Abstract
Background
Trichophyton tonsurans is the foremost fungal pathogen of minority children in the U.S. Despite overwhelming infection rates, it does not appear that this fungus infects children in a non-specific manner.
Objective
This study was designed to identify genes that may predispose or protect a child from T. tonsurans infection.
Methods
Children participating in an earlier longitudinal study wherein infection rates could be reliably determined were eligible for inclusion. DNA from a subset (n=40) of these children at the population extremes underwent whole genome genotyping (WGG). Allele frequencies between cases and controls were examined and significant SNPs were used to develop a candidate gene list for which the remainder of the cohort (n=115) were genotyped. Cumulative infection rate was examined by genotype and the ability of selected genotypes to predict the likelihood of infection explored by multivariable analysis.
Results
23 genes with a putative mechanistic role in cutaneous infection were selected for evaluation. Of these, 21 demonstrated significant differences in infection rate between genotypes. A risk index assigned to genotypes in the 21 genes accounted for over 60% of the variability observed in infection rate (adjusted r2=0.665, p<0.001). Among these, 8 appeared to account for the majority of variability that was observed (r2=0.603, p<0.001). These included genes involved in: leukocyte activation and migration, extracellular matrix integrity and remodeling, epidermal maintenance and wound repair, and cutaneous permeability.
Conclusions
Applying WGG to individuals at the extremes of phenotype can help to guide the selection of candidate genes in populations of small cohorts where disease etiology is likely polygenic in nature.
Introduction
Trichophyton tonsurans is the foremost fungal pathogen of children in the United States. Approximately 1 in 8 African American (AA) children in elementary school and as many as 1 in 2 AA preschoolers bear the fungus on their scalp.[1,2] While such overwhelming infection rates might suggest that T. tonsurans has the ability to broadly and non-specifically infect children, this does not appear to be the case. In the U.S., infections are almost exclusively restricted to AA children, yet every child of African ancestry does not appear to be equally susceptible to infection. In a recent 2-year longitudinal study of daycare center attendees we observed that infections did not occur haphazardly. Approximately 1/3rd of children were chronic carriers of T. tonsurans, as many as 25% were only transiently infected, and another 1/3rd never demonstrated evidence of infection despite the shared environment.[1] In fact, there were even cases of sibling pairs sharing both the home and school environment that demonstrated different infection patterns.[1]
The data from this earlier study offered compelling evidence that unique host factors mitigate the interaction between the host and the infecting pathogen. This theory is consistent with the reports of others which offer limited evidence that inherited traits influence susceptibility to dermatophyte infections.[3–8] Given the complexity of the human immune system and the involvement of different elements of innate and acquired immunity at various stages of infection, a complement of sequence variations in several genes may well account for differences in susceptibility between children. The current study was designed to identify genes which could predispose a child to, or protect him/her from, infection. Application of a preliminary “extreme phenotype” whole genome genotyping approach was used to inform subsequent candidate gene selection and offer an unbiased look at potential host factors that might influence infection.
Materials and Methods
Study Design
This was a single-center cohort study designed to generate preliminary data on genes or gene families that could influence predisposition to cutaneous fungal infections. Children were eligible for inclusion if they participated in the earlier longitudinal investigation that afforded determination of their infection frequency.[1] Infection frequency was defined by a continuous variable representing the percent of total daycare center visits during the 2 year period in which the participant’s scalp culture was positive for T. tonsurans irrespective of whether the child was clinically symptomatic. Only children with visits on 6 or more occasions over 2 years were included in the analysis to ensure the most reliable and conservative estimate of infection rate. Children were excluded from participation if they no longer attended the daycare center at the time of follow-up and/or if their parents did not consent to their participation in the study. All children were enrolled with informed parental permission under a protocol that was reviewed and approved by the Institutional Review Board of Children’s Mercy Hospital’s and Clinics.
Samples
Saliva (200–500 μL) was obtained from each child using Dacron swabs that were placed under the tongue for 20–30 seconds. The swabs were immediately placed into 200 μL of stabilizing solution and transported to the laboratory on ice. The composition of the stabilizing solution included; 93.75 μL PBS, 93.75 μL AL buffer (Qiagen, Valencia, CA), 5 μL proteinase K, 5 μL solution S (0.25 mM EDTA, 2% SDS) and 2.5 μL RNase. Additionally, 2 buccal swabs were obtained from each child at the time of collection and similarly kept on ice during transport. All specimens were processed within 2 hr of collection. Genomic DNA (gDNA) was extracted and purified using the column-based QIAamp DNA Blood Mini Kit (Qiagen) with slight modification. DNA was quantitated using the Quant-it double stranded assay kit (Invitrogen, Carlsbad, CA). DNA from the buccal specimens used to supplement the salivary DNA as needed to ensure enough nucleic acid for genotyping.
Whole Genome Genotyping
Cases and controls (n=20 each) were selected from among the children at the population extremes. Cases were required to demonstrate an infection frequency of ≥ 90% (i.e. always or almost always infected). Controls were required to demonstrate an infection rate of ≤ 10% (i.e. never or almost never infected). Samples were genotyped using the Sentrix® HumanHap650 genotyping bead chip (Illumina, San Diego, CA). In total, 750 ng of gDNA was subjected to whole genome amplification, fragmented, precipitated with isopropanol and subsequently resuspended in formamide-containing hybridization buffer. Reconstituted samples were denatured and allowed to hybridize to the capture probes overnight (48°C) after which the chips were washed, single-base extension performed, the chip stained and subsequently imaged on the Illumina BeadArray Reader. Image intensities were extracted with the manufacturers BeadScan software and normalized according to algorithms nested in the program.
