Abstract
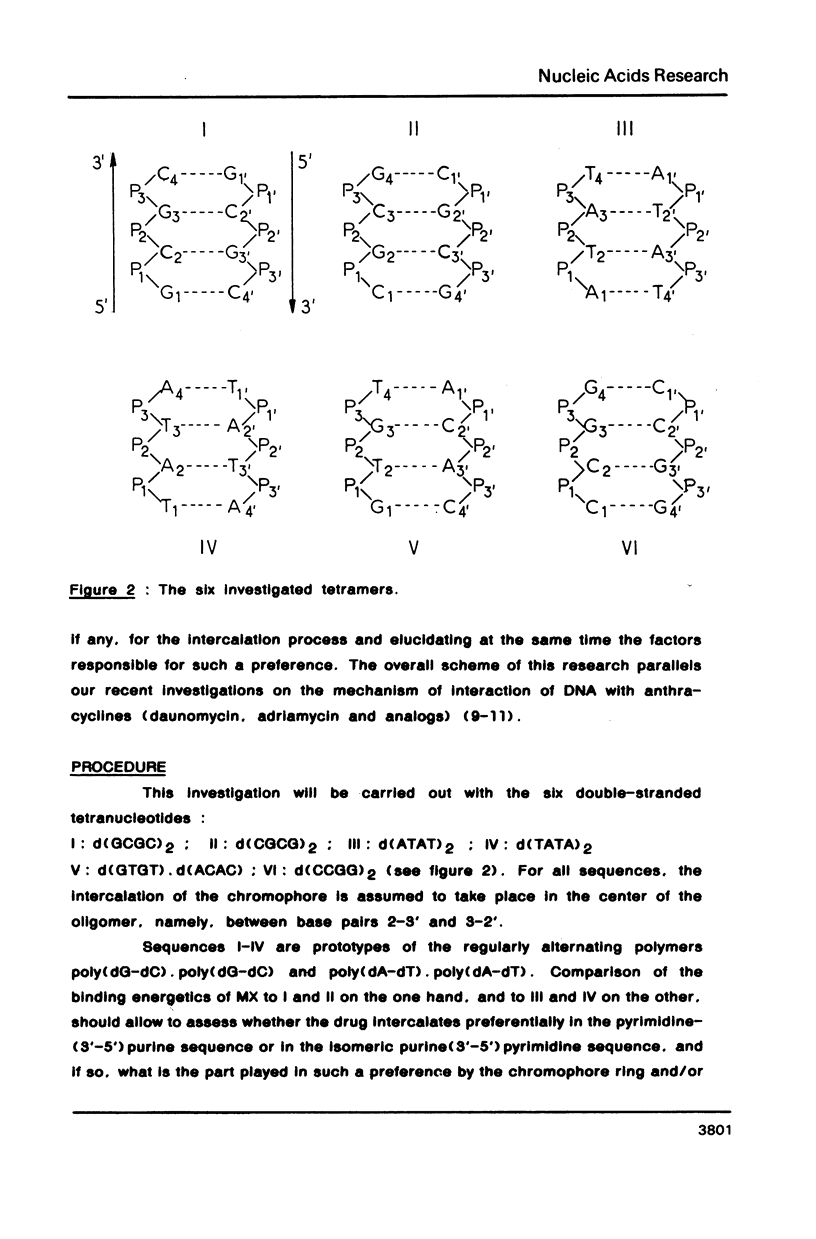
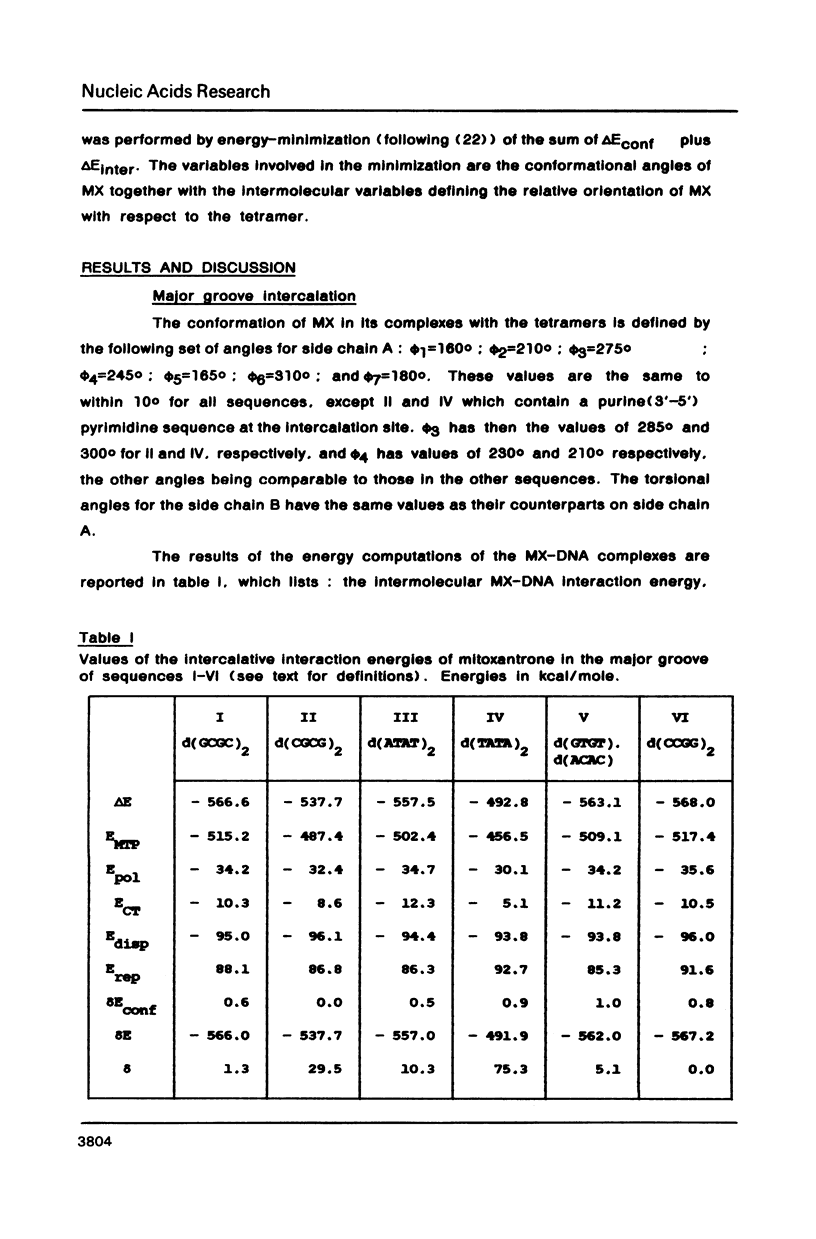
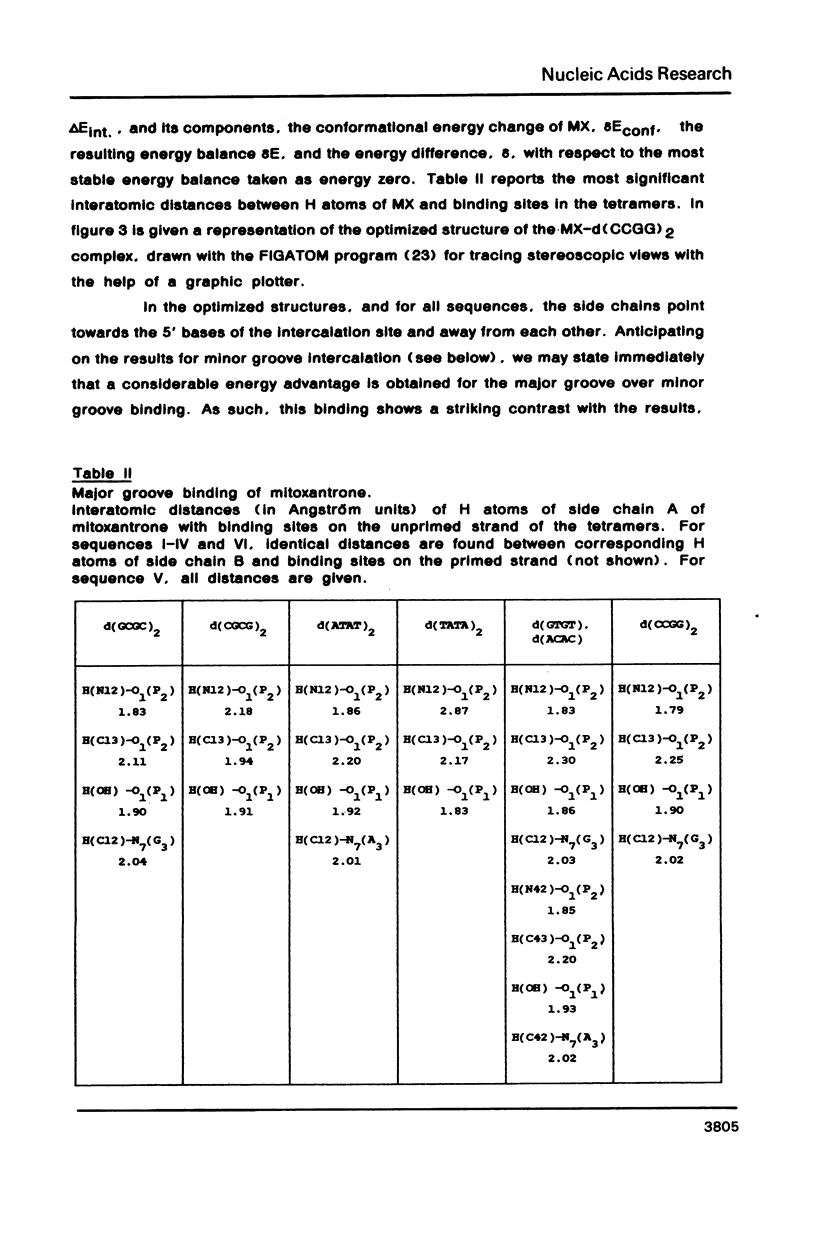
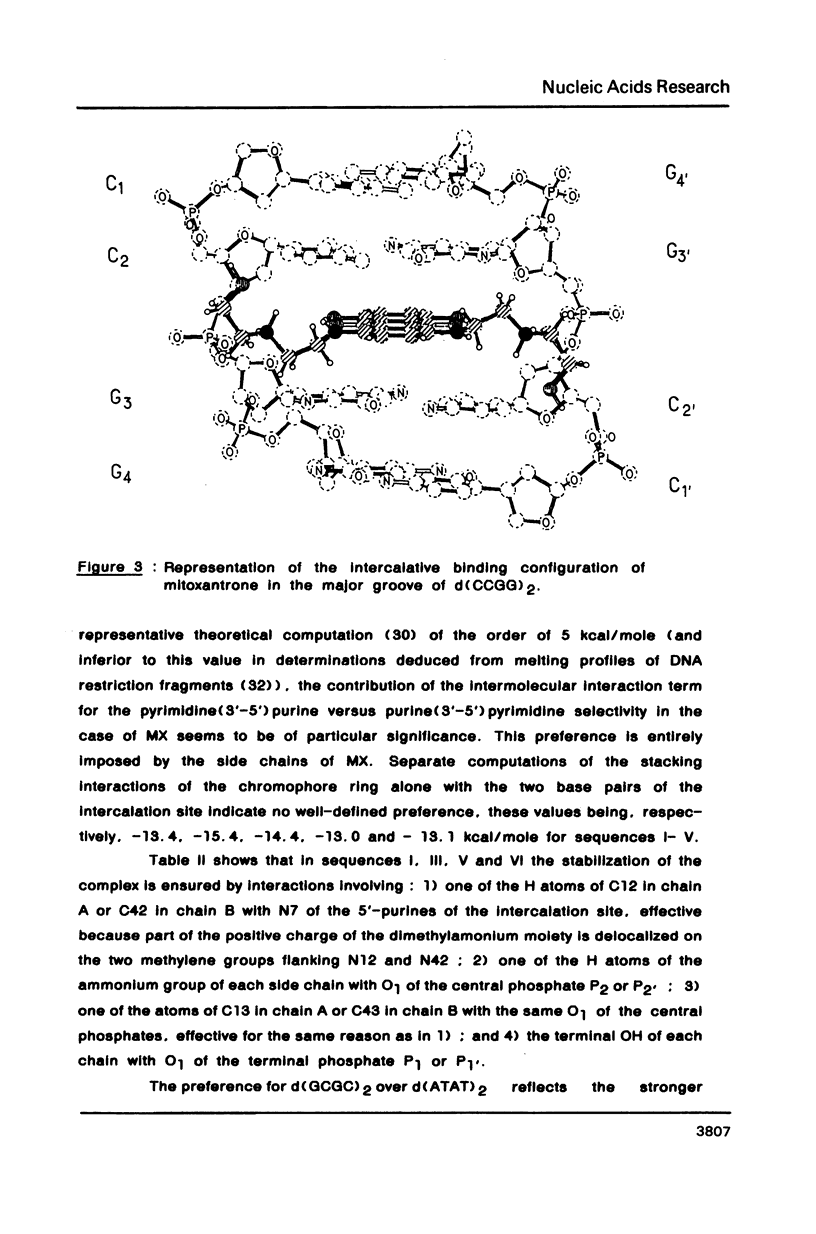
Theoretical computations are performed on the comparative binding energetics of mitoxantrone (MX), a newly synthesized intercalating anthraquinone antitumor drug, to six representative double-stranded tetranucleotides: d(GCGC)2, d(CGCG)2, d(ATAT)2, d(TATA)2, d(GTGT), d(ACAC), and d(CCGG)2. The computations are performed with the SIBFA procedure, which uses empirical formulas based on ab initio SCF computations. The best binding configuration of mitoxantrone locates its two side chains in the major groove. A considerable preference is elicited for intercalation of the chromophore ring in a pyrimidine (3'-5') purine sequence rather than the isomeric purine (3'-5') pyrimidine sequence. Contrary to the situation encountered with "simple" intercalators, in which this preference is generally attributed solely to differences in the energies of unstacking necessary to generate the intercalation sites, the preference is dictated in MX to a large extent by the intermolecular interaction energy term. This result is imposed by the interactions of the side chains of MX with the oligonucleotide.
