Abstract
Background
Rift Valley fever (RVF) is an emerging arthropod-borne zoonoses of global agricultural and public health importance. In December 2006, an RVF outbreak was recognized in Kenya which led to the deployment of international response laboratory teams to the area.
Objectives
A field laboratory was operated in Malindi, Kenya to provide safe sample handling and molecular testing for RVF virus (RVFV) as well as selected other pathogens for differential diagnosis.
Study Design
Safe sample handling was carried out using a negative pressure flexible film isolator (glovebox) and commercial reagents to inactivate clinical specimens and purify nucleic acid. Whole blood was routinely used for diagnostic testing although paired plasma samples were also tested in select cases. Subsequently, human macrophages were tested in vitro for their susceptibility to RVFV.
Results
The field laboratory received samples from 33 individuals and a definite laboratory diagnosis was provided in 16 of these cases. Using molecular diagnostic techniques, RVFV was more consistently detected in whole blood than in plasma samples most likely due to association of RVFV with blood cells. Subsequent in vitro studies identified macrophages as a target cell for RVFV replication.
Conclusions
RVFV appears to replicate in blood cells such as macrophages. Thus, the sensitivity of molecular diagnostic testing is improved if whole blood is used as the clinical specimen rather than plasma or serum.
1. BACKGROUND
Rift Valley fever virus (RVFV) has been responsible for extensive epizootics of disease in domestic animals and subsequent epidemics in the human population throughout Africa and the Arabian Peninsula.1,2 A member of the family Bunyaviridae, genus Phlebovirus, RVFV infection of livestock normally occurs via floodwater Aedes mosquitoes, producing high viremia and inducing abortions in susceptible animals; and epizootics can affect hundreds of thousands of animals. Humans are infected through the bites of infected mosquitoes or by direct contact with blood or tissues from infected animals. Although epidemics can extend to tens of thousands of infected people, most are asymptomatic or only develop mild illness. However, 0.5 to 1% of infected individuals will develop hemorrhagic fever or encephalitis with an additional 1 to 10% developing retinitis.3 In December 2006, a RVF outbreak was identified in the Garissa District of Kenya and virus activity was subsequently identified in other parts of Kenya as well as Somalia and Tanzania.4 Garissa District experienced a similar RVF outbreak in 1997-1998 when an estimated 27,500 persons were infected and 170 hemorrhagic fever-associated deaths occurred.5
2. OBJECTIVES
The main objective was to assist international response teams with laboratory diagnostics for RVFV and selected other pathogens of differential diagnostic concern during the 2007 outbreak of RVF in Kenya. We further aimed to advance RVFV diagnostics, in particular on-site field diagnostics, through enhanced molecular detection.
3. STUDY DESIGN
3.1.Biosafety and laboratory set-up
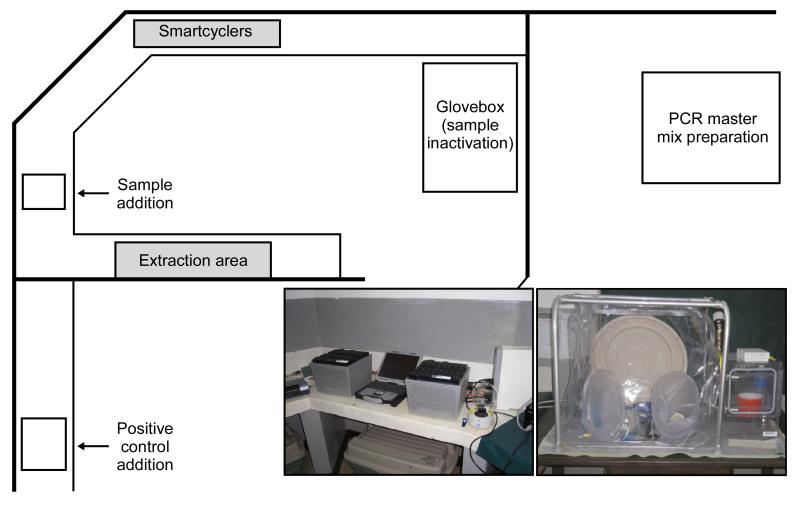
The laboratory set-up is schematically illustrated in Figure 1. All clinical specimens were handled in a High-Efficiency Particulate Air (HEPA) filtered, flexible film isolator which maintained negative pressure isolation (Figure1). The system consisted of a mini vinyl glovebox (Coy Lab Products, Grass Lake, MI) equipped with a pass-through chamber and air pump (Hagen, Baie d’Urfé, QC). The pump was used to evacuate the isolator’s internal air through two HEPA filters (Whatman, Toronto, ON) piped to the outside through a port. When the isolator became sufficiently negative, outside air passively entered the unit through a third HEPA filter (located in a separate port) allowing the unit to maintain a pressure of −30 pascals. Hypochlorite solution (0.5-1%) was used as a disinfectant and waste was incinerated daily
Figure 1. Laboratory set-up.