Candidate Gene Selection
Allele and genotype frequencies generated by WGG were examined between the cases and controls using a chi-square test. Independent SNPs demonstrating an unadjusted p-value of <0.001 were eligible for consideration of evaluation in the remaining children. SNPbrowser software (v. 4.0, Applied Biosystems, Carlsbad, CA) was used to examine the proximity of each SNP to a functional gene region. Synonymous and nonsynonymous sequence variations nested within functional gene regions were prioritized further to identify candidate genes with a putative mechanistic rationale for their contribution to the cutaneous infection process.
PCR based assays with allele specific fluorescent probes (TaqMan, Applied Biosystems, Carlsbad, CA) were identified or designed for SNP analysis of the DNA from the remaining children who did not undergo WGG. All reactions were performed in 8 μL and cycled on a Dyad DNA engine (Bio-Rad, Hercules, CA). An end point assay was used to measure fluorescence on an ABI Prism 7900HT (Applied Biosystems). Infection rate was compared across genotypes by ANOVA with a Tukey post-hoc test. The influence of demographic covariates on infection phenotype was also assessed in the same manner. All statistical analyses were performed with SPSS v. 12 (SPSS, Chicago, IL).
Risk Index Assignment
Individual genotypes were assigned a risk index score if the mean infection rate differed significantly from the mean infection rate for the other genotypes at that locus (e.g. infection rate among children with an AA genotype significantly different from children with an AG genotype). Genotypes for which the mean infection rate was less than 20% were ascribed a value of 0. Genotypes for which the mean infection rate ranged from 20–40% were assigned a 1. A value of 2 was used to designate genotypes for which the mean infection rate lay between 40–60%. Finally, genotypes for which the mean infection rate was greater than 60% were assigned a 3. By way of example; mean infection rates were 19.9%, 33.3% and 57.8% among children with an AA, AG and GG, respectively at rs154576 in the semaphorin 6A gene. Consequently, risk values of 0, 1 and 2 were assigned to the AA, AG and GG genotypes, respectively. Multivariable analysis of variance was used to build the initial regression model using all geneotypes as independent variables and stepwise linear regression (backward elimination) was used to identify the combination of genes whose combined risk index demonstrated the greatest association with infection rate.
To explore the validity of a risk-index based strategy for prioritizing gene candidates, a separate set of analyses were performed using the raw genotypes as independent variables. Standardized regression (i.e. beta) coefficients were calculated according to the following: beta = b * sx/sy, where b represents the raw regression coefficient, sx represents the standard deviation of the independent variable (x), and sy represents the standard deviation of the dependent variable (y). These beta coefficients were used to identify the genotypes that were incorporated into the final model. As above, all statistical analyses were performed in SPSS and the significance limit accepted for all analyses was α=0.05.
Results
Of the 446 children in the original longitudinal investigation, 163 remained in attendance at the participating daycare center and 161 were given parental permission to participate. Of these, 155 qualified for inclusion based on the number of visit dates for which data were available (i.e.≥6). Participating children were evenly distributed by gender (male-50.3%) and the majority were African-American (90.7%) with the remainder Caucasian (6.8%), mixed African-European ancestry (2.5%) or other (<1%). Among the subset of children selected as cases and controls, 100% were African-American with an even split between males and females (19 vs. 21, respectively). In all participants, there was a sufficient quantity of high quality DNA to permit genotyping.
WGG revealed 1,013 unique sequence variations (693 when analyzed by allele, 320 when analyzed by genotype) that met the a priori threshold cutoff defined in the methods. Approximately 40% of these SNPs (n=410) were distributed among 275 known or predicted genes. After reviewing existing data on the functionality of each the 275 genes, 23 were prioritized for subsequent analysis in the remaining study cohort based on a putative mechanistic rationale for involvement with cutaneous infection processes (Table 1). When the remaining 115 participants were genotyped at these loci, 21 of the 23 genes originally significant by WGG retained statistical significance for their association between genotype and infection frequency.
Table 1.
| gene name | gene abbreviation | rs | ANOVA p-value | Putative Mechanistic Role |
|---|---|---|---|---|
| Semaphorin 6A | SEMA6A | rs154576 | 2.03E-07 | LRAM |
| microfibril-associated protein 4 | MFAP4 | rs6587082 | 9.18E-05 | ECM |
| Fibrinogen C domain containing 1 | FibCD1 | rs12343454 | 3.27E-04 | PB |
| Kallmann syndrome 1 | KAL1 | rs12007434 | 7.50E-04 | APG |
| galanin-like peptide | GALP | rs16987032 | 1.02E-03 | CHP |
