Time quantitative RT-PCRs specific for the positive or negative strands of a bisegmented dsRNA viral genome. J. Virol. Methods. 2010;170:1–8. doi: 10.1016/j.jviromet.2010.07.009. [DOI] [PubMed] [Google Scholar]
- Goodfellow I, Chaudhry Y, Gioldasi I, Gerondopoulos A, Natoni A, Labrie L, Laliberte JF, Roberts L. Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO Rep. 2005;6:968–72. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunji T, Kato N, Hijikata M, Hayashi K, Saitoh S, Shimotohno K. Specific detection of positive and negative stranded hepatitis C viral RNA using chemical RNA modification. Arch. Virol. 1994;134:293–302. doi: 10.1007/BF01310568. [DOI] [PubMed] [Google Scholar]
- Haddad F, Qin AX, Giger JM, Guo H, Baldwin KM. Potential pitfalls in the accuracy of analysis of natural sense-antisense RNA pairs by reverse transcription-PCR. BMC Biotechnol. 2007;7:21. doi: 10.1186/1472-6750-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy ME. Norovirus protein structure and function. FEMS Microbiol. Lett. 2005;253:1–8. doi: 10.1016/j.femsle.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Horsington J, Zhang Z. Analysis of foot-and-mouth disease virus replication using strand-specific quantitative RT-PCR. J. Virol. Methods. 2007;144:149–55. doi: 10.1016/j.jviromet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW. STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299:1575–8. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- Kawakami E, Watanabe T, Fujii K, Goto H, Watanabe S, Noda T, Kawaoka Y. Strand-specific real-time RT-PCR for distinguishing infl uenza vRNA, cRNA, and mRNA. J. Virol. Methods. 2011;173:1–6. doi: 10.1016/j.jviromet.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komurian-Pradel F, Perret M, Deiman B, Sodoyer M, Lotteau V, Paranhos-Baccala G, Andre P. Strand specific quantitative real-time PCR to study replication of hepatitis C virus genome. J. Virol. Methods. 2004;116:103–6. doi: 10.1016/j.jviromet.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Konarska MM, Sharp PA. Replication of RNA by the DNA-dependent RNA polymerase of phage T7. Cell. 1989;57:423–31. doi: 10.1016/0092-8674(89)90917-3. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Sureau C, Jacob JR, White R, Fuerst TR. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology. 1994;202:606–14. doi: 10.1006/viro.1994.1381. [DOI] [PubMed] [Google Scholar]
- McFadden N, Bailey D, Carrara G, Benson A, Chaudhry Y, Shortland A, Heeney J, Yarovinsky F, Simmonds P, Macdonald A, Goodfellow I. Norovirus Regulation of the Innate Immune Response and Apoptosis Occurs via the Product of the Alternative Open Reading Frame 4. PLoS Pathog. 2011;7:e1002413. doi: 10.1371/journal.ppat.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton DA, Krieg PA, Rebagliati MR, Maniatis T, Zinn K, Green MR. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984;12:7035–56. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moison C, Arimondo PB, Guieysse-Peugeot AL. Commercial reverse transcriptase as source of false-positive strand-specific RNA detection in human cells. Biochimie. 2011;93:1731–7. doi: 10.1016/j.biochi.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Novak JE, Kirkegaard K. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J. Virol. 1991;65:3384–7. doi: 10.1128/jvi.65.6.3384-3387.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrefitte CN, Pastorino B, Bessaud M, Tolou HJ, Couissinier-Paris P. Evidence for in vitro falsely-primed cDNAs that prevent specific detection of virus negative strand RNAs in dengue-infected cells: improvement by tagged RT-PCR. J. Virol. Methods. 2003;113:19–28. doi: 10.1016/s0166-0934(03)00218-0. [DOI] [PubMed] [Google Scholar]
- Piche C, Schernthaner JP. Background priming during reverse transcription by oligo(dT) carried over from mRNA isolation. Biotechniques. 2003;34:720–2. 724. doi: 10.2144/03344bm08. [DOI] [PubMed] [Google Scholar]
- Plaskon NE, Adelman ZN, Myles KM. Accurate strand-specific quantification of viral RNA. PLoS One. 2009;4:e7468. doi: 10.1371/journal.pone.0007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell MK, Hart SA, Kurath G, Winton JR. Strand-specific, real-time RT-PCR assays for quantification of genomic and positive-sense RNAs of the fish rhabdovirus, Infectious hematopoietic necrosis virus. J. Virol. Methods. 2006;132:18–24. doi: 10.1016/j.jviromet.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Schenborn ET, Mierendorf RC., Jr. A novel transcription property of SP6 and T7 RNA polymerases: dependence on template structure. Nucleic Acids Res. 1985;13:6223–36. doi: 10.1093/nar/13.17.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stals A, Baert L, Botteldoorn N, Werbrouck H, Herman L, Uyttendaele M, Van Coillie E. Multiplex real-time RT-PCR for simultaneous detection of GI/GII noroviruses and murine norovirus 1. J. Virol. Methods. 2009;161:247–53. doi: 10.1016/j.jviromet.2009.06.019. [DOI] [PubMed] [Google Scholar]
- Timofeeva AV, Skrypina NA. Background activity of reverse transcriptases. Biotechniques. 2001;30:22–4. 26, 28. doi: 10.2144/01301bm02. [DOI] [PubMed] [Google Scholar]
- Tuiskunen A, Leparc-Goffart I, Boubis L, Monteil V, Klingstrom J, Tolou HJ, Lundkvist A, Plumet S. Self-priming of reverse transcriptase impairs strand-specific detection of dengue virus RNA. J. Gen. Virol. 2010;91:1019–27. doi: 10.1099/vir.0.016667-0. [DOI] [PubMed] [Google Scholar]
- Vashist S, Bailey D, Putics A, Goodfellow I. Model systems for the study of human norovirus biology. Future Virology. 2009;4:353–367. doi: 10.2217/fvl.09.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward VK, McCormick CJ, Clarke IN, Salim O, Wobus CE, Thackray LB, Virgin HW, Lambden PR. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11050–5. doi: 10.1073/pnas.0700336104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus CE, Karst SM, Thackray LB, Chang K, Sosnovtsev SV, Belliot G, Krug A, Mackenzie JM, Green KY, Virgin HW. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004;2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus MA, Chung LM, Chaudhry Y, Bailey D, Goodfellow I. Development of an optimized RNA-based murine norovirus reverse genetics system. J. Virol. Methods. 2010;169:112–8. doi: 10.1016/j.jviromet.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]