Laboratory was established in space provided at the Malindi District Hospital. Patient samples were handled inside the flexible film isolator (glovebox) (bottom right photo) to aliquot for inactivation for testing in Malindi and repacking for forwarding to the Kenya Medical Research Institute. Spatial separation of procedures was maximized to avoid the potential for contamination in PCR based testing using the SmartCycler platform (bottom left photo).
3.2. Clinical specimens
Blood samples were collected in ethylenediaminetetraacetic acid (EDTA) containing blood tubes and plasma was separated after allowing settling of the EDTA blood sample at 4°C. Centrifugation was avoided to minimize exposure of laboratory personnel. Whole blood and plasma were used for molecular diagnostics.
3.3. Virus inactivation and nucleic acid extraction
Whole blood or plasma (140uL) was used for virus inactivation and RNA extraction using the QIAamp Viral RNA Mini kit (Qiagen, Mississauga, ON) following the manufacturer’s instructions with the exception that a second AW1 wash step was added.
3.4. Molecular detection
Assays were performed on the SmartCycler platform (Cepheid, Sunnyvale, CA) utilizing Quantitect Probe RT-PCR and PCR kits (Qiagen, Mississauga, ON) for RVFV targets and bacterial/parasitic targets, respectively. Primary testing focused on RVFV diagnosis and used two quantitative real-time PCR (qRT-PCR) based tests6,7 using cut-off cycle threshold (Ct) values for positive, equivocal and negative of <40, 40 to 45 and >45 respectively. The assays were designed and validated by the original authors and verified by us to detect RVFV at 20 genome equivalents (g.e.) or 0.5 plaque forming units (pfu)/reaction. For differential diagnostics, the laboratory offered qRT-PCR tests for Leptospira spp.,8 Brucella spp.,9 Plasmodium spp.,10 Salmonella typhi11 and Vibrio cholerae12 using primers and conditions as published and cut-off Ct values for positive, equivocal and negative of <35, 35 to 40 and >40 respectively. As an internal control for extraction and amplification procedures, inactivated vesicular stomatitis virus (VSV), Indiana serotype (courtesy J. Rose, Yale University), was added to the lysis buffer with each extraction containing 12,000 and each amplification reactions containing 1,000 g.e.. Subsequent detection in each sample was done by qRT-PCR using VSVF (5′-caggacttacaggcaatatgataatga), VSVR (5′-tctatgctcaaattaccaccactgtt) and VSVP (5′ TET-caagagacggatggatcaccagttgtacat-TAMRA). Routinely CT values for VSV detection of 30 are obtained with this assay. Samples that provide CT values that are ≥ 2 cycles higher than the extraction control are considered to show inhibition and are re-extracted with additional washes until inhibition is no longer observed.