| BH-protocadherin | PCDH7 | rs11935515 | 1.41E-03 | ECM, EDMW |
| Fibulin5 | FBLN5 | rs741198 | 1.42E-03 | ECM |
| Slit homolog 3 | SLIT3 | rs11134534 | 1.55E-03 | LRAM |
| Marapsin-2 | MPN2 | rs12079887 | 1.76E-03 | EDMW |
| Cub and Sushi Multiple Domains-1 | CsmDI | rs6986644 | 1.93E-03 | LRAM |
| Roundabout receptor 1 | ROBOI | rs498805 | 2.24E-03 | LRAM |
| Mitogen-activated protein kinase | MAPK8 | rs7075976 | 5.13E-03 | EDMW |
| Secreted modular calcium-binding protein-2 | SMOC2 | rs747994 | 5.20E-03 | ECM, MF |
| Longevity assurance gene 4 | Lass4 | rs11666866 | 5.64E-03 | APG, CHP |
| CD99-Like 2 | cd99L2 | rs2142003 | 9.04E-03 | LRAM |
| Insulin-like Growth Factor 1 Receptor | IGF1R | rs11633294 | 1.31E-02 | EDMW |
| Grb2-associated binder 2 | GAB2 | rs2450129 | 2.37E-02 | LRAM |
| Matrix metalinoproteinase 3 | MMP3 | rs522616 | 2.78E-02 | ECM |
| Fibroblast growth factor 1 | FGF1 | rs2299020 | 2.93E-02 | EDMW |
| Fibrillin-2 | FBN2 | rs27855 | 3.38E-02 | ECM |
| A disintegrin and metalloprotease 12 | ADAM12 | rs17154587 | 4.11E-02 | ECM, EDMW |
| Matrillin-2 | MTR2 | rs4360265 | 6.76E-02 | ECM |
| Tribbles homolog 1 | TribI | rs2235108 | 1.53E-01 | APG, EDMW |
APG- antimicrobial peptides/glycopeptides, CHP- cutaneous hydration and permeability, ECM- extracellular matrix formation/integrity/remodeling, EDMW- epidermal development/maintenance/wound repair, LRAM- leukocte recruitment/activation/migration, MF- melanocyte function, PB- parasite binding
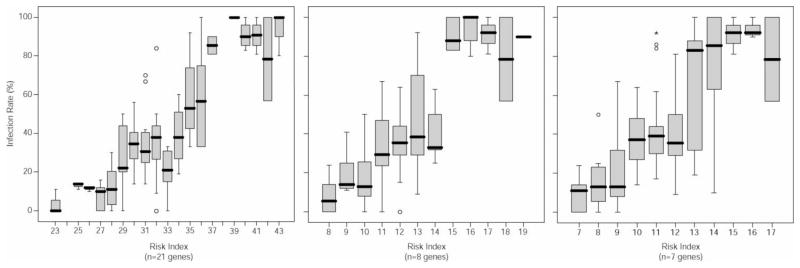
The cumulative risk index assigned to the genotypes of these 21 genes accounted for more than 60% of the variability observed in infection rate (Figure 1). Stepwise regression revealed that 8 genes; CsmD1, FibCD1, Fibulin 5, LASS4, MAPK8, SEMA6A, SLIT3 and SMOC2 appeared to accounted for the majority of variability that was observed in infection rate (Table 2, Figure 1). The inclusion of gender and race in the regression model offered a slight but significant increase in correlation coefficient between infection rate and risk index (Table 2). Slightly less variability (58%) was accounted for when using the β-coefficient based approach (Table 2, Figure 1). Notably, 6 of the genes identified by risk index were also identified by β-coefficient. SLIT3 and SMOC2 were not significant using the latter approach; however, GALP emerged as significant in this model.
Figure 1.
Box plots of risk index vs. infection phenotype with all genes considered (left), with the subset of genes demonstrating the greatest impact on phenotype (center) and with the subset of genes demonstrating the greatest impact on phenotype based on β-coefficient (right).
Table 2.
Statistical parameters for multivariable regression
| Model | df | r2 | adjusted r2 | SE of the estimate | F | p-value |
|---|---|---|---|---|---|---|
| Risk-index based regression | ||||||
| 21 genes | 21 | 0.704 | 0.635 | 17.930 | 10.178 | <0.0001 |
| 21 genes + gender | 22 | 0.724 | 0.656 | 17.406 | 10.604 | <0.0001 |
| 21 genes + gender + race | 23 | 0.758 | 0.695 | 16.390 | 11.979 | <0.0001 |
| 8 significant genesa | 8 | 0.631 | 0.603 | 18.696 | 22.454 | <0.0001 |
| 8 genes + gender | 9 | 0.661 | 0.632 | 18.011 | 22.523 | <0.0001 |
| 8 genes + gender + race | 10 | 0.698 | 0.669 | 17.081 | 23.800 | <0.0001 |
| β-coefficient based regression | ||||||
| 21 genes | 21 | 0.656 | 0.577 | 19.214 | 8.269 | <0.0001 |
| 21 genes + gender | 22 | 0.683 | 0.605 | 18.565 | 8.795 | <0.0001 |
| 21 genes + gender + race | 23 | 0.707 | 0.631 | 17.945 | 9.322 | <0.0001 |
| 7 significant genesb | 7 | 0.561 | 0.533 | 20.289 | 19.570 | <0.0001 |
| 7 genes + gender | 8 | 0.588 | 0.557 | 19.759 | 18.909 | <0.0001 |
| 7 genes + gender + race | 9 | 0.612 | 0.579 | 19.262 | 18.411 | <0.0001 |
df- degrees of freedom, r- multiple correlation coefficient, SE- standard error, F- F-statistic
CsmD1, FibCD1, Fibulin 5, LASS4, MAPK8, SEMA6A, SLIT3 and SMOC2
CsmD1, FibCD1, Fibulin 5, GALP, LASS4, MAPK8, SEMA6A
Discussion
Identifying individuals for inclusion in a genetic association study of cutaneous infection risk can be a challenge. Adopting a cross-sectional, case-control study design introduces the problems of selection and information bias and necessitate the need for very large sample sizes. Cohort-designs are more powerful; however, they are resource-intensive, and some might argue impractical, as they require large population-based cohorts to be followed for long periods of time while tracking the appearance and disappearance of infections.[9]
The pilot study described herein attempted to capitalize on the advantages of both study designs. The longitudinal assessment of a high-risk cohort deriving from a singular environment allowed us to define infection rates over time in children with presumably similar non-genetic risk factors. Using a subset of the population for a case-control WGG analysis permitted the identification of unique gene candidates that could be further refined for subsequent evaluation in the remainder of the population. With this approach we identified an association between T. tonsurans infection rate and genotype in 21 genes for which a plausible mechanism in the development of cutaneous infections could be constructed.