Full text
PDF





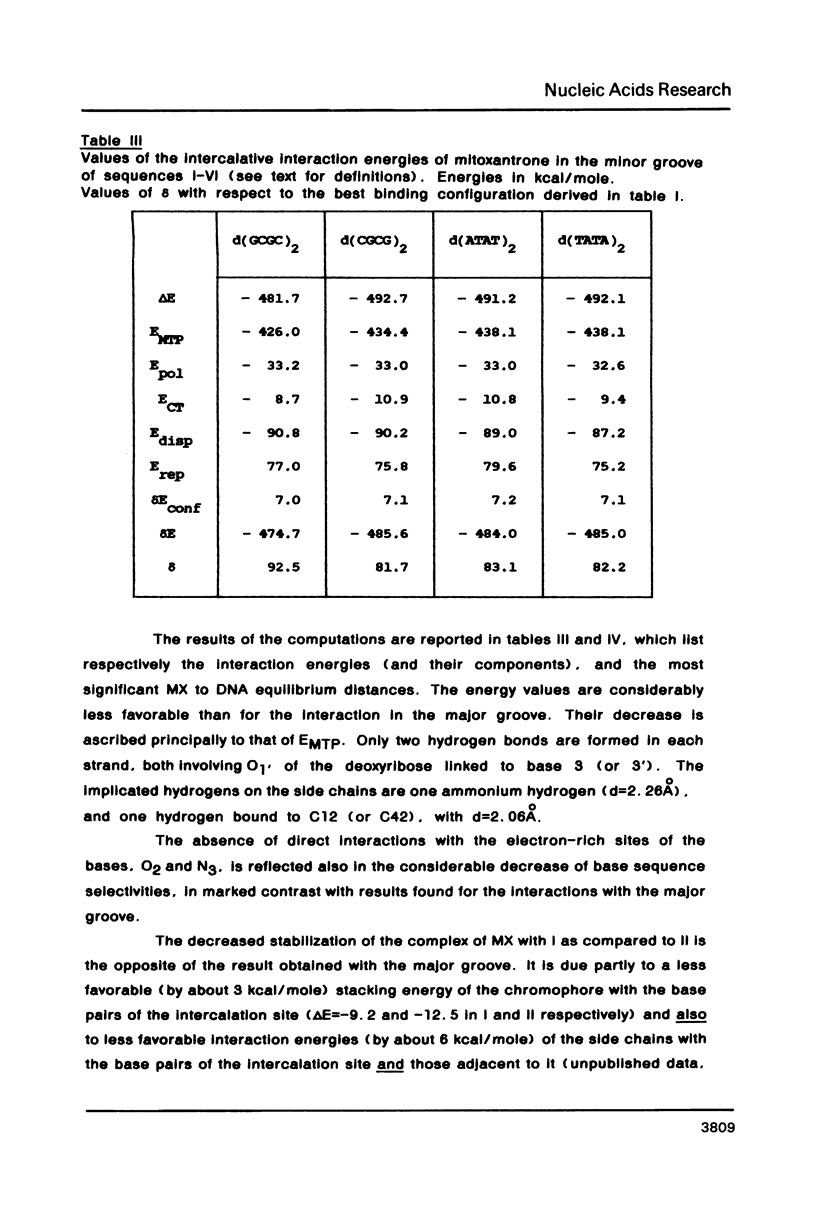
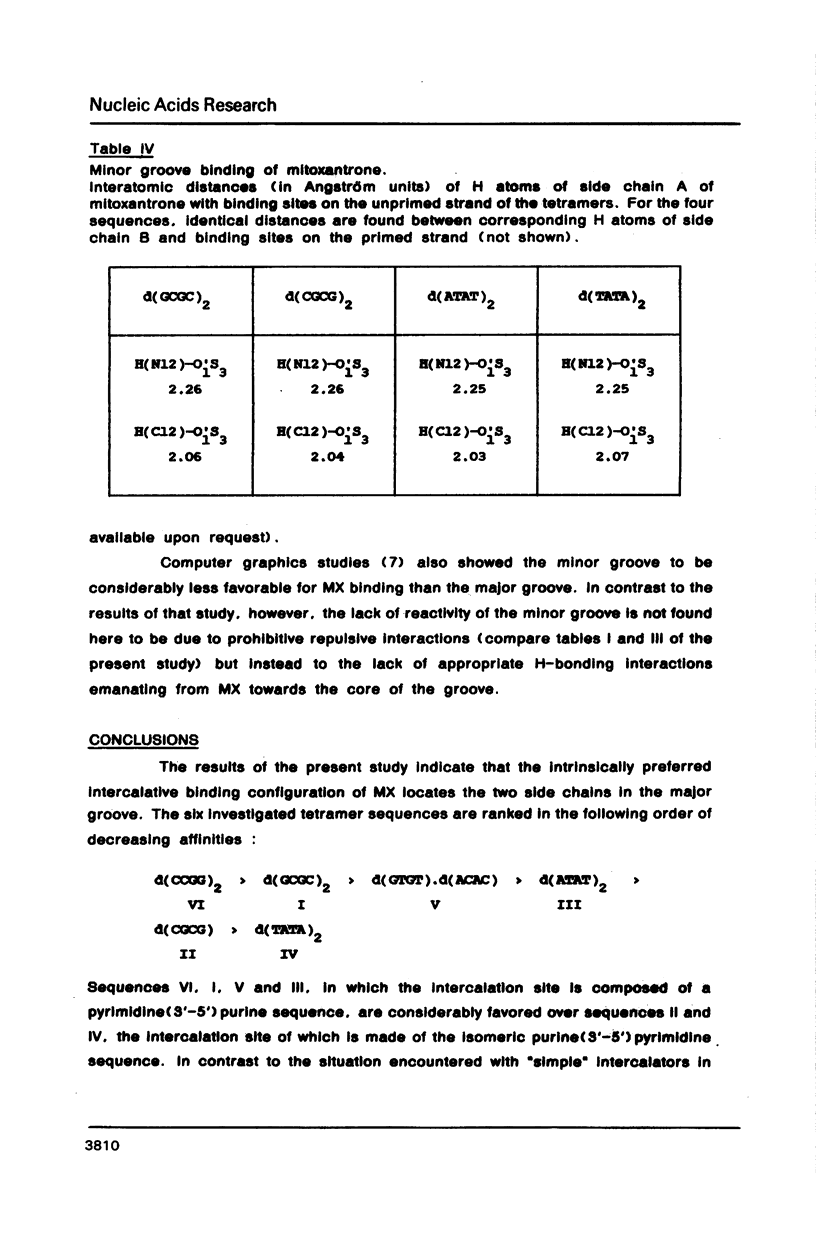


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Hukins D. W. Refinement of the structure of B-DNA and implications for the analysis of x-ray diffraction data from fibers of biopolymers. J Mol Biol. 1973 Dec 5;81(2):93–105. doi: 10.1016/0022-2836(73)90182-4. [DOI] [PubMed] [Google Scholar]
- Chen K. X., Gresh N., Pullman B. A theoretical investigation on the sequence selective binding of daunomycin to double-stranded polynucleotides. J Biomol Struct Dyn. 1985 Dec;3(3):445–466. doi: 10.1080/07391102.1985.10508434. [DOI] [PubMed] [Google Scholar]
- Foye W. O., Vajragupta O., Sengupta S. K. DNA-binding specificity and RNA polymerase inhibitory activity of bis(aminoalkyl)anthraquinones and bis(methylthio)vinylquinolinium iodides. J Pharm Sci. 1982 Feb;71(2):253–257. doi: 10.1002/jps.2600710228. [DOI] [PubMed] [Google Scholar]
- Islam S. A., Neidle S., Gandecha B. M., Partridge M., Patterson L. H., Brown J. R. Comparative computer graphics and solution studies of the DNA interaction of substituted anthraquinones based on doxorubicin and mitoxantrone. J Med Chem. 1985 Jul;28(7):857–864. doi: 10.1021/jm00145a003. [DOI] [PubMed] [Google Scholar]
- Kapuscinski J., Darzynkiewicz Z., Traganos F., Melamed M. R. Interactions of a new antitumor agent, 1,4-dihydroxy-5,8-bis[[2-[(2-hydroxyethyl)amino]-ethyl]amino]-9,10-anthracenedione, with nucleic acids. Biochem Pharmacol. 1981 Feb 1;30(3):231–240. doi: 10.1016/0006-2952(81)90083-6. [DOI] [PubMed] [Google Scholar]