3.5. In vitro infection of human macrophages
Human blood was collected from anonymous volunteers under a protocol reviewed and approved by the NIH Institutional Biosafety Committee. Human monocytes were obtained from peripheral blood provided by the Department for Transfusion Medicine and the NIH Clinical Center at the National Institutes of Health (Bethesda, MD) and enriched by apheresis and Ficoll-paque (GE Healthcare) as previously described.13 Monocytes were seeded at 2 × 107 cells/20mls RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum (FCS), 0.2 mM L-glutamine, 1 mM HEPES buffer and 0.1 mM nonessential amino acids (all from Invitrogen) (complete RPMI [cRPMI]) in T-75cm2 flask. Cells were incubated for 1 hour at 37°C/5% CO2 and non-adherent cells were removed by gently washing cells with PBS supplemented with 2% FBS. Following washing, cells were incubated for 3 days in 20 mls of cRPMI supplemented with 100 ng/ml GM-CSF (Peprotech) at 37°C/5% CO2. On day 3 of culture, half of the medium was replaced with fresh cRPMI supplemented with 100% of cytokine. On day 4 of culture, the medium was removed, cells were washed once with PBS and removed from the flask using Trypsin-EDTA (Invitrogen). Cells were pelleted by centrifugation at 1200 rpm for 5 minutes and resuspended at 5×105/ml in the reserved cRPMI. Cells were seeded 1ml per well of a 24 well plate and incubated at 37°C/5% CO2. On day 5, half of the medium and 100% of GM-CSF was replaced with fresh reagents. The resulting differentiated macrophages were used on day 6 of culture. Cells were infected for 1 hour with RVFV, strain ZH501 (multiplicity of infection (MOI) = 0.1),14 washed twice with RPMI followed by incubation in growth medium. Cell supernatants were collected at 0, 6, 12, 24, 48, 72 and 96 hours post infection. Vero cells, which are permissive for RVFV, were used as a control for RVFV growth.15 Cell supernatants were titered for infectious virus using the TCID50 (tissue culture dose that leads to 50% cytopathogenicity) method as previously derscribed.15 Standard deviations were calculated based on three independent experiments for each growth kinetic and time point. The infectious work with RVFV was conducted in the high containment laboratory of the Integrated Research Facility at the Rocky Mountain Laboratories, Hamilton, Montana.
4. Results and Discussion
As part of the request for field laboratory assistance by the Global Outbreak and Alert Response Network (GOARN); the Public Health Agency of Canada provided mobile molecular diagnostics to the District Hospital in Malindi, Coastal Province, Kenya. The laboratory was in operation from January 15-28, 2007 and provided qRT-PCR based tests for RVFV diagnosis and differential diagnostics for Leptospira spp. , Brucella spp., Plasmodia spp., Salmonella typhi and Vibrio cholerae.6-12 Laboratory results were routinely available within 3 hours of sample receipt. As per the outbreak protocol, following collection and on-site testing all samples were shipped to the Kenyan Medical Research Institute (KEMRI), Nairobi for long term storage and independent analysis.
In total, the laboratory received 47 clinical specimens from 33 patients (Table 1). Of these, 20 patients were from Malindi District Hospital, 11 from Kilifi District Hospital and 2 from Ngao Hospital, Tana River District. A laboratory diagnosis was achieved in 16 out of 33 cases based on molecular testing. This testing identified 10 cases of RVF, 3 cases of malaria, and a single case each of leptospirosis, cholera and typhoid fever (Table 1).
Table 1.
Summary of testing performed in Malindi, Kenya.