Several of the genes for which a significant association was demonstrated are involved in the recruitment, activation, and migration of leukoctes. Though not detected in normal skin epithelium, semaphorin 6A (SEMA6A) is strongly expressed on Langerhan’s cells (LC) residing in the dermis and draining lymphnodes of patients with LC histiocytosis and dermatopathic lymphadenopathy. High levels of expression under conditions of inflammation suggest that SEMA6A may be involved in guiding the migration of these antigen-presenting cells out of the skin.[10,11] Roundabout receptor 1 (ROBO1) and the corresponding SLIT gene products function cooperatively to influence the directional migration of numerous cell types including monocytes and lymphocytes.[12–14]. Though SLIT3 is present in skin, most of the data on ROBO/SLIT guided migration of dendritic cells is described for a related gene family member (SLIT2).[15,16] CD99-Like 2 (cd99L2) was also identified as a candidate and the product of this gene is involved with the extravasation of neutrophils and monocytes into inflamed tissue including skin.[17,18] The Cub and Sushi Multiple Domains-1 (CSMD1) gene has been implicated in inflammatory processes via the activation of complement and was recently identified as candidate gene associated with psoriasis.[19,20] Finally, Grb2-associated binder 2 (GAB2) demonstrates a polyfunctional role in the process of inflammation. This gene appears to be a major effector for IgE-dependent allergic reactions, it participates in the activation of T- and B-cells via cytokine- and growth factor-mediated signaling and it is involved in mast cell signaling and function in response to pathogenic stimuli. Importantly, Gab2 knockout mice demonstrate reduced mast cell recruitment into the skin.[21,22]
A large proportion of the genes in our dataset are involved in the extracellular matrix formation, integrity and remodeling. Fibulin 5 (FBLN5), fibrillin 2 (FBN2) and microfibrillar-associated protein 4 (MFAP4) are all essential components of the extracellular matrix (ECM) and elastin fiber network under normal homeostatic conditions and during periods remodeling and wound healing.[23–25] Further, FBN2 expression is markedly upregulated in sclerotic skin diseases,[26,27]and mutations in FBLN5 disrupt the elastic properties of the skin.[28,29] Secreted modular calcium-binding protein-2 (SMOC2) appears to be involved with melanocyte function and genome wide studies have identified an association between SMOC2 and the autoimmune disorder vitiligo in selected populations.[30,31] Importantly, vitiligo is also associated with lower hydration and altered permeability in the stratum corneum.[32] BH-protocadherin (PCDH7), a member of the cadherin superfamily of genes, is involved with cell-cell recognition and is upregulated during keratinocyte differentiation.[33,34] Finally, the enzymes encoded by matrix metalinoproteinase 3 (MMP3) and a disintegrin and metalloprotease 12 (ADAM 12) are involved with remodeling of the ECM under normal and pathological conditions.[35–39]
A smaller subset of the genes identified in this study are involved with epidermal development, maintenance, and wound repair the most obvious of which is fibroblast growth factor 1 (FGF1). Others include mitogen-activated protein kinase 8 (MAPK8) which is involved in toll-like receptor signaling in the skin and implicated in the pathogenesis of skin diseases (e.g. psoriasis);[40] and insulin-like Growth Factor 1 Receptor (IGF1R) which is expressed in keratinocytes and responds to IGF1 secreted from dermal fibroblasts.[41] Notably, dermatologic changes stimulated by Propionibacterium acnes appear to be mediated through the IGF/IGF1R system [42,43] Other genes in our dataset that can be grouped in this functional class include PCDH7 and ADAM-12.
Among the genes with a more unique role in skin homeostasis and host-pathogen interactions is longevity assurance gene 4 (LASS4). With its involvement in the ceramide synthesis pathway (i.e. the formation of n-acetylsphinganine from sphinganine), LASS4 is involved in maintaining the water permeability and barrier function of the skin. [44–46] In addition, the substrate on which LASS4 acts (sphinganine) is capable of inhibiting mycelial development and growth in a number of cutaneous pathogens including T. tonsurans which appears to be highly sensitive to this lipid [47] Another unique gene is galanin-like peptide (GALP), a neuropeptide that is expressed in dermal microvasculature. Although there is no direct evidence for GALP, related members of this gene family regulate cutaneous blood flow and demonstrate the ability to inhibit the budding and growth of the cutaneous yeast Candida albicans.[48–50] The Kallmann syndrome 1 (KAL1) gene innervates itch perception and is differentially regulated in atopic dermatitis.[51] Interestingly, it is othrologous to a canine antimicrobial peptide that is active on skin.[52] Finally, Fibrinogen C domain containing 1 (FibCD1) functions primarily to bind chitin from invading fungi and parasites. While interesting with respect to a putative role in cutaneous infections, FibCD1 is highly expressed in the gastrointestinal tract and it is unclear whether it is expressed in the skin. [53,54].
This pilot analysis offers some intriguing gene candidates that may be associated with a child’s susceptibility to superficial dermatophyte infections. Interestingly, genes that appear to play a role in susceptibility to chronic mucocutaneous candidiasis (e.g. STAT1 and 3, CARD9, AIRE, CLEC4E and 7A, CD209, IL-17) were not represented among the preliminary 275 genes identified by WGG in this study [55–60]. The recessive nature of the implicated sequence variations for some genes (AIRE, CARD9, IL-17RA) and the low allele frequency of other sequence variations in individuals of African ancestry (CLEC7A) may explain why they were not detected in our population. However, the role of other gene families (e.g. STAT) in the susceptibility to dermatophyte infections remains unknown.
Though the genes reported herein appear to have reasonable predictive power to discriminate children at risk for chronic T. tonsurans infection, it is important to clarify that we were simply able to detect an association and cannot infer a causal relationship. It is also important to emphasize that the study is limited by a small sample. It is possible that some of these associations we observed reflect false positive results and equally possible that we failed to identify associations with genes that play a role in disease susceptibility. Equally important is the fact that we restricted our analyses to functional gene regions. Intergenic sequence variations that could influence the regulation and/or expression of relevant genes would have been overlooked unless they exist in linkage disequilibrium with the SNPs that were evaluated.