- Lown J. W., Hanstock C. C., Bradley R. D., Scraba D. G. Interactions of the antitumor agents mitoxantrone and bisantrene with deoxyribonucleic acids studied by electron microscopy. Mol Pharmacol. 1984 Jan;25(1):178–184. [PubMed] [Google Scholar]
- Lown J. W., Hanstock C. C. High field 1H-NMR analysis of the 1:1 intercalation complex of the antitumor agent mitoxantrone and the DNA duplex [d(CpGpCpG)]. J Biomol Struct Dyn. 1985 Jun;2(6):1097–1106. doi: 10.1080/07391102.1985.10507626. [DOI] [PubMed] [Google Scholar]
- Lown J. W., Morgan A. R., Yen S. F., Wang Y. H., Wilson W. D. Characteristics of the binding of the anticancer agents mitoxantrone and ametantrone and related structures to deoxyribonucleic acids. Biochemistry. 1985 Jul 16;24(15):4028–4035. doi: 10.1021/bi00336a034. [DOI] [PubMed] [Google Scholar]
- Nakata Y., Hopfinger A. J. Predicted mode of intercalation of doxorubicin with dinucleotide dimers. Biochem Biophys Res Commun. 1980 Jul 31;95(2):583–588. doi: 10.1016/0006-291x(80)90824-4. [DOI] [PubMed] [Google Scholar]
- Newlin D. D., Miller K. J., Pilch D. F. Interactions of molecules with nucleic acids. VII. Intercalation and T.A specificity of daunomycin in DNA. Biopolymers. 1984 Jan;23(1):139–158. doi: 10.1002/bip.360230111. [DOI] [PubMed] [Google Scholar]
- Ornstein R. L., Rein R. Energetic and structural aspects of ethidium cation intercalation into DNA minihelices. Biopolymers. 1979 Nov;18(11):2821–2847. doi: 10.1002/bip.1979.360181112. [DOI] [PubMed] [Google Scholar]