| Pathogen | Test (No.) |
Pos. | Neg. | Equiv. | Patients (No.) |
Pos. | Neg. | Equiv. |
|---|---|---|---|---|---|---|---|---|
| Rift Valley fever | 47 | 18 | 29 | 0 | 33 | 10 | 23 | 0 |
| Brucella | 39 | 0 | 39 | 0 | 33 | 0 | 33 | 0 |
| Leptospira | 40 | 1 | 38 | 1 | 33 | 1 | 32 | 0 |
| Malaria | 39 | 4 | 35 | 0 | 33 | 3 | 30 | 0 |
| Typhoid fever | 33 | 1 | 32 | 0 | 33 | 1 | 32 | 0 |
| Cholera | 30 | 1 | 29 | 0 | 30* | 1 | 29 | 0 |
| Total | 228 | 25 | 202 | 1 | 33 | 16 | 17 | 0 |
Result of diagnostic testing on all samples and patients performed by the field laboratory for 6 different pathogens. A definitive laboratory diagnosis was achieved for 16 of the 33 cases. Key:
Only 30 patients tested; Equiv. = equivocal; Neg. = negative; No. = number; Pos. = positive
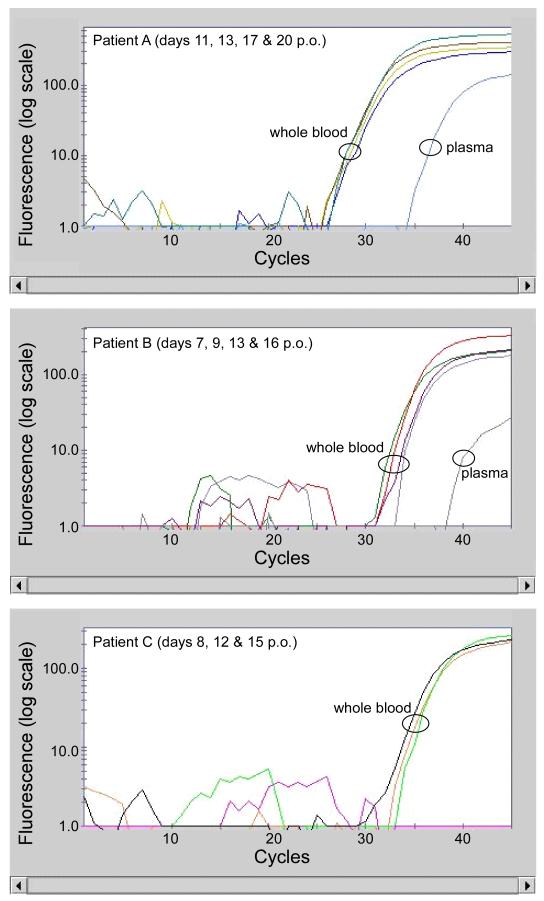
Three patients (patients A, B and C), positive for RVFV, were admitted to the Malindi District Hospital, and repeated whole blood samples from each of these individuals were available for testing. The Ct values of the qRT-PCR assays for RVFV remained essentially constant over a period of 9 days and up to 20 days post onset of disease (Figure 2). This was surprising as we expected Ct values to steadily increase reflecting the clearance of viremia over time. Most reports indicate that RVFV is quite transitory and by 10 days post onset of symptoms viral RNA is rarely detected.16 Molecular diagnostics to detect RVFV are routinely performed on serum or plasma samples. Therefore, the unexpected persistence of RVFV RNA found in whole blood may be due to the type of sample collected. Thus, we also assessed plasma collected on the last day of sampling from each donor for the presence of RVFV RNA. Interestingly, we observed that the Ct values for RVFV RNA were higher, indicative of lower viral load in plasma samples than that in the paired blood from each patient (Figure 2). A similar trend was observed in paired whole blood and plasma specimens from additional five patients (Table 2). In the two individuals (patients D and E) where Ct values were below 25, both specimen sources produced essentially identical viremia levels (similar Ct values). In contrast, samples from the remaining six patients (patients A, B, C, F, G and H) produced Ct values that were greater than 25 and in all of these instances we began to see a disparity between Ct values from whole blood versus plasma samples. Whole blood samples consistently produced lower Ct values when compared to plasma from the same patient, indeed plasma samples from two patients (C and H) tested negative. Presumably these data suggest that higher quantities of RVFV RNA are associated with whole blood compared to plasma.
Figure 2. Detection of RVFV in serial samples.
Amplification curves for RVFV (single target shown) from patients from whom serial samples were available. Day post onset of clinical symptoms (p.o.) for each sample is shown in the panels; the plasma sample was run on each final p.o. sample available. RVFV was not detected in day 9 p.o. sample for Patient C.
Table 2.