Given the extent to which infections are observed in the urban pediatric community, numerous genes likely contribute to a child’s overall risk of acquiring and sustaining dermatophyte infections following contact with the pathogen. Multiple pathways, rather than a singular common biological pathway likely contribute to the shared phenotype of infection risk. It is conceivable that pathogen, in addition to host, genetics also play a role in the long-term relationship between host and organism; however, we observed too many genetic strain types in our community to identify a relationship between fungal genotype and pathogenicity. Undoubtedly, it is the combination of host and pathogen traits that permit a long-term relationship between the two species.
When demographics and host genetics were considered together, we were able to account for approximately 55 to 65% of the variability observed in infection rate. Though other genetic factors may play a role, it is also important to consider host-environment interactions that may influence infection status. By enrolling children who shared a common daytime environment, we attempted to minimize the contribution of environment. However, the influence of the environment in which the children spend the remainder of their time cannot be discounted. Notably, there were several sibling sets in our earlier investigation wherein the children shared both a home and school environment yet demonstrated different infection patterns. This was the first cue that a unique host-pathogen relationship may exist.
Evidence for the contribution of any of the genes described herein will require replication in an independent cohort of children. Despite the intensive nature of the longitudinal study design that would be required, there is no shortage of populations (U.S. and international) in whom dermatophyte infection rates equal or exceed those observed in our community. Our small cohort is unlikely to provide all of the answers as to what factors predispose or prevent infection but our initial observations suggest a much larger effect on total heritability of the trait than we initially anticipated. Subsequent studies with dermatophytes, or other chronic cutaneous pathogens, that clarify the role of selected gene products in the infection process may facilitate the identification of therapeutic targets that restrict host-pathogen interactions in high-risk populations.
Acknowledgments
This work was supported by grants from the NIH-National Institute of Arthritis and Musculoskeletal and Skin Diseases (1R21 AR053234) and the Henson Endowed Fund for Pediatric Research.
Footnotes
The authors have no conflict of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abdel-Rahman SM, Simon S, Wright KJ, Ndjountche L, Gaedigk A. Tracking Trichophyton tonsurans through a large urban childcare center: defining infection prevalence and transmission patterns by molecular stain typing. Pediatrics. 2006;118:2365–2373. doi: 10.1542/peds.2006-2065. [DOI] [PubMed] [Google Scholar]
- 2.Abdel-Rahman SM, Farrand N, Schuenemann E, Stering TK, Preuett B, Magie R, Campbell A. Prevalence of Infections with Trichophyton tonsurans in School Children (the CAPITIS Study) Pediatrics. 2010;125:966–973. doi: 10.1542/peds.2009-2522. [DOI] [PubMed] [Google Scholar]
- 3.Hay RJ. Genetic susceptibility to dermatophytosis. Eur J Epidemiol. 1992;8:346–349. doi: 10.1007/BF00158566. [DOI] [PubMed] [Google Scholar]
- 4.Serjeantson S, Lawrence G. Autosomal recessive inheritance of susceptibility to tinea imbricata. Lancet. 1977;i:13–15. doi: 10.1016/s0140-6736(77)91653-1. [DOI] [PubMed] [Google Scholar]
- 5.Jones HE, Reinhardt JH, Rinaldi MG. Immunologic susceptibility to chronic dermatophytosis. Arch Dermatol. 1974;110:213–220. [PubMed] [Google Scholar]
- 6.Ravine D, Turner KJ, Alpers MO. Genetic inheritance of susceptibility to tinea imbricata. J Med Genet. 1980;17:342–348. doi: 10.1136/jmg.17.5.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svejgaard E, Jakobsen B, Svejgaard A. HLA studies in chronic dermatophytosis caused by Trichophyton rubrum. Acta Derm Venereol. 1983;63:254–255. [PubMed] [Google Scholar]
- 8.Elmros T, Liden S. Hereditary palmo-plantar keratoderma: incidence of dermatophyte infections and the results of topical treatment with retinoic acid. Acta Derm Venereol. 1981;61:453–455. [PubMed] [Google Scholar]
- 9.Yende S, Kammerer CM, Angus DC. Bench-to-bedside review: genetics and proteomics: deciphering gene association studies in critical illness. Critical Care. 2006;10:227. doi: 10.1186/cc5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautier G, de Saint-Vis B, Senechal B, Pin JJ, Bates EE, Caux C, Geissmann F, Garrone P. The class 6 semaphorin SEMA6A is induced by interferon-gamma and defines an activation status of langerhans cells observed in pathological situations. Am J Pathol. 2006;168:453–465. doi: 10.2353/ajpath.2006.050288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klostermann A, Lutz B, Gertler F, Behl C. The orthologous human and murine semaphorin 6A-1 proteins (SEMA6A-1/Sema6A-1) bind to the enabled/vasodilator-stimulated phosphoprotein-like protein (EVL) via a novel carboxyl-terminal zyxin-like domain. J Biol Chem. 2000;275:39647–39653. doi: 10.1074/jbc.M006316200. [DOI] [PubMed] [Google Scholar]