- Ornstein R. L., Rein R. Energetics of intercalation specificity. I. Backbone unwinding. Biopolymers. 1979 May;18(5):1277–1291. doi: 10.1002/bip.1979.360180517. [DOI] [PubMed] [Google Scholar]
- Pack G. R., Loew G. Origins of the specificity in the intercalation of ethidium into nucleic acids. A theoretical analysis. Biochim Biophys Acta. 1978 Jun 22;519(1):163–172. doi: 10.1016/0005-2787(78)90070-9. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Wang A. H., Ughetto G., van der Marel G., van Boom J. H., Rich A. Molecular structure of an anticancer drug-DNA complex: daunomycin plus d(CpGpTpApCpG). Proc Natl Acad Sci U S A. 1980 Dec;77(12):7204–7208. doi: 10.1073/pnas.77.12.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell H. M., Jain S. C. Stereochemistry of actinomycin binding to DNA. II. Detailed molecular model of actinomycin-DNA complex and its implications. J Mol Biol. 1972 Jul 14;68(1):21–34. doi: 10.1016/0022-2836(72)90259-8. [DOI] [PubMed] [Google Scholar]
- Sobell H. M., Sakore T. D., Jain S. C., Banerjee A., Bhandary K. K., Reddy B. S., Lozansky E. D. beta-kinked DNA--a structure that gives rise to drug intercalation and DNA breathing--and its wider significance in determining the premelting and melting behavior of DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):293–314. doi: 10.1101/sqb.1983.047.01.035. [DOI] [PubMed] [Google Scholar]