Comparison of RVFV in whole blood and plasma samples.
| Patient | Days (p.o.) |
Whole Blood (Ct) |
Plasma (Ct) |
|---|---|---|---|
| A | 20 | 29.5 | 37.8 |
| B | 16 | 35.0 | >45.0 |
| C | 15 | 35.1 | n.d. |
| D | 4 | 16.8 | 16.5 |
| E | 7 | 23.4 | 23.8 |
| F | 11 | 32.1 | 36.1 |
| G | 6 | 30.0 | 40.0 |
| H | 10 | 36.5 | n.d. |
Apparent viral genome loads in whole blood and plasma are equivalent for high titer patients (Ct values <25) but become increasingly disparate as viral genome titers decrease (Ct values >25). Key: Ct = cycle threshold; n.d. = not detectable (no amplification present - negative); >45 = amplification was present but did not reach threshold within 45 cycles, considered negative; p.o. = post onset of clinical symptoms
One explanation for the increased concentration of RVFV RNA in whole blood samples is that the virus is associated with blood cells such as mononuclear cells. Infection of monocytes and macrophages has been observed in samples obtained from experimentally and naturally infected animals as well as clinical samples from humans.17,18 Furthermore, in a recent study human CD14+ cells supported replication of RVFV.19 Thus, we proceeded to determine whether RVFV could infect and replicate in fully differentiated human macrophages. Cultures of primary human macrophages were infected with RVFV, strain ZH-501,14 using a MOI of 0.1. Human macrophages exhibited similar titers of RVFV as those observed in VERO cells during the first 6 hours after infection (Figure 3). Over the next 18 hours the virus appeared to replicate in human macrophages, although more slowly than VERO cells, and reached maximum titers 24 hours after infection (Figure 3). Thus, our data demonstrates that RVFV can infect and replicate in human blood cells. Furthermore, our data support the contention that the increased RVFV RNA loads detected in whole blood may be due to the presence of virus in mononuclear cells when compared to plasma samples.
Figure 3. Growth kinetics of RVFV in primary human macrophages.
Differentiated human macrophages and Vero cells were infected with RVFV, washed and the media monitored for release of infectious virus over 96 hours. Peak titers were reached by 24 hours in macrophages and remained steady for several days. Error bars represent the standard error of mean from three independent experiments.
Previous investigators have reported PCR data complemented with results from serology, virus isolation and/or antigen detection techniques.3,5,20 In these studies, no single methodology was able to consistently detect every RVFV-positive case. The utility of PCR testing is greatest during acute infection, before RVFV-specific IgM responses are detectable, or in fatal cases where significant antibody responses do not always develop.6 These previous investigations all used serum as test samples, and some of them have used less sensitive PCR approaches for detection. With the application of highly sensitive qRT-PCR to the detection of RVFV as done here, coupled with whole blood as the test sample, we believe that this algorithm would allow the detection of RVFV viremia in the vast majority, if not all, active RVFV cases.
While the provision of serological testing along with molecular detection is currently recognized as ideal, qRT-PCR from whole blood samples may prove molecular detection of RVFV to be sufficient in identifying active cases to enable outbreak control measures and effective case patient management. This approach also has the potential benefit of minimizing the biosafety risk associated with RVFV serologic tests. Laboratory-acquired infections of RVFV have occurred following exposure to infectious material directly or through the generation of aerosols and appropriate biocontainment measures and procedures are necessary to mitigate the risk of infection in a laboratory or clinical environment.21, 22 Laboratory diagnostic techniques including antigen and antibody detection, virus isolation and PCR-based assays each have risks associated with the handling of potentially infectious samples. These risks relate to the level of inactivation possible for a given sample type, the potential for generation of aerosols in the testing procedure, and the amount of manipulation required in each procedure. Minimizing the manipulation of potentially infectious samples and completely inactivating subsamples for testing minimize the efforts needed to ensure safe handling of patient samples. As extensive manipulation of infectious samples, especially centrifugation and aspiration, is not required for nucleic acid isolation from whole blood, potential sources of laboratory-acquired infection are minimal. This can be of importance especially in local clinics and field sites where centrifugation of serum samples is routinely performed prior to shipment to laboratories for testing.