- 12.Geutskens SB, Hordijk PL, van Hennik PB. The chemorepellent Slit3 promotes monocyte migration. J Immunol. 2010;185:7691–7698. doi: 10.4049/jimmunol.0903898. [DOI] [PubMed] [Google Scholar]
- 13.Wu JY, Feng L, Park HT, Havlioglu N, Wen L, Tang H, Bacon KB, Jiang Z, Zhang X, Rao Y. The neuronal repellent Slit inhibits leukocyte chemotaxis induced by chemotactic factors. Nature. 2001;410:948–952. doi: 10.1038/35073616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denk AE, Braig S, Schubert T, Bosserhoff AK. Slit3 inhibits activator protein 1-mediated migration of malignant melanoma cells. Int J Mol Med. 2011 Jul 8; doi: 10.3892/ijmm.2011.742. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 15.Guan H, Zu G, Xie Y, Tang H, Johnson M, Xu X, Kevil C, Xiong WC, Elmets C, Rao Y, Wu JY, Xu H. Neuronal repellent Slit2 inhibits dendritic cell migration and the development of immune responses. J Immunol. 2003;171:6519–6526. doi: 10.4049/jimmunol.171.12.6519. [DOI] [PubMed] [Google Scholar]
- 16.Yang XM, Han HX, Sui F, Dai YM, Chen M, Geng JG. Slit-Robo signaling mediates lymphangiogenesis and promotes tumor lymphatic metastasis. Biochem Biophys Res Commun. 2010;396:571–577. doi: 10.1016/j.bbrc.2010.04.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schenkel AR, Dufour EM, Chew TW, Sorg E, Muller WA. The murine CD99-related molecule CD99-like 2 (CD99L2) is an adhesion molecule involved in the inflammatory response. Cell Commun Adhes. 2007;14:227–237. doi: 10.1080/15419060701755966. [DOI] [PubMed] [Google Scholar]
- 18.Bixel MG, Li H, Petri B, Khandoga AG, Khandoga A, Zarbock A, Wolburg-Buchholz K, Wolburg H, Sorokin L, Zeuschner D, Maerz S, Butz S, Krombach F, Vestweber D. CD99 and CD99L2 act at the same site as, but independently of, PECAM-1 during leukocyte diapedesis. Blood. 2010;116:1172–1184. doi: 10.1182/blood-2009-12-256388. [DOI] [PubMed] [Google Scholar]
- 19.Kraus DM, Elliott GS, Chute H, Horan T, Pfenninger KH, Sanford SD, Foster S, Scully S, Welcher AA, Holers VM. CSMD1 is a novel multiple domain complement-regulatory protein highly expressed in the central nervous system and epithelial tissues. J Immunol. 2006;176:4419–4430. doi: 10.4049/jimmunol.176.7.4419. [DOI] [PubMed] [Google Scholar]
- 20.Sun LD, Cheng H, Wang ZX, Zhang AP, Wang PG, Xu JH, Zhu QX, Zhou HS, Ellinghaus E, Zhang FR, Pu XM, Yang XQ, Zhang JZ, Xu AE, Wu RN, Xu LM, Peng L, Helms CA, Ren YQ, Zhang C, Zhang SM, Nair RP, Wang HY, Lin GS, Stuart PE, Fan X, Chen G, Tejasvi T, Li P, Zhu J, Li ZM, Ge HM, Weichenthal M, Ye WZ, Zhang C, Shen SK, Yang BQ, Sun YY, Li SS, Lin Y, Jiang JH, Li CT, Chen RX, Cheng J, Jiang X, Zhang P, Song WM, Tang J, Zhang HQ, Sun L, Cui J, Zhang LJ, Tang B, Huang F, Qin Q, Pei XP, Zhou AM, Shao LM, Liu JL, Zhang FY, Du WD, Franke A, Bowcock AM, Elder JT, Liu JJ, Yang S, Zhang XJ. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet. 2010;42:1005–1009. doi: 10.1038/ng.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie ZH, Ambudkar I, Siraganian RP. The adapter molecule Gab2 regulates Fc epsilon RI-mediated signal transduction in mast cells. J Immunol. 2002;168:4682–4691. doi: 10.4049/jimmunol.168.9.4682. [DOI] [PubMed] [Google Scholar]
- 22.Nishida K, Yamasaki S, Hasegawa A, Iwamatsu A, Koseki H, Hirano T. Gab2, via PI-3K, regulates ARF1 in FcεRI-mediated granule translocation and mast cell degranulation. J Immunol. 2011;187:932–941. doi: 10.4049/jimmunol.1100360. [DOI] [PubMed] [Google Scholar]
- 23.Yanagisawa H, Schluterman MK, Brekken RA. Fibulin-5, an integrin-binding matricellular protein: its function in development and disease. J Cell Commun Signal. 2009;3:337–347. doi: 10.1007/s12079-009-0065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olivieri J, Smaldone S, Ramirez F. Fibrillin assemblies: extracellular determinants of tissue formation and fibrosis. Fibrogenesis Tissue Repair. 2010;3:24. doi: 10.1186/1755-1536-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plasari G, Calabrese A, Dusserre Y, Gronostajski RM, Mcnair A, Michalik L, Mermod N. Nuclear Factor I-C Links Platelet-Derived Growth Factor and Transforming Growth Factor β1 Signaling to Skin Wound Healing Progression. Mol Cell Biol. 2009;29:6006–6017. doi: 10.1128/MCB.01921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brinckmann J, Hunzelmann N, Kahle B, Rohwedel J, Kramer J, Gibson MA, Hubmacher D, Reinhardt DP. Enhanced fibrillin-2 expression is a general feature of wound healing and sclerosis: potential alteration of cell attachment and storage of TGF-beta. Lab Invest. 2010;90:739–752. doi: 10.1038/labinvest.2010.49. [DOI] [PubMed] [Google Scholar]
- 27.Samuel CS, Sakai LY, Amento EP. Relaxin regulates fibrillin 2, but not fibrillin 1, mRNA and protein expression by human dermal fibroblasts and murine fetal skin. Arch Biochem Biophys. 2003;411:47–55. doi: 10.1016/s0003-9861(02)00710-5. [DOI] [PubMed] [Google Scholar]
- 28.Auer-Grumbach M, Weger M, Fink-Puches R, Papi L, Fröhlich E, Auer-Grumbach P, El Shabrawi-Caelen L, Schabhüttl M, Windpassinger C, Senderek J, Budka H, Trajanoski S, Janecke AR, Haas A, Metze D, Pieber TR, Guelly C. Fibulin-5 mutations link inherited neuropathies, age-related macular degeneration and hyperelastic skin. Brain. 2011;134:1839–1852. doi: 10.1093/brain/awr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langton AK, Sherratt MJ, Griffiths CE, Watson RE. Differential expression of elastic fibre components in intrinsically aged skin. Biogerontology. 2011 Apr 2; doi: 10.1007/s10522-011-9332-9. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 30.Birlea SA, Gowan K, Fain PR, Spritz RA. Genome-wide association study of generalized vitiligo in an isolated European founder population identifies SMOC2, in close proximity to IDDM8. J Invest Dermatol. 2010;130:798–803. doi: 10.1038/jid.2009.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alkhateeb A, Al-Dain Marzouka N, Qarqaz F. SMOC2 gene variant and the risk of vitiligo in Jordanian Arabs. Eur J Dermatol. 2010;20:701–704. doi: 10.1684/ejd.2010.1095. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Man WY, Lv CZ, Song SP, Shi YJ, Elias PM, Manb MQ. Epidermal Permeability Barrier Recovery Is Delayed in Vitiligo-Involved Sites. Skin Pharmacol Physiol. 2010;23:193–200. doi: 10.1159/000288166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banno T, Adachi M, Mukkamala L, Blumberg M. Unique keratinocyte-specific effects of interferon-γ that protect skin from viruses, identified using transcriptional profiling. Antiviral Therapy. 2003;8:541–554. [PubMed] [Google Scholar]
- 34.Taylor JM, Street TL, Hao L, Copely R, Taylor MS, Hayden PJ, Stopler G, Mott R, Moffatt MF, Cookson WOCM. Dynamic and Physical Clustering of Gene Expression during Epidermal Barrier Formation in Differentiating Keratinocytes. PLoS ONE. 2009:e7651. doi: 10.1371/journal.pone.0007651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harsha A, Stojadinovic O, Brem H, Sehara-Fujisawa A, Wewer U, Loomis CA, Blobel CP, Tomic-Canic M. ADAM12: a potential target for the treatment of chronic wounds. J Mol Med. 2008;86:961–969. doi: 10.1007/s00109-008-0353-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seifert O, Bayat A, Geffers R, Dienus K, Buer J, Löfgren S, Matussek A. Identification of unique gene expression patterns within different lesional sites of keloids. Wound Repair Regen. 2008;16:254–265. doi: 10.1111/j.1524-475X.2007.00343.x. [DOI] [PubMed] [Google Scholar]
- 37.Clemmensen A, Andersen KE, Clemmensen O, Tan Q, Petersen TK, Kruse TA, Thomassen M. Genome-wide expression analysis of human in vivo irritated epidermis: differential profiles induced by sodium lauryl sulfate and nonanoic acid. J Invest Dermatol. 2010;130:2201–2210. doi: 10.1038/jid.2010.102. [DOI] [PubMed] [Google Scholar]
- 38.McFarland KL, Glaser K, Hahn JM, Boyce ST, Supp DM. Culture medium and cell density impact gene expression in normal skin and abnormal scar-derived fibroblasts. J Burn Care Res. 2011;32:498–508. doi: 10.1097/BCR.0b013e3182223cb1. [DOI] [PubMed] [Google Scholar]
- 39.Russell SB, Russell JD, Trupin KM, Gayden AE, Opalenik SR, Nanney LB, Broquist AH, Raju L, Williams SM. Epigenetically altered wound healing in keloid fibroblasts. J Invest Dermatol. 2010;130:2489–2496. doi: 10.1038/jid.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogawa E, Okuyama R, Egawa T, Nagoshi H, Obinata M, Tagami H, Ikawa S, Aiba S. p63/p51-induced onset of keratinocyte differentiation via the c-Jun N-terminal kinase pathway is counteracted by keratinocyte growth factor. J Biol Chem. 2008;283:34241–34249. doi: 10.1074/jbc.M804101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis DA, Travers JB, Somani AK, Spandau DF. The IGF-1/IGF-1R signaling axis in the skin: a new role for the dermis in aging-associated skin cancer. Oncogene. 2010;29:1475–1485. doi: 10.1038/onc.2009.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Isard O, Knol AC, Ariès MF, Nguyen JM, Khammari A, Castex-Rizzi N, Dréno B. Propionibacterium acnes activates the IGF-1/IGF-1R system in the epidermis and induces keratinocyte proliferation. J Invest Dermatol. 2011;131:59–66. doi: 10.1038/jid.2010.281. [DOI] [PubMed] [Google Scholar]
- 43.Miura M, Sasaki M, Mizukoshi K, Shibasaki M, Izumi Y, Shimosato G, Amaya F. Peripheral sensitization caused by insulin-like growth factor 1 contributes to pain hypersensitivity after tissue injury. Pain 2011. 2011;152:888–895. doi: 10.1016/j.pain.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 44.Hicks AA, Pramstaller PP, Johansson A, Vitart V, Rudan I, Ugocsai P, Aulchenko Y, Franklin CS, Liebisch G, Erdmann J, Jonasson I, Zorkoltseva IV, Pattaro C, Hayward C, Isaacs A, Hengstenberg C, Campbell S, Gnewuch C, Janssens AC, Kirichenko AV, König IR, Marroni F, Polasek O, Demirkan A, Kolcic I, Schwienbacher C, Igl W, Biloglav Z, Witteman JC, Pichler I, Zaboli G, Axenovich TI, Peters A, Schreiber S, Wichmann HE, Schunkert H, Hastie N, Oostra BA, Wild SH, Meitinger T, Gyllensten U, van Duijn CM, Wilson JF, Wright A, Schmitz G, Campbell H. Genetic determinants of circulating sphingolipid concentrations in European populations. PLoS Genet. 2009:e1000672. doi: 10.1371/journal.pgen.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jarrold BB, Bimder RL, Robinson MK, Tiesman JP, Juhlin KD, Finlay DR, Osborne R. Expression Profiles of Stratum Corneum Lipid Metabolism Pathways Associated with Intrinsic and Extrinsic Aging (Abstract P824) J Am Acad Dermatol. 2009;60:AB28. [Google Scholar]