Acknowledgements
The Mobile Laboratory Unit was supported by the National Microbiology Laboratory of the Public Health Agency of Canada. The outbreak response mission was supported by the World Health Organization. The authors would like to thank the following agencies/institutions for their support and assistance: Ministry of Health, Nairobi, Kenya; World Health Organization, Kenya Country Office, Nairobi, Kenya, and Geneva, Switzerland; Malindi District Hospital, Malindi, Kenya; Kenya Medical Research Institute, Nairobi, Kenya; Centers for Disease Control and Prevention, Nairobi, Kenya, Atlanta, USA, Fort Collins, USA; Institute of Tropical and Infectious Diseases, University of Nairobi, Nairobi, Kenya; National Institute of Communicable Diseases, Sandringham, South Africa. The authors are grateful to local hospital and public health staff for their help and support. We highly appreciated the collaboration by the patients and their family members as well as the communities. We also would like to thank Anita Mora (Division of Intramural Research, NIAID, NIH) for assistance with the graphical work.
Funding
The field component of this work was funded by the World Health organization and the National Microbiology Laboratory of the Public Health Agency of Canada while the studies on in vitro infection of human macrophages was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The authors declare that they have no conflict of interest.
Ethical Approval
Peripheral blood mononuclear cells (PBMCs) were isolated from venous blood of human donors on a protocol was approved by the Institutional Review Board for Human Subjects, NIAID, NIH. Collection of human specimens during the Rift Valley fever outbreak occurred on a World Health Organization outbreak response protocol.
References
- 1.Hoogstraal H, Meegan JM, Khalil GM, Adham FK. The Rift Valley fever epizootic in Egypt 1977-78. 2. Ecological and entomological studies. (Translated from eng) Trans R Soc Trop Med Hyg. 1979;73:624–29. doi: 10.1016/0035-9203(79)90005-1. [DOI] [PubMed] [Google Scholar]
- 2.Bird BH, Khristova ML, Rollin PE, Ksiazek TG, Nichol ST. Complete genome analysis of 33 ecologically and biologically diverse Rift Valley fever virus strains reveals widespread virus movement and low genetic diversity due to recent common ancestry. J Virol. 2007;81:2805–16. doi: 10.1128/JVI.02095-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoemaker T, Boulianne C, Vincent MJ, Pezzanite L, Al-Qahtani MM, Al-Mazrou Y, et al. Genetic analysis of viruses associated with emergence of Rift Valley fever in Saudi Arabia and Yemen, 2000-2001. Emerg Infect Dis. 2002;8:1415–20. doi: 10.3201/eid0812.020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention Rift Valley fever outbreak - Kenya, November 2006-January 2007. Morb Mortal Wkly Rep. 2007;56:73–6. [PubMed] [Google Scholar]
- 5.Woods CW, Karpati AM, Grein T, McCarthy N, Gaturuku P, Muchiri E, et al. World Health Organization Hemorrhagic Fever Task Force, 2002 An outbreak of Rift Valley fever in northeastern Kenya, 1997-1998. Emerg Infect Dis. 2002;8:138–44. doi: 10.3201/eid0802.010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird BH, Bawiec DA, Ksiazek TG, Shoemaker TR, Nichol ST. Highly Sensitive and Broadly Reactive Quantitative Reverse Transcription-PCR Assay for High-Throughput Detection of Rift Valley Fever Virus. J Clin Microbiol. 2007;45:350613. doi: 10.1128/JCM.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drosten C, Gottig S, Schilling S, Asper M, Panning M, Schmitz H, et al. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Microbiol. 2002;40:2323–30. doi: 10.1128/JCM.40.7.2323-2330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smythe LD, Smith IL, Smith GA, Dohnt MF, Symonds ML, Barnett LJ, McKay DB. A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infect Dis. 2002;2:13. doi: 10.1186/1471-2334-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Probert WS, Schrader KN, Khuong NY, Bystrom SL, Graves MH. Real-time multiplex PCR assay for detection of Brucella spp., B. abortus, and B. melitensis. J Clin Microbiol. 2004;42:1290–93. doi: 10.1128/JCM.42.3.1290-1293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee MA, Tan CH, Aw LT, Tang CS, Singh M, Lee SH, et al. Real-time fluorescence-based PCR for detection of malaria parasites. J Clin Microbiol. 2002;40:4343–45. doi: 10.1128/JCM.40.11.4343-4345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tracz DM, Tabor H, Jerome M, Ng LK, Gilmour MW. Genetic determinants and polymorphisms specific for human-adapted serovars of Salmonella enterica that cause enteric fever. J Clin Microbiol. 2006;44:2007–18. doi: 10.1128/JCM.02630-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyon WJ. TaqMan PCR for detection of Vibrio cholerae O1, O139, non-O1, and non-O139 in pure cultures, raw oysters, and synthetic seawater. Appl Environ Microbiol. 2001;67:4685–93. doi: 10.1128/AEM.67.10.4685-4693.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chase JC, Celli J, Bosio CM. Direct and indirect impairment of human dendritic cell function by virulent Francisella tularensis Schu S4. Infect Immun. 2009;77:180–95. doi: 10.1128/IAI.00879-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meegan JM. The Rift Valley fever epizootic in Egypt 1977-1978. I. Description of the epizootic and virological studies. Trans R Soc Trop Med Hyg. 1979;73:618–23. doi: 10.1016/0035-9203(79)90004-x. [DOI] [PubMed] [Google Scholar]
- 15.Ellis DS, Shirodaria PV, Fleming E, Simpson DIH. Morphology and Development of Rift Valley Fever Virus in Vero Cell Cultures. J Med Virology. 1988;24:161–74. doi: 10.1002/jmv.1890240205. [DOI] [PubMed] [Google Scholar]
- 16.Bird BH, Ksiazek TG, Nichol ST, Maclachlan NJ. Rift Valley fever virus. J Am Vet Med Assoc. 2009;234:883–93. doi: 10.2460/javma.234.7.883. [DOI] [PubMed] [Google Scholar]
- 17.Shieh WJ, Paddock CD, Lederman E, Rao CY, Gould LH, Mohamed M, Mosha F, Mghamba J, Bloland P, Njenga MK, Mutonga D, Samuel AA, Guarner J, Breiman RF, Zaki SR. Pathologic studies on suspect animal and human cases of Rift Valley fever from an outbreak in Eastern Africa, 2006-2007. Am J Trop Med Hyg. 2010;83(2 Suppl):38–42. doi: 10.4269/ajtmh.2010.09-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith DR, Steele KE, Shamblin J, Honko A, Johnson J, Reed C, Kennedy M, Chapman JL, Hensley LE. The pathogenesis of Rift Valley fever virus in the mouse model. Virology. 2010;407:256–67. doi: 10.1016/j.virol.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 19.McElroy AK, Nichol ST. Rift Valley fever virus inhibits a pro-inflammatory response in experimentally infected human monocyte derived macrophages and a pro-inflammatory cytokine response may be associated with patient survival during natural infection. Virology. 2012;422:6–12. doi: 10.1016/j.virol.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Njenga MK, Paweska J, Wanjala R, Rao CY, Weiner M, Omballa V, et al. Using a Field Quantitative Real-Time PCR Test To Rapidly Identify Highly Viremic Rift Valley Fever Cases. J Clin Microbiol. 2009;47:1166–71. doi: 10.1128/JCM.01905-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins CH, Kennedy DA. Laboratory acquired infections. In: Laboratory acquired infections: History, incidence, causes and prevention. 4th ed Butterworth-Heinemann; Oxford UK: 1999. pp. 1–37. [Google Scholar]
- 22.Paragas J, Endy TP. Viral Agents of Human Disease: Biosafety Concerns. In: Fleming DO, Hunt DL, editors. Biological Safety: Principles and Practices. ASM Press; Washington (DC): 2006. pp. 179–207. [Google Scholar]