- 46.Jarrold BB, Bimder RL, Robinson MK, Tiesman JP, Juhlin KD, Finlay DR, Osborne R. Hexamidine, a Protease Inhibitor, Promotes Stratum Corneum Lipid Biomarkers In Vitro (Abstract) J Am Acad Dermatol. 2010;62:AB1. [Google Scholar]
- 47.Bibel DJ, Aly R, Shah S, Shinefield HR. Sphingosines: antimicrobial barriers of the skin. Acta Derm Venereol. 1993;73:407–411. doi: 10.2340/0001555573407411. [DOI] [PubMed] [Google Scholar]
- 48.Schmidhuber SM, Santic R, Tam CW, Bauer JW, Kofler B, Brain SD. Galanin-like peptides exert potent vasoactive functions in vivo. J Invest Dermatol. 2007;127:716–721. doi: 10.1038/sj.jid.5700569. [DOI] [PubMed] [Google Scholar]
- 49.Kofler B, Berger A, Santic R, Moritz K, Almer D, Tuechler C, Lang R, Emberger M, Klausegger A, Sperl W, Bauer JW. Expression of neuropeptide galanin and galanin receptors in human skin. J Invest Dermatol. 2004;122:1050–1053. doi: 10.1111/j.0022-202X.2004.22418.x. [DOI] [PubMed] [Google Scholar]
- 50.Lang R, Gundlach AL, Kofler B. The galanin peptide family: Receptor pharmacology, pleiotropic biological actions, and implications in health and disease. Pharmacol and Ther. 2007;115:177–207. doi: 10.1016/j.pharmthera.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Tengara S, Tominaga M, Kamo A, Taneda K, Negi O, Ogawa H, Takamori K. Keratinocyte-derived anosmin-1, an extracellular glycoprotein encoded by the X-linked Kallmann syndrome gene, is involved in modulation of epidermal nerve density in atopic dermatitis. J Dermatol Sci. 2010;58:64–71. doi: 10.1016/j.jdermsci.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 52.Wingate KV, Torres SM, Silverstein KAT, Hendrickson JA, Rutherford MS. Expression of endogenous antimicrobial peptides in normal canine skin. Vet Dermatol. 2009;20:19–26. doi: 10.1111/j.1365-3164.2008.00707.x. [DOI] [PubMed] [Google Scholar]
- 53.Schlosser A, Thomsen T, Moeller JB, Nielsen O, Torn⊘e I, Mollenhauer J, Moestrup SK, Holmskov U. Characterization of FIBCD1 as an acetyl group-binding receptor that binds chitin. J Immunol. 2009;183:3800–3809. doi: 10.4049/jimmunol.0901526. [DOI] [PubMed] [Google Scholar]
- 54.Thomsen T, Moeller JB, Schlosser A, Sorensen GL, Moestrup SK, Palaniyar N, Wallis R, Mollenhauer J, Holmskov U. The recognition unit of FIBCD1 organizes into a noncovalently linked tetrameric structure and uses a hydrophobic funnel (S1) for acetyl group recognition. J Biol Chem. 2010;285:1229–1238. doi: 10.1074/jbc.M109.061523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van de Veerdonk FL, Plantinga TS, Hoischen A, Smeekens SP, Joosten LA, Gilissen C, Arts P, Rosentul DC, Carmichael AJ, Smits-van der Graaf CA, Kullberg BJ, van der Meer JW, Lilic D, Veltman JA, Netea MG. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011;365:54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 56.Ma CS, Chew GY, Simpson N, Priyadarshi A, Wong M, Grimbacher B, Fulcher DA, Tangye SG, Cook MC. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med. 2008;205:1551–1557. doi: 10.1084/jem.20080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glocker EO, Hennigs A, Nabavi M, Schäffer AA, Woellner C, Salzer U, Pfeifer D, Veelken H, Warnatz K, Tahami F, Jamal S, Manguiat A, Rezaei N, Amirzargar AA, Plebani A, Hannesschläger N, Gross O, Ruland J, Grimbacher B. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361:1727–35. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Björses P, Halonen M, Palvimo JJ, Kolmer M, Aaltonen J, Ellonen P, Perheentupa J, Ulmanen I, Peltonen L. Mutations in the AIRE gene: effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy protein. Am J Hum Genet. 2000;66:378–392. doi: 10.1086/302765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferwerda B, Ferwerda G, Plantinga TS, Willment JA, van Spriel AB, Venselaar H, Elbers CC, Johnson MD, Cambi A, Huysamen C, Jacobs L, Jansen T, Verheijen K, Masthoff L, Morré SA, Vriend G, Williams DL, Perfect JR, Joosten LA, Wijmenga C, van der Meer JW, Adema GJ, Kullberg BJ, Brown GD, Netea MG. Human dectin-1 deficiency and mucocutaneous fungal infections. N Engl J Med. 2009;361:1760–1767. doi: 10.1056/NEJMoa0901053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, Migaud M, Israel L, Chrabieh M, Audry M, Gumbleton M, Toulon A, Bodemer C, El-Baghdadi J, Whitters M, Paradis T, Brooks J, Collins M, Wolfman NM, Al-Muhsen S, Galicchio M, Abel L, Picard C, Casanova JL. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332:65–68. doi: 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]